Abstract
Host cell factor-1(HCF-1) was first discovered as a cellular cofactor in the VP16-induced complex, a multi-protein DNA complex that forms on immediate early gene promoters of herpes simplex virus (HSV) to activate viral gene transcription. Subsequent research has revealed HCF-1 to be an abundant chromatin-associated protein that regulates various stages of the cell cycle. Recent reports show that HCF-1 interacts with diverse E2F proteins to induce cell-cycle-specific transcription. HCF-1 can act as a scaffold to a variety of histone-modifying proteins and these HCF-1-E2F-containing multi-protein complexes can bring about context-dependent activation or repression of transcription. In this review we examine the diversity of HCF-E2F interactions and the variety of multi-protein complexes it occurs in, to influence the local chromatin landscape at the E2F-promoters.
Keywords: E2F, H3K4 HMT, HCF-1, histone modification, transcription
Introduction
Since its discovery as a Herpes Simplex virus host cell factor of unknown cellular functions more than two decades ago, Host cell factor-1 or HCF-1 has emerged as an important regulator of multiple steps in the cell cycle progression. Recent studies implicate HCF-1 as a key regulatory protein in many important and diverse pathways like embryonic stem cell pluripotency, stress responses and development.1-4 Importantly, the discovery that gene-encoding HCF-1 is overexpressed in tumors, and this overexpression is a marker that predicts poor prognosis in cancer treatment proves that the functions of HCF-1 are more complex than previously appreciated.5
In regulation of cell cycle, one of the well-studied roles of HCF-1, it acts as a cofactor to various members of the E2F proteins. The E2F family is one of the best-studied networks of transcriptional regulators. They regulate human-cell proliferation by repressing and activating the transcription of genes required for cell cycle progression, particularly the S phase. In this review we will focus on how HCF-1 is intimately linked with transcriptional regulation of E2F-responsive promoters and discuss how HCF-1 is central to assembly of different chromatin-modifying protein complexes on these promoters.
Two subunits of HCF-1 regulate the cell cycle
HCF-1 is a heterodimeric complex of two subunits—namely N—(HCF-1N) and C—(HCF-1C) subunits. These subunits arise from the proteolytic cleavage of the 2035 amino acid precursor protein.6,7 Although HCF-1 does not bind to DNA directly, both N and C subunits can associate with DNA. In N-terminal subunit, the Kelch domain (so named because of its similarity to drosophila protein Kelch) recognizes and associates with a short tetrapeptide HCF-1-binding motif (HBM) present in many sequence-specific DNA-binding factors like VP16. A single point mutation in the Kelch domain (called P134S) causes a temperature-induced cell-proliferation arrest and cytokinesis defect, while disrupting HCF-1 chromatin association in the temperature-sensitive baby hamster kidney cell line tsBN67.8,9 siRNA experiments have subsequently revealed that these cell cycle defects can be segregated into two subunits where the N subunit is required for G1 phase progression while the C subunit effects proper cytokinesis and M phase progression.10
HCF-1 associates with different chromatin modifiers
HCF-1 cannot bind directly to the DNA but is known to have domains that bind to many chromatin-binding proteins. Indeed, HCF-1 has been shown to interact with a number of proteins. These include transcription factors (LZIP/Luman, Zhangfei, HPIP, Sp1, GABP, YY1, FOXO3 and members of THAP zinc finger protein family), protein phosphatase PP1, cell-death protein PDCD2, deubiquitinating enzyme BAP1, and O-GlcNAc transferase (OGT).6,11-16 Besides these interactions, HCF-1 has also been found to occur in many different multi-protein complexes. First such proteomic analysis revealed, among other proteins, an interaction with Sin3 histone deacetylase (HDAC) complex, and a previously uncharacterized human trithorax- related Set1 histone H3 lysine 4 methyltranseferase (H3K4 HMT) complex.16 Subsequently, HCF-1 has been found associated with related mixed lineage leukemia (MLL) H3K4 HMT complex and the MOF histone acetyltransferase (HAT) complex and other unrelated chromatin remodeler proteins like histone demethylase PHF8, histone deacetylase SIRT1, and histone chaperone Asf1b.2,17-20 Double immunoprecipitation experiments have shown that HCF-1 can bind to the Sin3 HDAC and Set1 H3K4 HMT complex simultaneously, even though these complexes are associated with opposite transcriptional outcomes: repression and activation respectively.16 However, HCF-1 associates with only Set1 H3K4 HMT but not Sin3 HDAC when bound to viral transcriptional activator VP16. These results have suggested that HCF-1 can selectively modulate chromatin structure to broadly regulate transcription.
