Abstract
Background. Hematopoietic progenitor cells (HPCs) in the bone marrow of human immunodeficiency virus (HIV)–infected individuals have been proposed as a persistent reservoir of virus. However, some studies have suggested that HIV genomes detected in HPCs arise from T-cell contamination.
Methods. CD133-sorted HPCs and CD133-depleted bone marrow cells were purified from bone marrow specimens obtained from 11 antiretroviral-treated donors in whom the HIV load had been <48 copies/mL for at least 6 months. CD133 and CD3 expression on the cells was assessed by flow cytometry. HIV DNA was quantified by real-time polymerase chain reaction analysis.
Results. HIV genomes were detected in CD133-sorted samples from 6 donors, including 2 in whom viral loads were undetectable for >8 years. CD3+ T cells represented <1% of cells in all CD133-sorted samples. For 5 of 6 CD133-sorted samples with detectable HIV DNA, the HIV genomes could not be explained by contaminating CD3+ T cells. Donors with detectable HIV DNA in HPCs received their diagnosis significantly more recently than the remaining donors but had had undetectable viral loads for similar periods.
Conclusions. HIV genomes can be detected in CD133-sorted cells from a subset of donors with long-term viral suppression and, in most cases, cannot be explained by contamination with CD3+ T cells.
Keywords: HIV, hematopoietic progenitor cells, reservoirs
(See the editorial commentary by Pace and O'Doherty on pages 1790–2.)
Latent human immunodeficiency virus (HIV) infection represents a major barrier to curing HIV [1, 2]. When HIV establishes a latent infection within a cell, the DNA provirus integrates into the host cell's genome, but viral genes are not transcribed [3]. The latently infected cell is thus indistinguishable from an uninfected cell and cannot be targeted by the immune system or current antiretroviral therapies. The HIV provirus can persist in this state for the lifetime of the cell; however, it can also be reactivated if cellular conditions change, leading to the production of new virions and potentially new infection events [4]. Thus, HIV replication will resume if antiretroviral therapy is discontinued, unless all latent reservoirs of virus are eliminated.
Although resting memory CD4+ T cells are a well-studied reservoir for latent HIV, not all HIV sequences in the plasma of many successfully treated HIV-positive donors can be matched to sequences in peripheral blood CD4+ T cells [5–7]. These data may suggest that additional reservoirs of virus exist and contribute to residual viremia during treatment, as well as to viral rebound upon treatment interruption [7].
Recently, we proposed that hematopoietic progenitor cells (HPCs) in the bone marrow serve as a reservoir for latent HIV. We assessed HIV type 1 (HIV-1) infection in CD34+ HPCs from 9 HIV-infected donors receiving highly active antiretroviral therapy (HAART) in whom viral loads had been undetectable for at least 6 months [8]. In 4 of 9 donors, we detected HIV-1 proviral genomes in CD34-sorted cells at a frequency of 3-40 genomes per 10 000 cells [8], suggesting that HPCs might serve as a reservoir of virus. Comparable amounts of HIV DNA were not observed in bone marrow cells immunodepleted for CD34 [8]. However, subsequent studies have not detected HIV genomes in CD34+ HPCs from donors with undetectable viral loads [9, 10] and have suggested that HIV genomes in CD34+ samples may be due to contaminating CD3+ T cells [9].
In addition to detecting HIV genomes in CD34+ cells ex vivo, we have shown that CD34+ cells can be infected by CCR5- and CXCR4-tropic HIV in vitro [8]. We also demonstrated that HPCs expressing CD133, a marker for an immature subset of CD34+ HPCs, are only infected by CXCR4-using HIV-1 in vitro [11]. However, HIV infection of CD133+ cells in vivo has not been assessed.
To investigate whether CD133+ HPCs harbor HIV-1 in vivo, we quantified HIV proviral genomes in CD133+ HPCs from 11 HIV-positive donors in whom plasma viral loads had been <48 copies/mL for at least 6 months. We furthermore analyzed the frequency of CD3+ T cells in each sample to assess whether HIV genomes could arise from contamination with CD3+ T cells.
METHODS
Clinical Samples
The donors who provided samples analyzed in this study are a consecutive subset of our cohort, excluding 2 donors for whom the CD133-sorted sample was <80% pure and 2 donors for whom samples had been used up in prior experiments (1 donor) or lost (1 donor). We recruited HIV-positive donors currently receiving antiretroviral therapy from the University of Michigan HIV-AIDS Treatment Program and obtained informed consent according to a protocol approved by the University of Michigan Institutional Review Board. At the time of aspiration, all donors were >18 years old, had normal white blood cell counts, and had had plasma viral loads of <48 copies/mL for at least 6 months. Twenty milliliters of bone marrow was aspirated from the posterior iliac crest, collected in preservative-free heparin, and processed immediately.
Isolation of CD133-Sorted and CD133-Depleted Cells
Bone marrow mononuclear cells (BMMCs) were isolated by Ficoll-Paque density separation (GE Healthcare). Adherent cells were depleted by incubation in serum-free StemSpan media (StemCell Technologies) for 2 hours at 37°C, and then CD133+ cells were isolated by magnetic sorting (Miltenyi Biotec). Cells were sequentially sorted on 2 columns to increase purity. BMMCs that flowed through the first column were collected as the CD133-depleted fraction. Samples were cryopreserved in 10% dimethyl sulfoxide in fetal bovine serum until analysis.
