Abstract
Summary: Chromatin immunoprecipitation and DNase I hypersensitivity assays with high-throughput sequencing have greatly accelerated the understanding of transcriptional and epigenetic regulation, although data reuse for the community of experimental biologists has been challenging. We created a data portal CistromeFinder that can help query, evaluate and visualize publicly available Chromatin immunoprecipitation and DNase I hypersensitivity assays with high-throughput sequencing data in human and mouse. The database currently contains 6378 samples over 4391 datasets, 313 factors and 102 cell lines or cell populations. Each dataset has gone through a consistent analysis and quality control pipeline; therefore, users could evaluate the overall quality of each dataset before examining binding sites near their genes of interest. CistromeFinder is integrated with UCSC genome browser for visualization, Primer3Plus for ChIP-qPCR primer design and CistromeMap for submitting newly available datasets. It also allows users to leave comments to facilitate data evaluation and update.
Availability: http://cistrome.org/finder.
Contact: xsliu@jimmy.harvard.edu or henry_long@dfci.harvard.edu
1 Introduction
Genome-wide mapping of transcription factor and chromatin regulator binding, histone modifications and chromatin accessibility is essential for elucidating the regulatory network of various biological processes. Chromatin immunoprecipitation and DNase I hypersensitivity assays with high-throughput sequencing (ChIP-seq and DNase-seq) have become essential techniques for studying transcriptional and epigenetic gene regulation. Most ChIP-seq and DNase-seq studies have raw data available from public repositories, such as NCBI Sequence Read Archive. However, the raw sequencing data are usually in the order of gigabyte size per sample, and data analysis is not only time-consuming but also technically inhibitive for most experimental biologists. Even if investigators only want to check a few specific binding sites near their genes of interest, they need the computational resources and expertise to download the raw data, conduct data analysis such as read alignment, peak calling, nearby gene assignment and many others. In addition, these public datasets are often of variable quality, and bioinformatics novices do not often have the expertise to evaluate and compare different datasets. Although projects like ENCODE (ENCODE Project Consortium, 2011) and Epigenome Roadmap (Bernstein et al., 2010) provide processed data online, they only cover data from these projects. We developed a web portal called CistromeFinder, which contains the most comprehensive collection to date of public ChIP-seq and DNase-seq data in human and mouse that have gone through a uniform data analysis and quality control pipeline. It enables investigators to query, evaluate, visualize and compare public ChIP-seq and DNase-seq data and select specific binding sites near genes of interest for functional studies.
2 Design AND Implementation
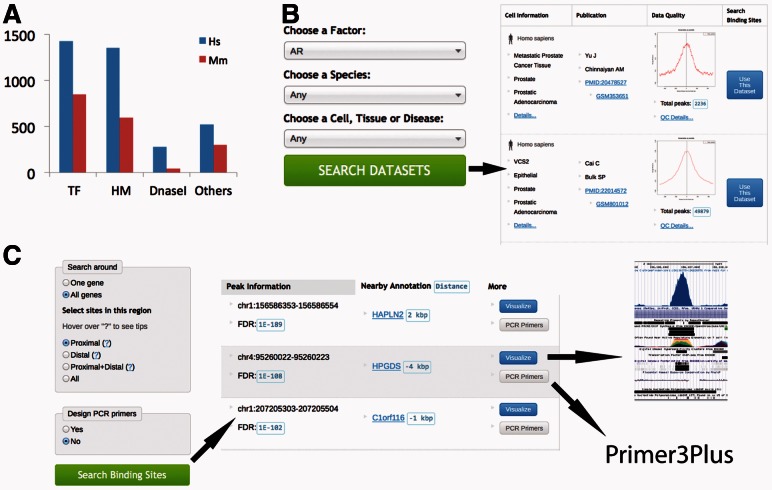
We downloaded public ChIP-seq and DNase-seq datasets for Homo sapiens and Mus musculus from high-throughput sequencing data repositories such as Sequence Read Archive and ENA (Leinonen et al., 2010), ENCODE and Epigenome Roadmap projects, and data websites linked to relevant papers. The collection has 3887 human and 2491 mouse samples, including ChIP-seq of 260 transcription and chromatin factors, 53 histone modifications and variants and 326 DNase-seq samples (Fig. 1A). When relevant replicates and corresponding controls are available, we combined them to form a dataset. All associated metadata were manually curated and standardized in our previous work CistromeMap (Qin et al., 2012). They include information on factor symbol, species, cell line, cell population, cell type, tissue, disease state, authors, paper and data source, to give users the flexibility to search for and view data of interest. All the raw data were processed in a standard pipeline using bowtie (Langmead et al., 2009) for read mapping, MACS (Zhang et al., 2008) for peak calling, CEAS (Shin et al., 2009) for genome binding distribution and several other quality control scripts. The details of the pipeline protocol, including algorithms and parameters used, as well as guidelines for interpreting the Quality Control (QC) reports can be viewed on the help page of CistromeFinder. The CistromeFinder web interface allows users to examine the pre-processed ChIP/DNase-seq datasets in the following manner:
Fig. 1.