Regulating the Cell Cycle
HCF-1 may be involved in the regulation of cell cycle at different levels. For instance, HCF-1 may directly interact with proteins involved in cell cycle (e.g., PP1). Or it may regulate the transcription of proteins that regulate the cell cycle (e.g., Pr-Set7/SETD8).21 However, till date the most diverse and most abundant interactions of HCF-1 are with the E2F family, where it interacts with not only different members of the E2F family but also with proteins associated intimately with them.
Interactions with E2Fs
The G1 to S phase transition in mammalian cells is largely achieved by E2F transcriptional factors, which not only regulate the expression of cyclins and genes involved in DNA synthesis but also mitosis, DNA damage and apoptosis pathways. Recent studies show how HCF-1 acts in concert with E2F1 to activate transcription of S phase genes and places HCF-1 right at the heart of cell-cycle transcriptional regulation.22,23
The E2F protein family has currently eight members, called E2F1–E2F8. Of these, E2F1–E2F5 represent a subfamily that shares the property of binding one or more members of the pRb family namely —pRb, p107, and p130. The activation or repression specificity of E2F factors is largely conferred by the E2F-protein subunit. Among the E2F1–E2F5 proteins, E2F1, E2F2, and E2F3a primarily activate transcription (“activator E2Fs”), and E2F3b, E2F4, and E2F5 primarily repress transcription (“repressor E2Fs”).
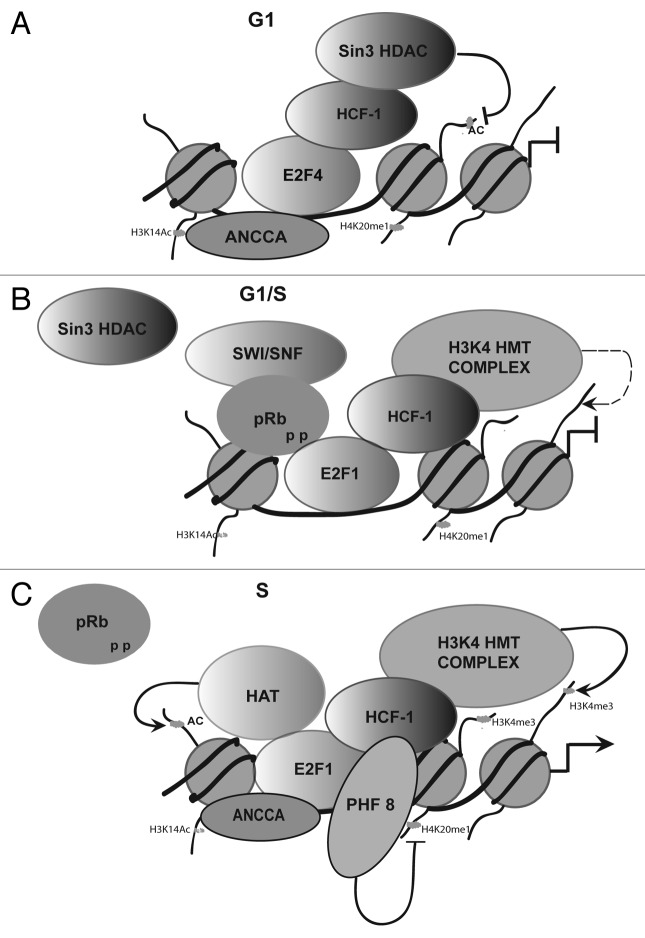
Using a number of techniques, Tyagi et al., demonstrated that HCF-1 associates with both kinds of E2Fs namely activator (i.e., E2F1 and E2F3a) and repressor (i.e., E2F4) E2F proteins in multi-protein complexes.23 Association of HCF-1 with both activator as well as repressor E2Fs gave the first indication that HCF-1 may have a broad role in regulation of genes throughout the cell cycle and that the associations of HCF-1 with different E2F proteins may be cell cycle dependent.23,24 Indeed, the HCF-1—E2F interactions are dynamic and cell cycle selective. Thus, in the early G1-phase when, the E2F responsive promoters are repressed by E2F4 proteins, HCF-1 associates with this repressor protein. The E2F4—HCF-1 complex transitions to the activator E2F1—HCF-1 complex during the G1/S phase progression. E2F4 represses transcription by associating with pocket proteins and recruitment of a Sin3 HDAC co-repressor complex in resting and early G1 phase cells.25 Interestingly, the E2F4-containing HCF-1 complex in early G1 phase contains the Sin3A co-repressor protein, but no pocket protein (see Figure 1A). These results support the idea that E2F4 can regulate genes independent of pocket proteins by recruiting proteins like HCF-1.