Flow Cytometry
A fraction of each clinical sample was stained with R-phycoerythrin–conjugated anti-CD133 (Miltenyi Biotec), allophycocyanin-conjugated anti-CD3 (eBioscience), and 7-aminoactinomycin D (7-AAD). Healthy donor BMMCs (AllCells) were stained with R-phycoerythrin–conjugated anti-CD133, R-phycoerythrin-Cy7–conjugated anti-CD34 (BD), and 7-AAD. Samples were analyzed on a BD FACSCanto.
Polymerase Chain Reaction (PCR)
Cells were lysed in MagNA Pure DNA Lysis/Binding Buffer (Roche), and DNA was extracted using a MagNA Pure Compact System (Roche). HIV-1 DNA was quantified by 2-step quantitative PCR (qPCR). In the first round, 5 µL of DNA was amplified in each of 6–18 25-µL reactions containing 2.5 µL of 10× Expand Long Template Buffer 2, 1.875 U of Expand Long Template Enzyme mix (Roche), 400 nM of primers 1st-Gag-R (5′-CAATATCATACGCCGAGAGTGCGCGCTTCAGCAAG-3′; HXB2 702–718) and 2nd-LTR-F-univ (5′-GTGTIGAAAATCTCTAGCAGTGGC-3′; HXB2 616–639), and 500 µM deoxyribonucleotide triphosphates. In some reactions, 400 nM of β-actin primers β-actin-F (5′-CCTTTTTTGTCCCCCAACTTG-3′) and β-actin-R (5′-TGGCTGCCTCCACCCA-3′) were also added. The 5′ 18 bases of 1st-Gag-R are a tag used in the second round of PCR.
ACH-2 [12] cell DNA was diluted in DNA from uninfected primary HPCs or peripheral blood mononuclear cells (PBMCs) to serve as a positive control as a positive control (10 HIV genomes per μl) or control for sensitivity (0.2 HIV genomes per μl). DNA from uninfected HPCs or PBMCs was used as a negative control. Thermocycling was conducted using a thermocycler preheated to 93°C, with the following cycling conditions: 93°C for 2 minutes; 12 cycles of 93°C for 15 seconds, 60°C for 30 seconds, 68°C for 1 minute; and a final cycle of 68°C for 1 minute.
Second-round qPCR reactions were conducted in triplicate, each using 2 µL of the first-round sample in a 50-µL reaction. Reactions contained 25 µL of FastStart TaqMan Probe Master 2× Master Mix (Roche), 1 µM each of primers 2nd-LTR-F-univ and 2nd-Tag-R (5′-CAATATCATACGCCGAGAGTGC-3′), and 250 nM Gag-probe-2 (5′-FAM-CGCTTCAGCAAGCCGAGTCCTGC-BHQ-1-3′; Biosearch Technologies). Reactions were run and analyzed on an Applied Biosystems 7300 thermocycler, with the following cycling conditions: 95°C for 10 minutes, then 45 cycles of 95°C for 15 seconds followed by 60°C for 60 seconds.
Second-round qPCR reactions to amplify β-actin were conducted to validate cell counts. Conditions were identical to those listed for HIV-1 qPCR except that the β-actin-F and β-actin-R primers and a β-actin probe (5′-FAM-CCCAGGGAGACCAAAAGCCTTCATACA-BHQ-1-3′; Biosearch Technologies) were used.
DNA Sequencing
Positive qPCR reactions were run on a 2% agarose gel, the amplicon excised, and the DNA extracted (QIAquick Gel Extraction Kit, Qiagen). Amplicons were sequenced by Sanger dideoxy sequencing and analyzed using 4Peaks (Mekentosj), EditSeq (DNAStar), and MEGA 5.05.
Statistical Analysis
Excel 2004 (Microsoft) was used to calculate 95% confidence intervals (CIs) for cell counts. We calculated 95% CIs for the fraction of cells that were CD133+ and CD3+, using a 95% CI generator for binomial distributions (available at: http://statpages.org/confint.html). Fisher's exact test was performed using an online calculator (available at: http://www.langsrud.com/fisher.htm). The Mann–Whitney U test and t test were performed using GraphPad Prism 5.0a.
RESULTS
Donor Characteristics
For the 11 donors in this study, the mean time (±SD) since diagnosis of HIV infection was 11.9 ± 8.1 years (range, 3–24 years; Table 1). Donors had had viral loads of <48 copies/mL for at least 6 months (mean duration [±SD], 4.1 ± 2.7 years; range, <1–8.4 years) and were being treated with at least 3 active antiretroviral agents at the time of bone marrow aspiration (Table 1).
Table 1.
Donor Characteristics
| Donor | Year of Diagnosis | Years of ART Use With a VL of <50 Copies/mL | CD4+ T-Cell Count, Cells/µL | WBC Count, ×109 Cells/L | BMMC Count, ×106 Cells/mL | Repeat Donor?a |
|---|---|---|---|---|---|---|
| 303000 | 1990 | 1.0 | 1026 | 4.5 | 15.9 | No |
| 304000 | 2004 | 3.3 | 533 | 4.8 | 9.35 | No |
| 305000 | 2007 | >0.5 | 1421 | 6.4 | 15.4 | No |
| 306000 | 1986/1987 | 3.4 | 1039 | 9.4 | 11.4 | No |
| 307000 | 1992 | 5.8 | 829 | 8.0 | 20.7 | No |
| 308103 | 2008 | 1.6 | 852 | 5.3 | 5.05 | Yes (3) |
| 311000 | 1999 | 8.2 | 564 | 7.2 | 6.25 | No |
| 312101 | 2006 | 2.2 | 812 | 7.5 | 8.75 | Yes (1) |
| 313212 | 2002 | 8.4 | 466 | 4.0 | 9.00 | Yes (12) |
| 314000 | Late 1980s | 5.1 | 543 | 7.6 | 4.22 | No |
| 315214 | 2006 | 5.0 | 850 | 6.3 | 4.11 | Yes (14) |
Abbreviations: BMMC, bone marrow mononuclear cell; VL, viral load; WBC, white blood cells.
a Number in parentheses indicates the donor number in the study by Carter et al [8].