(A) Statistics of datasets in CistromeFinder, the height of bar represents the number of datasets under each category: transcription factor (TF), histone modification (HM), DNaseI and others (such as chromatin regulators), of Homo sapiens (Hs) and Mus musculus (Mm). (B) Screenshot of dataset pages. (C) Screenshot for binding sites pages
2.1 Search datasets
On the front page of CistromeFinder, users can search for datasets by species and factor name or select from the full list of available data by factor name. Additional options also allow users to specify cell, tissue or disease name to narrow down search results, which are useful for factors such as CTCF with many different ChIP-seq datasets. The result page displays a table of datasets with information on the cell, paper, reference genome, data source, quick data summary, as well as links to more detailed metadata and quality control measures (Fig. 1B).
2.2 Evaluate datasets
Quality control measurements for each dataset include a summary of total read count, mapping rate, number of peaks, overlap with union of DNaseI hypersensitive sites (Thurman et al., 2012), cross-species conservation (Pollard et al., 2010) and genome-wide distribution of enrichment from CEAS. CistromeFinder also allows users to leave comments to any dataset to help other researchers in the community. If users are satisfied with the data quality, they can use the data to select binding sites.
2.3 Search binding sites
On average, each ChIP-seq dataset has ∼10 000 binding sites, and each DNase-seq dataset has >100 000 sites. Users have several options to select binding sites: all binding sites (no limit), those located in gene proximal regions [within ±5 kb of any transcription start site (TSS)], distal regions (from ±5 kb to ±50 kb of any TSS) or proximal plus distal regions (within ±50 kb of any TSS). In addition, users could select binding sites near a specific gene and further filter binding sites that overlap with repeat-masked sequences. After filtering, the top 200 qualified binding sites are displayed in order of enrichment false discovery rate.
2.4 Evaluate binding sites
Each selected peak in Section 2.3 will have information such as genomic coordinates, ChIP-seq significance score, distance to nearby genes and DNA sequences centred at the peak summit (Fig. 1C). Users can ‘view binding site’ signal profile from UCSC genome browser (Kent et al., 2002) or use Primer3Plus (Untergasser et al., 2012) to design qPCR primers to validate the binding of a factor near genes of interest in a specific cell context.
3 Conclusion
CistromeFinder provides a comprehensive catalogue and uniform processing of published ChIP-seq and DNase-seq datasets. The web interface provides flexibility to search and visualize the binding of specified factors in various conditions near genes of interest. Investigators can use CistromeFinder to check whether a gene could be the potential target of a transcription or chromatin factor in a specific conditions and design ChIP-qPCR primers to validate the binding in other conditions. We also give the users options to leave comments to help us evaluate datasets and submit new data through CistromeMap to help keep the database up-to-date. We will continue to maintain and improve CistromeFinder as more ChIP/DNase-seq data become available to facilitate the community in their transcriptional and epigenetic gene regulation studies. We keep the raw data and the pipeline script in case we need to rerun all the data with newest algorithms; we also use a version system for the pipeline to reproduce legacy results. In the future, we plan to combine Cistrome analysis pipeline (Liu et al., 2011) with our ChIP/DNase-seq data collection; therefore, users could conduct integrative analysis of their own data and public data.
Funding: National Basic Research (973) Program of China [2010CB944904]; National Natural Science Foundation of China [31028011]; and National Institutes of Health [HG4069].
Conflict of Interest: none declared.
References
- Bernstein BE, et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, et al. The European Nucleotide Archive. Nucleic Acids Res. 2010;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, et al. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B, et al. CistromeMap: a knowledgebase and web server for ChIP-Seq and DNase-Seq studies in mouse and human. Bioinformatics. 2012;28:1411–1412. doi: 10.1093/bioinformatics/bts157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, et al. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]