Figure 1. Model for E2F-HCF-1 complexes during the cell cycle. This model illustrates the interactions of HCF-1 with various members of E2F family during the cell cycle and the different co-factors associated with them. (A) During early G1 phase, HCF-1 interacts with E2F4 and recruits Sin3 HDAC to E2F-responsive promoters. We speculate that this mechanism of repression is independent of pocket proteins. ANCCA binds to E2F-promoters in late mitosis. However, we do not know if HCF-1 and ANCCA co-occupy these promoters at this time. (B) This model proposes that HCF-1 forms an intermediate complex with pRB-E2F, where pRb has undergone phosphorylation by cyclin D /CDK4 but still remains bound to E2F. As the G1 to S phase transition progresses, this intermediate complex is freed of pRb by subsequent phosphorylation by cyclin/CDK complex thereby switching from repressive (pRb) to activating (HCF-1) E2F-complex.23,37 (C) Here we speculate the co-factors associated with E2F1 during S phase. ANCCA gets recruited to chromatin by binding to H3K14ac mark. Here it facilitates the loading of E2F to the chromatin. HCF-1 binds to E2F to promote the activation of S-phase genes by recruiting H3K4 HMT (MLL /Set1) and stimulating H3K4 trimethylation of gene promoters. PHF8 binds to H3K4 me2/3 mark and interacts with HCF-1. It removes the repressive H4K20 me1 marks at these promoters. Previous reports have shown that several histone acetyltransferase (HAT) are required for E2F activity.38-41
During the G1 to S phase transition, when the cell prepares for active S-phase gene expression, a different HCF-1 complex binds to E2F-responsive promoters. Not only does HCF-1 associate with activator E2F1 but double immunoprecipitation experiments revealed that the E2F1 containing HCF-1 complex selectively associate with H3K4 HMT activator (and not Sin3A repressor complex). The authors proposed that HCF-1 might activate transcription of E2F-responsive promoters by recruiting MLL and Set1 complex to these promoters (Fig. 1C). In support of this model, experiments in which HCF-1 was depleted using siRNA, did not affect promoter-binding property of E2F1 protein, but drastically diminished transcription and the presence HMTs and H3K4 trimethylation on these promoters. Thus E2F1 affects G1 to S phase activation of E2F-responsive promoters in concert with HCF-1 and its associated HMT complexes.23
The HCF-1 and E2F interactions may not be as straightforward as they appear here because HCF-1 also interacts with E2F3, an E2F, which exists in two forms (with two different functions!)—E2F3a the longer isoform acts as an activator while E2F3b the shorter isoform acts as a repressor. Unlike E2F1 and E2F4, E2F3 does not possess a canonical HBM. However, a non-canonical E2F3 HBM was sufficient to interact with HCF-1. Even more intriguing is the fact that both E2F3a and E2F3b possess this non-canonical HBM. It will be interesting to find out the nature of HCF-1 interactions with both forms of E2F3 and how these interactions influence the cell cycle.
Interactions with pRb
Previous studies have shown that the temperature-induced cell-proliferation arrest caused by loss of HCF-1 activity in hamster-cell line tsBN67 can be bypassed through the inactivation of pRb by the adenovirus E1A oncoprotein and SV40 large T antigen, indicating that one role of HCF-1 is to counter the activity of pRb in cell-cycle inhibition.8,9 Recent findings indicate that this opposition may be direct as pRb and HCF-1 can bind to E2F1 simultaneously in what is proposed as a repressive (pRb) to activating (HCF-1) transition complex during the G1 to S phase progression (Fig. 1B).23 These studies led Mani and Fay to hypothesize that a similar antagonistic regulatory relationship may exist in worms.26 They identified SUP-35, a new member of the RMD (regulator of microtubule dynamics), as the transcriptional regulator of novel protein PHA-1. PHA-1 regulates morphogenesis of pharynx early on in development. They proposed that C. elegans E2F ortholog, EFL-1, may partner with LIN-35/pRb in the regulation of SUP-35. Using the conserved ortholog of HCF-1 in C. elegans, they showed that HCF-1 functionally antagonizes LIN-35/pRb in the regulation of sup-35 in a complex regulatory network, which functions to control organ morphogenesis. Therefore, the regulatory relationship between pRb and HCF-1 that exists in mammals seems to be conserved in worms.