Magnetic Sorting for CD133 Minimizes T-Cell Contamination
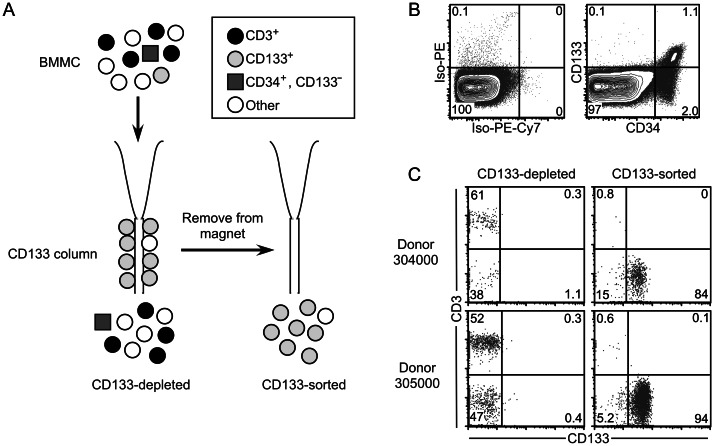
BMMCs from each donor were subjected to magnetic sorting for CD133 (Figure 1A), a surface marker found on a subset of CD34+ cells (Figure 1B). CD133 and CD3 expression on CD133-sorted and CD133-depleted populations was analyzed by flow cytometry (Figure 1C). Although the purity of the CD133-sorted populations varied from 84.4% to 98.9% among donors, <1% of the cells in each CD133-sorted sample were CD3+ T cells (Figure 1C and Table 2). By contrast, 36%–82% of cells in CD133-depleted samples were CD3+ T cells.
Figure 1.
CD133+ cells isolated by magnetic sorting are minimally contaminated with CD3+ T cells. A, Purification protocol. CD133+ hematopoietic progenitor cells (HPCs) were isolated from total bone marrow mononuclear cells (BMMCs) by using anti-CD133-conjugated magnetic beads. Cells were sequentially sorted on 2 columns to maximize the purity of the CD133-sorted population. Bone marrow cells not expressing CD133, including more mature CD34+CD133− HPCs, were collected in the CD133-depleted fraction. B, Example of CD133 and CD34 staining on adherence-depleted bone marrow cells from a healthy donor. Live cells were gated on the basis of forward scatter (FSC), side scatter (SSC), and 7-aminoactinomycin D (7-AAD) uptake. Numbers indicate the percentage of the population falling into each quadrant. The percentage of BMMCs that are CD34+ ranges from 0.1% to 5% between donors. C, Flow cytometric analysis of CD133 and CD3 expression in CD133-sorted and CD133-depleted samples. Live cells were gated based on FSC, SSC, and 7-AAD uptake. Numbers indicate the percentage of the population falling into each quadrant.
Table 2.
Purity of Samples and Frequency of Human Immunodeficiency Virus (HIV) Genomes Detected
| Donor | 104 Cells Analyzed (95% CI) | Percentage of CD133+ Cells (95% CI) | Percentage of CD3+ T Cells (95% CI) | HIV Genomes Detected, No. | Frequency of HIV Genomes per 105 Cells In Vivo (95% CI) |
|---|---|---|---|---|---|
| CD133-sorted samples | |||||
| 303000 | 14 (9.7–17) | 98.8 (98.4–99.0) | .05 (.01–.14) | 0 | <.71 (0–3.8) |
| 304000 | 2.7 (2.4–3.0) | 84.4 (81.6–86.9) | .80 (.29–1.7) | 1 | 3.7 (.084–23) |
| 305000 | 6.0 (5.6–6.4) | 94.1 (93.3–94.9) | .59 (.36–.89) | 1 | 1.7 (.040–9.9) |
| 306000 | 5.4 (4.8–6.0) | 94.3 (93.5–95.0) | .62 (.39–.94) | 0 | <1.9 (0–7.7) |
| 307000 | 12 (11–13) | 93.2 (92.5–93.8) | .22 (.12–.38) | 0 | <.83 (0–3.3) |
| 308103 | 1.4 (1.0–1.7) | 92.1 (90.2–93.8) | .23 (.03–.82) | 0 | <7.1 (0–37) |
| 311000 | 4.1 (3.6–4.5) | 97.7 (97.1–98.3) | .83 (.51–1.3) | 1 | 2.5 (.057–15) |
| 312101 | 8.4 (5.9–11) | 98.9 (98.7–99.1) | .06 (.02–.15) | 1 | 1.2 (.023–9.5) |
| 313212 | 5.4 (4.3–6.5) | 98.4 (98.0–98.7) | .29 (.16–.48) | 1 | 1.9 (.039–13) |
| 314000 | .96 (.83–1.1) | 85.8 (81.4–89.5) | .65 (.08–2.3) | 0 | <10.4 (0–44) |
| 315214 | .48 (.35–.61) | 92.4 (89.4–94.8) | .76 (.16–2.2) | 3 | 63 (10–250) |
| CD133-depleted samples | |||||
| 303000 | 18 (13–23) | 5.0 (4.0–6.2) | 36 (34–38) | 2 | 1.1 (.11–5.7) |
| 304000 | 4.0 (3.6–4.3) | .18 (.01–1.5) | 61 (56–66) | 0 | <2.5 (0–10) |
| 305000 | 17 (14–20) | .13 (.03–.43) | 52 (50–54) | 0 | <.59 (0–2.7) |
| 306000 | 1.8 (1.7–2.0) | .31 (.08–1.1) | 73 (70–76) | 0 | <5.6 (0–22) |
| 307000 | 13 (12–14) | 1.6 (1.0–2.4) | 43 (40–46) | 1 | .77 (.017–4.8) |
| 308103 | 1.1 (.90–1.2) | <.19 (0–.69) | 75 (71–79) | 2 | 19 (2.0–80) |
| 311000 | 2.5 (2.1–2.9) | <.20 (0–.74) | 58 (54–63) | 2 | 8.1 (.84–35) |
| 312101 | 5.4 (4.7–6.0) | <.30 (0–1.1) | 44 (38–49) | 0 | <1.9 (0–7.8) |
| 313212 | 3.1 (2.4–3.8) | <.15 (0–.55) | 59 (55–63) | 0 | <3.2 (0–15) |
| 314000 | .54 (.42–.66) | <.21 (0–.76) | 82 (78–85) | 1 | 19 (.38–130) |
| 315214 | .41 (.32–.50) | <.22 (0–.79) | 81 (77–84) | 2 | 49 (4.9–224) |
Abbreviation: CI, confidence interval.