HCF-1 may be involved in regulation of pRb activity through another pathway involving Miz-1.27 Miz-1 can activate the promoter of the cyclin-dependent kinase inhibitor p15INK4b, which inhibits pRb inactivation by phosphorylation. HCF-1 can repress Miz-1 activation of p15INK4b gene transcription, suggesting that HCF-1 indirectly promotes pRb inactivation by phosphorylation.27 Interestingly, HCF-1 is also involved in regulation of pRb expression.28 During myogenesis, upon induction of differentiation, HCF-1 is recruited to the pRb promoter and upregulates the expression of pRb by coactivating GABP.28 RNAi-mediated knock-down of HCF-1 results in inhibition of pRb upregulation as well as myotube formation. Therefore HCF-1 may be involved in regulating pRb at several levels, suppressing the function of this tumor suppressor in cycling cells and promoting its expression during differentiation.
Even though, pRb family of pocket proteins has three members—pRb, p107, and p130—the regulatory relationship of HCF-1 may be partial to pRb as HCF-1 has not been found in complex with other pocket proteins like p130.23,29 On the contrary, HCF-1 binding site in E2F4 overlaps the pocket-protein binding site, suggesting that pocket-protein and HCF-1 association with E2F4 is mutually exclusive. Although pRb has been best characterized for its repressive E2F-dependent functions that contribute to cell cycle control, there is emerging evidence that pRb has additional targets and performs multiple functions not common to other pocket proteins.30 Given the multi-dimension nature of HCF-1-pRb relationship, it is tempting to speculate that HCF-1 may also participate in such pRb functions.
Interactions with PHF8
Recently, HCF-1 was found in another multi-protein complex containing E2F1, Set1 and PHF8.31 PHF8 contains a Plant Homeo domain (PHD) and Jumonji C domain, and functions as a demethylase for H3K9 mono- and di- (H3K9me1/2), H3K27 di- (H3K27me2)(see reference 32), and H4K20 mono- (H4K20me1) methylated lysines (see reference 31).31,32 PHF8 interacts with the HCF-1N subunit and PHF8 siRNA, like HCF-1 siRNA, induces cell-proliferation arrest. During the G1 to S phase transition, PHF8 is recruited to selected E2F-responsive promoters where it removes the repressive H4K20me1 mark. PHF8 recruitment to these promoters coincides with decrease in H4K20me1 levels and binding of L3MBTL1 — a repressor protein, which has been shown to associate with H4K20me1.33 Further, PHF8 is recruited to its promoters via the interaction of its PHD domain to H3K4me2/3. PHF8 knockdown was accompanied with impaired recruitment of HCF-1 and Set1 along with decreased levels of H3K4 trimethylation on E2F-responsive promoters, whereas HCF-1 knockdown did not impair PHF8 recruitment. As PHD finger proteins strongly bind H3K4me3 and knockdown of WDR5 led to displacement of PHF8 from rDNA, it is not very clear how in this case HCF-1 knockout, which has been shown to recruit H3K4 complex to these promoters, did not impair PHF8 recruitment.32 Nevertheless, this study shows how the same protein complexes (E2F1/HCF-1/Set1/PHF8) may be modulating chromatin at various levels (H3K4 methylation and H4K20 demethylation) to activate transcription (see Table 1).23,31
Table 1. Table summarizing histone residues and modifications which affect the E2F-responsive promoters. Modifying complexes and their co-factors on E2F promoters are listed.