CD133-Sorted BMMCs Contain HIV DNA
We used qPCR to determine the frequency of HIV genomes in CD133-sorted and CD133-depleted samples. Samples were at limiting dilution as prepared, with no more than one-third of reactions yielding HIV amplification for any sample. Between 4800 and 180 000 cells were analyzed from each sample. HIV-1 DNA was detected in CD133-sorted samples for 6 of 11 donors and in CD133-depleted samples for 6 of 11 donors, with 0–3 total copies of HIV DNA detected per sample (Table 2). The frequency of cells containing HIV DNA varied from <0.71 to 63 per 100 000 cells for CD133-sorted samples and from <0.59 to 49 per 100 000 cells for CD133-depleted samples (Table 2).
CD3+ T cells Are Unlikely to Account for HIV DNA in CD133-Sorted Samples
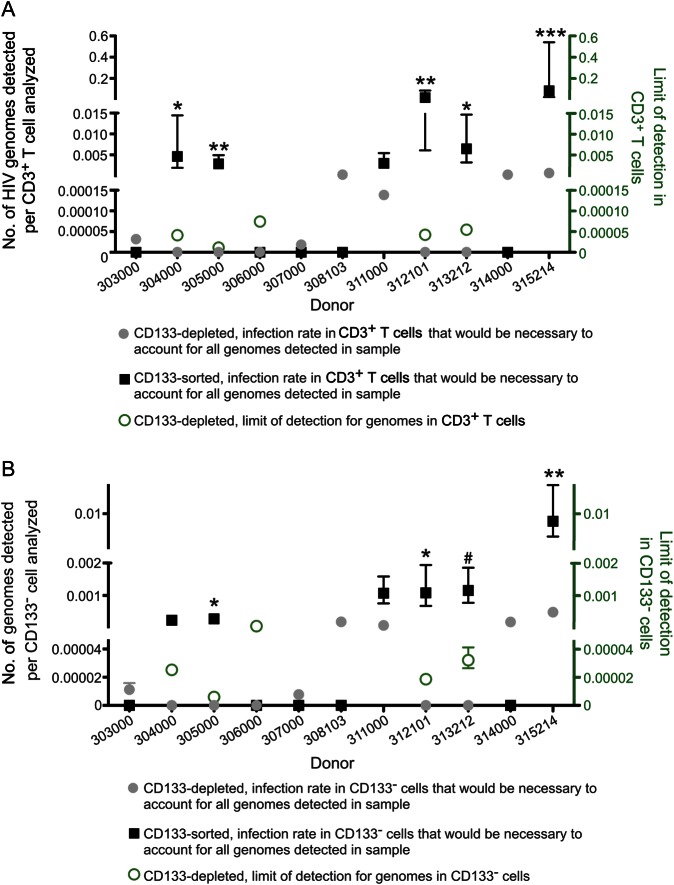
If the HIV genomes detected in our samples derived from CD3+ T cells, we would expect to observe many more HIV genomes in the CD133-depleted samples, which are composed of 36%–82% are CD3+ T cells, than in the CD133-sorted samples, which are composed of <1% are CD3+ T cells. However, we instead observed that the frequency of HIV genomes in CD133-sorted samples was higher than the frequency of HIV genomes in CD133-depleted samples for 4 of 11 donors (Table 2). To further assess the possibility that CD3+ T-cell contamination accounted for the HIV genomes observed in CD133-sorted samples, we calculated the necessary rate of CD3+ T-cell infection to account for all of the genomes detected in each sample (Figure 2A). If all of the HIV genomes were derived from CD3+ T cells, we would expect these calculated frequencies to be similar in the CD133-depleted and CD133-sorted samples for each donor. However, we found that, for 5 of the 6 positive samples, the frequency of HIV genomes in the CD3+ T cells from the CD133-depleted sample was approximately 100-fold too low to support this conclusion (Figure 2A). Using Fisher's exact test, we obtained significantly discordant calculated rates of CD3+ T-cell infection in the 2 samples for donors 304000, 305000, 312101, 313212, and 315214 (P < .0001 [for donor 315214] or P < .01 [for other donors], using mean estimates of CD3+ cell number; P < .01 [for donors 305000, 312101, and 315214] or P < .05 [for donors 304000 and 313212], using conservative estimates; Figure 2A). Donor 311000 demonstrated the same trend, but statistical significance was not achieved (P = .066, using mean estimates; Figure 2A). We therefore conclude that for at least 5 of the 6 donors with detectable HIV DNA in CD133-sorted cells, it is unlikely that the HIV DNA detected in the sorted samples comes from T-cell contamination.
Figure 2.
CD3+ T cells are unlikely to account for human immunodeficiency virus (HIV) DNA in CD133-sorted samples. A, Necessary rate of infection in CD3+ T cells to account for all HIV genomes detected in CD133-depleted samples (gray circles) and CD133-sorted samples (black squares). Error bars indicate 95% confidence intervals. For these calculations, the total number of HIV genomes detected in each sample was divided by the total number of CD3+ T cells analyzed, which was equal to [total number of cells analyzed] × [fraction of cells that are CD3+]. Fisher's exact test was used to compare these calculated frequencies using (1) a mean estimate of the number of CD3+ T cells analyzed, as well as (2) a conservative estimate. The conservative estimate compared the top of the 95% confidence interval for the calculated infection rate in CD3+ T cells in the CD133-depleted sample with the bottom of the 95% confidence interval for the calculated infection rate in CD3+ T cells in the CD133-sorted sample to minimize the difference between these calculated infection rates . *P < .01 by (1), P < .05 by (2); **P < .01 by (1) and (2); ***P < .0001 by (1), P < .01 by (2). For CD133-depleted samples, the limit of detection (green circles) shows the frequency of HIV genomes in CD3+ T cells that would have been calculated on the basis of an observation of 1 HIV genome in the sample. B, Data are as in panel A, except that it was assumed that all genomes were found in total CD133− cells. Mean (1) and conservative (2) estimates of the total number of CD133− cells in each sample were calculated as in panel A. *P < .05 by (1) and (2); **P < .01 by (1), P < .05 by (2); #P < .05 by (1) but not by (2).
It is also theoretically possible that CD133−, non-T cell contaminants could account for the genomes detected in the CD133-sorted cells. However, a calculation analogous to the one described above revealed that CD133− contaminants were unlikely to account for the genomes present in CD133-sorted samples for donors 305000, 312101, and 315214 (P < .05; Figure 2B). For donor 313212, a significant difference was observed using mean values (P < .05) but not conservative estimates (P = .0506) of CD133− cell number (Figure 2B). Donors 304000 and 311000 demonstrated the same trend, but statistical significance was not achieved (P = .096 [for donor 304000] and P = .105 [for donor 311000], using mean estimates; Figure 2B). We therefore conclude that CD133− contaminants are an unlikely source of HIV genomes in the CD133-sorted samples from 3 of 6 positive donors. Importantly, the converse is also true: CD133+ cells are an unlikely source of genomes detected in CD133-depleted samples (Table 2 and data not shown). Thus, these data suggest that at least 2 separable cellular sources can contain HIV DNA in the bone marrow.
HIV Sequencing and Assessment of Contamination
To assess whether the HIV DNA detected arose from laboratory contamination, qPCR amplicons were separated from the primers and probe by gel electrophoresis. In all cases, a distinct band of approximately 120 base pairs was observed, confirming amplification (Figure 3A). Amplicons were extracted from the gel, sequenced, and aligned to the positive control (HXB2; Figure 3B). Analysis of HXB2 DNA from 3 single copy reactions amplified alongside donor samples revealed that all 3 sequences were identical and agreed with the HXB2 reference sequence. In contrast, no 2 donors’ samples yielded identical sequences. Unsurprisingly, we observed identical sequences within the CD133-sorted and CD133-depleted fractions from the same donor (donors 311000 and 315214). Compared with the HXB2 sequence, the numbers of differences observed were similar to those observed in samples from the Los Alamos database (Figure 3C). Thus, it is unlikely that the positive results we obtained are due to contamination by the positive control.
Figure 3.
Sequence analysis of polymerase chain reaction (PCR) products does not suggest contamination with HXB2 DNA. A, Example of agarose gel analysis and purification of quantitative PCR (qPCR) products. S, CD133-sorted sample from the listed donor; FT, CD133-depleted (flow through); NT, no-template PCR control; HXB2, single copy of HXB2 human immunodeficiency virus (HIV) DNA amplified from ACH-2 cells. B, Alignment of donor sequences with HXB2. Numbers indicate coordinates in the HXB2 reference sequence. The HXB2 sequence was obtained by sequencing 3 single-copy qPCR reactions of HXB2 genomes from ACH-2 cells. All 3 sequences were identical to the HXB2 reference sequence. Because the original sequencing chromatogram revealed evidence of 2 different amplicons in the sample from 315214 sort 2, the amplicons were cloned using standard protocols, and sequencing of multiple clones confirmed the presence of 2 genomes (A and B). C, Comparison of the number of differences from HXB2 in our donor sequences and the number of differences from HXB2 in all subtype B isolates in the Los Alamos database that have been sequenced through the region of our 61-nucleotide qPCR amplicon (n = 378; 1 sequence per patient). Each base pair change, insertion, and deletion was counted as 1 difference. Box plots indicate median, first and third quartiles, and minimum and maximum excluding outliers; outliers are indicated with dots. The median number of differences from HXB2 is not significantly different between our samples and those in the Los Alamos database (P = .07) or between ours and Los Alamos subtype B sequences from the United States N = 98, (P = .27, by the Mann–Whitney U test).
Stability of HIV DNA in HPCs Over Time
As noted in Table 1, 4 donors had donated samples for prior studies [8, 11]. At the time of the previous donation, 2 of these donors (donors 308103 and 312101) had high viral loads (>50 000 copies/mL) and subsequently started therapy. The other repeat donors (donors 313212 and 315214) had undetectable viral loads at the time of the previous donation. In the prior study, HIV DNA was detected in HPC samples from both of these donors (previously referred to as donors 12 and 14, respectively [8]). In concordance with these results, we also detected HIV DNA in the current HPC samples, which were collected after an additional 3.3 years of suppressive therapy. Thus, HIV infection of HPCs can be consistently detected in the same donors after years of suppressed viral replication.
Year of Diagnosis Is Associated With Detection of HIV DNA in CD133-Sorted Cells
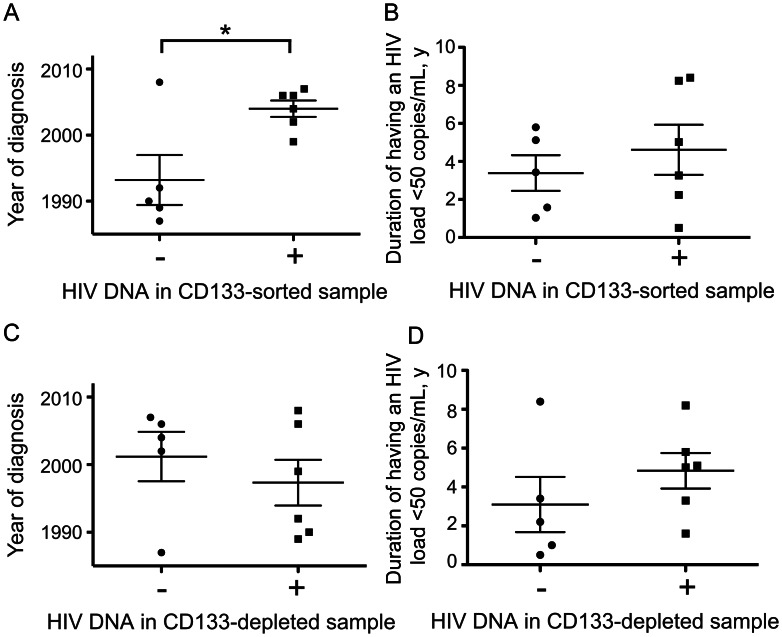
In our prior study we had noted that all 4 donors with detectable HIV DNA in progenitor cells received their diagnosis relatively recently (2001 or later), whereas the 3 donors in whom HIV infection was diagnosed prior to 2001 were PCR negative [8]. We assessed whether this trend held in our current cohort and observed that donors with positive PCR results received their diagnosis significantly more recently than donors without detectable provirus in CD133-sorted samples (P < .02, by the t test; Figure 4A). One sample from a donor with recently diagnosed HIV infection (donor 308103) was negative in the current study, but because only 14,000 cells could be analyzed from this donor (Table 2), this may be a false-negative result. The association between infection of HPCs and year of diagnosis does not result from differences in the duration of viral suppression (P = .49, by the t test; Table 1 and Figure 4B) or from differences in the purity of CD133-sorted samples or the percentage of T cells present (P = .65 or P = .29, respectively). Neither year of diagnosis nor duration of viral suppression was related to detection of HIV DNA in CD133-depleted samples (P = .46 or P = .32, respectively; Figure 4C and 4D).
Figure 4.
Donors with detectable human immunodeficiency virus (HIV) DNA in CD133-sorted samples received a diagnosis of HIV infection significantly more recently than donors without detectable HIV DNA in CD133-sorted cells. A, Comparison of the mean year of diagnosis of donors with or without detectable HIV DNA in CD133-sorted cells. *P < .02, by the t test. Lines indicate means and standard errors; symbols indicate individual values. Conservative estimates of the year of diagnosis were used in cases where the year of diagnosis was not known precisely (1987 for donor 306000 and 1989 for donor 314000). B, Comparison of the mean length of time that the plasma HIV load has been undetectable in donors with or without detectable HIV DNA in CD133-sorted cells. The difference between the 2 groups is not significant (P = .49, by the t test). Lines indicate means and standard errors; symbols indicate individual values. A conservative estimate of 0.5 years for the time that the plasma viral load had been undetectable was used for donor 305000. C, Data are as in panel A, but with donors grouped by whether HIV DNA was detected in CD133-depleted cells. The difference between the 2 groups is not significant (P = .46, by the t test). D, Data are as in B, but with donors grouped by whether HIV DNA was detected in CD133-depleted cells. The difference between the 2 groups is not significant (P = .32, by the t test).
DISCUSSION
Because reservoirs of latent virus represent a barrier to curing HIV infection, it is essential to identify all sources of persistent virus. We previously showed that CD34+ HPCs may harbor HIV genomes in donors with HIV loads of <48 copies/mL [8]; however, subsequent studies suggested that contamination with CD3+ T cells could explain our results [9] or that HIV genomes in HPCs may not persist during years of therapy [10]. Here, we extend our previous findings by showing that HIV can be detected in immature, CD133+ HPCs from donors in whom viral loads have been undetectable for up to 8 years, including 2 donors in whom we detected HIV DNA in CD34+ HPCs in samples donated for our previous study 3 years earlier [8]. We also demonstrate that, for 5 of 6 CD133-sorted samples in which HIV genomes were detected, CD3+ T-cell contamination is a poor explanation for our results. These findings demonstrate that HPCs, including CD133+ HPCs, can harbor HIV DNA during years of therapy.
We estimate that the frequency of HIV genomes in CD133+ HPCs in our donors is <0.71–63 genomes per 100 000 cells. These frequencies are similar to the reported frequency of HIV genomes in peripheral blood CD4+ T cells (1–100 genomes per 100 000 cells [13]). Consistent with reports that bone marrow CD4+ T cells can also harbor provirus [9], we observed HIV DNA in some samples depleted for CD133+ cells. However, our analysis was not designed to determine the type of CD133− cell that was infected.
For the 2 donors examined both here and in our prior study, we found higher frequencies of HIV DNA in HPCs in our prior study [8] than we did here. However, for both donors, the 95% CIs for the true frequency of genomes in these cell populations are overlapping (data not shown). These 95% CIs are very broad because of the low number of detectable genomes. Furthermore, our current study assesses the frequency of HIV DNA among CD133+ cells, whereas our previous study examined total CD34+ cells. It is not clear whether the frequency of HIV genomes in these 2 HPC subsets differs. Finally, the level of variation observed between the sequential measurements from these 2 donors is consistent with the variation among sequential measurements of HIV frequency in resting CD4+ T cells in donors with suppressed viral loads, even though this reservoir is known to decay very slowly, with a half-life of approximately 44 months [14]. To better compare the number of genomes in CD34+ HPCs, CD133+ HPCs, and peripheral blood resting memory T cells, additional studies that simultaneously compare HIV proviral DNA frequencies in all of these cell populations from the same donor are needed. Further studies examining the frequency of HIV genomes over time in the same HPC population from a larger cohort of donors are also required to understand the rate at which HIV genomes in HPCs decay over time and the contribution of these cells to long-term viral persistence.
We report here that donors with evidence of infected HPCs received their diagnosis significantly more recently than donors without evidence of infection. This result cannot be explained by a shorter period of suppressive therapy or by the number of contaminating CD3+ T cells. Instead, we hypothesize that individuals with high levels of HIV infection in HPCs are less likely to have survived or maintained low viral loads if they received their diagnosis prior to the advent of HAART. This reduced survival might be due to higher levels of CXCR4-tropic virus, which we have shown to be required for infection of immature HPCs in vitro [11] and which is associated with more rapid disease progression [15–21]. Alternatively, the inflammation associated with chronic high-level viremia may influence the stability of HIV genomes in HPCs. Further study is required to understand the connection between the frequency of HIV genomes found in HPCs and the year of diagnosis.
In a recent study by Durand et al, which failed to detect evidence of infection of HPCs in optimally treated donors, 10 of 11 total donors received a diagnosis of HIV infection prior to 2001, and 5 of 11 donors received their diagnosis during the 1980s [9]. On the basis of the results reported here, it is not surprising that positive results were not achieved in this study. In addition, the protocol used by Durand et al, included an overnight incubation in serum-containing media, whereas our 1-day protocol used media optimized to maintain progenitors in an undifferentiated state that preserves latent infection [22].
A second study authored by Josefsson et al also failed to detect evidence of HIV infection of HPCs in 8 optimally treated donors whose diagnosis was more recent. However, methodological differences may have contributed to these negative results. Josefsson and colleagues excluded CD4+CD34+ cells from the HPC population studied [10]. Because we have previously shown that CD4 expression on HPCs is required for infection of HPCs in vitro [11], the exclusion of CD4+CD34+ cells would exclude the CD34+ cell population most likely to contain HIV genomes. Furthermore, the primers used in our PCR analysis are substantially more conserved than those used by Josefsson and colleagues (data not shown), which could limit the sensitivity of their assay to detect variable donor HIV sequences.
In vitro, CD133+ HPCs are almost exclusively infected by HIV-1 envelopes that use CXCR4 as a coreceptor [11]. We would therefore expect that at least a minor population of CXCR4-tropic virus exists in the 6 donors in whom we detected HIV DNA in CD133-sorted cells. This is consistent with studies showing that isolates predicted to use CXCR4 can be detected as a minor population in 12%–50% of recently infected patients [23–25]; furthermore, CXCR4-using virus persists in patients receiving suppressive therapy [26, 27] and may become more prevalent during therapy in some patients [28, 29]. Further study to assess HIV envelope tropism in our cohort is needed to confirm the role of HIV envelope tropism in the infection of HPCs in vivo.
Our results demonstrate that HIV genomes can be detected in CD133+ HPCs from a subset of donors with undetectable viral loads and that the genomes detected are not explained by contamination with CD3+ T cells. These findings indicate that HPCs can retain HIV DNA during years of successful antiretroviral therapy. However, they do not prove that HPCs serve as a reservoir for HIV in these donors, as the genomes detected may be defective. We are currently investigating the contribution of HIV genomes in HPCs to residual viremia in treated donors. Meanwhile, strategies to reactivate latent virus from HPCs should be considered alongside strategies that reactivate virus in resting memory T cells to develop therapies with the best chance of eliminating all reservoirs of persistent HIV.
Notes
Acknowledgments. We thank the University of Michigan Center for Statistical Consultation, for services; Mary Reyes, for assistance with recruitment of donors to our study and for help with human subjects regulatory documentation; and Robert Siliciano, John Coffin, Steve King, and Frances Taschuk, for their careful reading of the manuscript.
Financial support. This work was supported by the National Institutes of Health (RO1AI096962), the Burroughs Wellcome Foundation, a National Science Graduate Research Fellowship (DGE 0718128 to L. A. M.), and the University of Michigan (Rackham Predoctoral Fellowship to L. A. M.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature Med. 1999;5:512–7. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Ramratnam B, Tenner-Racz K, et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. New Engl J Med. 1999;340:1605–13. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 3.Geeraert L, Kraus G, Pomerantz RJ. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu Rev Med. 2008;59:487–501. doi: 10.1146/annurev.med.59.062806.123001. [DOI] [PubMed] [Google Scholar]
- 4.Trono D, Van Lint C, Rouzioux C, et al. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;239:174–80. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 5.Bailey JR, Sedaghat AR, Kieffer T, et al. Residual Human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T Cells. J Virol. 2006;80:6441–57. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, Wilke CO. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J Virol. 2009;83:8470–81. doi: 10.1128/JVI.02568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahu GK, Paar D, Frost DW, Smith MM, Weaver S, Cloyd MW. Low-level plasma HIVs in patients on prolonged suppressive highly active antiretroviral therapy are produced mostly by cells other than CD4 T-cells. J Med Virol. 2009;81:9–15. doi: 10.1002/jmv.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter CC, Onafuwa-Nuga A, McNamara LA, et al. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nature Med. 2010;16:446–51. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand CM, Ghiaur G, Siliciano JD, et al. HIV-1 DNA is detected in bone marrow populations containing CD4+ T cells but is not found in purified CD34+ hematpoietic progenitor cells in most patients on antiretroviral therapy. J Infect Dis. 2012;205:1014–8. doi: 10.1093/infdis/jir884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josefsson L, Eriksson S, Sinclair E, et al. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J Infect Dis. 2012;206:28–34. doi: 10.1093/infdis/jis301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter CC, McNamara LA, Onafuwa-Nuga A, et al. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host & Microbe. 2011;9:223–34. doi: 10.1016/j.chom.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clouse KA, Powell D, Washington I, et al. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol. 1989;142:431–8. [PubMed] [Google Scholar]
- 13.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nature Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 15.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–8. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson A, Parsmyr K, Sandström E, Fenyö EM, Albert J. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol. 1994;32:364–70. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarlatti G, Tresoldi E, Björndal A, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nature Med. 1997;3:1259–65. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 18.Shepherd JC, Jacobson LP, Qiao W, et al. Emergence and persistence of CXCR4-Tropic HIV-1 in a population of men from the Multicenter AIDS Cohort Study. J Infect Dis. 2008;198:1104–12. doi: 10.1086/591623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters L, Mandalia S, Randell P, Wildfire A, Gazzard B, Moyle G. The impact of HIV tropism on decreases in CD4 cell count, clinical progression, and subsequent response to a first antiretroviral therapy regimen. Clin Infect Dis. 2008;46:1617–23. doi: 10.1086/587660. [DOI] [PubMed] [Google Scholar]
- 20.Weiser B, Philpott S, Klimkait T, et al. Swiss HIV Cohort Study. HIV-1 coreceptor usage and CXCR4-specific viral load predict clinical disease progression during combination antiretroviral therapy. AIDS. 2008;22:469–79. doi: 10.1097/QAD.0b013e3282f4196c. [DOI] [PubMed] [Google Scholar]
- 21.Yu XF, Wang Z, Vlahov D, Markham RB, Farzadegan H, Margolick JB. Infection with dual-tropic human immunodeficiency virus type 1 variants associated with rapid total T cell decline and disease progression in injection drug users. J Infect Dis. 1998;178:388–96. doi: 10.1086/515646. [DOI] [PubMed] [Google Scholar]
- 22.McNamara LA, Ganesh JA, Collins KL. Latent HIV-1 infection occurs in multiple subsets of hematopoietic progenitor cells and is reversed by NF-κB activation. J Virol. 2012;86:9337–50. doi: 10.1128/JVI.00895-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbate I, Vlassi C, Rozera G, et al. Detection of quasispecies variants predicted to use CXCR4 by ultra-deep pyrosequencing during early HIV infection. AIDS. 2011;25:611–7. doi: 10.1097/QAD.0b013e328343489e. [DOI] [PubMed] [Google Scholar]
- 24.Chalmet K, Dauwe K, Foquet L, et al. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J Infect Dis. 2012;205:174–84. doi: 10.1093/infdis/jir714. [DOI] [PubMed] [Google Scholar]
- 25.Daar ES, Kesler KL, Petropoulos CJ, et al. Baseline HIV type 1 coreceptor tropism predicts disease progression. Clin Infect Dis. 2007;45:643–9. doi: 10.1086/520650. [DOI] [PubMed] [Google Scholar]
- 26.Seclén E, del Mar González M, De Mendoza C, Soriano V, Poveda E. Dynamics of HIV tropism under suppressive antiretroviral therapy: implications for tropism testing in subjects with undetectable viraemia. J Antimicrob Chemother. 2010;65:1493–6. doi: 10.1093/jac/dkq156. [DOI] [PubMed] [Google Scholar]
- 27.Soulié C, Marcelin A-G, Ghosn J, et al. HIV-1 X4/R5 co-receptor in viral reservoir during suppressive HAART. AIDS. 2007;21:2243–5. doi: 10.1097/QAD.0b013e3282f0e3d0. [DOI] [PubMed] [Google Scholar]
- 28.Delobel P, Sandres-Sauné K, Cazabat M, et al. R5 to X4 switch of the predominant HIV-1 population in cellular reservoirs during effective highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2005;38:382–92. doi: 10.1097/01.qai.0000152835.17747.47. [DOI] [PubMed] [Google Scholar]
- 29.Hunt PW, Harrigan PR, Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194:926–30. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]