| Histone Residue | Type of modification | Modifying Protein Complex |
Cofactors | Effect on the E2F-responsive promoter | Associated with E2F protein |
|---|---|---|---|---|---|
| H3K4 |
Di- and Tri-methylation |
MLL |
WDR5, Ash2L, HCF-1 |
Activation/ recruitment of co-factors |
E2F1 |
| |
|
Set1 |
WDR5, Ash2L, HCF-1 |
Activation/ recruitment of co-factors |
E2F1 |
| H4K20 |
Demethylation |
PHF8 |
HCF-1 |
Activation |
E2F1 |
| H3K14 |
Acetylation |
HAT |
|
Activation/ recruitment of co-factors |
E2F1 |
| H3K9 | Acetylation |
HAT |
|
Activation/ recruitment of co-factors |
E2F1 |
| Deacetylation | Sin3/HDAC | HCF-1 | Repression | E2F4 |
HCF-1 and PHF8 may be involved in regulation of more than one phase of cell cycle. Liu et al., suggest that as cells enter mitosis, timely dissociation of PHF8, triggered by its phosphorylation by CDK1/cyclin B1 complex, and accumulation of H4K20 me1 primarily by increased activity of methyltranseferase Pr-Set7, facilitates loading of Condensin II complex on the DNA.34 Interestingly, loss of HCF-1 leads to upregulation of Pr-Set7 and perturbs the balance between H4K20 mono- and di-methylation mark.21 These results suggest that both proteins —HCF-1 and PHF8— play crucial role in maintaining proper levels of H4K20 mono methylation at the onset of mitosis. Only future experiments can reveal the extent of their involvement and if they regulate H4K20 mono methylation through the same pathway.
Interactions with ANCCA
The cell may start preparing for G1/S transition right after it exits mitosis. A recent report shows that, the AAA+ ATPase ANCCA (AAA nuclear co-regulator cancer-associated protein)/ATAD2 may be involved in such a function. ANCCA is recruited to the chromatin late in mitosis by interacting with acetylated lysine 14 of histone 3 (H3K14ac) via its bromodomain.35 The authors suggest that ANCCA facilitates in loading of E2F1 to the chromatin and subsequent assembly of the HCF-1-MLL complex on the E2F-responsive promoters that leads to gene activation. In support of this model, ANCCA interacts with E2F1, E2F2, E2F3 and E2F4 directly, and siRNA knockdown of ANCCA affects expression of key cell-cycle E2F target genes. ANCCA depletion results in a significant decrease of the H3K4me3 mark and chromatin occupancy of E2F1, HCF-1 and MLL complex, suggesting that ANCCA may be crucial for the assembly of E2F1 protein complex at G1 to S phase transition.35 However, recent reports from Farnham laboratory showed that N- and C-terminal domains of E2F1, involved in protein-protein interaction, do not participate in targeting E2F1 to the chromatin.36 Therefore, it remains unclear how ANCCA may be involved in loading of E2F1 to chromatin. Perhaps ANCCA does not recruit E2F1 to chromatin directly by protein-protein interactions but influences the local chromatin state rendering it favorable for E2F1 to bind to DNA. In any case, this model brings to lights the importance of distinctive histone modifications like H3K14ac and H3K9K14ac in recruitment of cofactors to E2F-responsive gene promoters (Table 1).
Conclusions
It is becoming exceedingly clear that E2F transcription is not as limited as it appears in our text-book models, but utilizes an array of proteins and histone modifications to activate or repress genes involved in cell proliferation. HCF-1 acts as a co-factor to E2F protein in distinct multi-protein complexes. HCF-1 containing E2F complexes can undergo a switch in composition to act as either an activator or a repressor making a clear classification of HCF-1 into either category difficult. The HCF-1 complexes can act as ‘writers’ and ‘readers’ to a variety of histone marks, adding another layer of regulation to the already complex E2F pathway. The role of E2F pathway is well recognized in cancer. Now, activities of HCF-1 in promoting oncogenesis are coming to light. Therefore, it is all the more essential to understand the full extent of HCF-1 involvement in E2F regulation, the range of such diverse E2F-HCF-1 complexes and their precise role during the cell cycle.
Acknowledgments
We thank W. Herr for his support in our new ventures. Z. Z. is supported by research fellowship from Council of Scientific and Industrial Research and S.T. by Ramalingswamy Fellowship, Department of Biotechnology.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/20711
References
- 1.Rodriguez-Jato S, Busturia A, Herr W. Drosophila melanogaster dHCF interacts with both PcG and TrxG epigenetic regulators. PLoS One. 2011;6:e27479. doi: 10.1371/journal.pone.0027479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, et al. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyagi S, Herr W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 2009;28:3185–95. doi: 10.1038/emboj.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LFF, Songyang Z, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–74. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wysocka J, Herr W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci. 2003;28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 7.Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–88. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Reilly PT, Wysocka J, Herr W. Inactivation of the retinoblastoma protein family can bypass the HCF-1 defect in tsBN67 cell proliferation and cytokinesis. Mol Cell Biol. 2002;22:6767–78. doi: 10.1128/MCB.22.19.6767-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, et al. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–37. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 10.Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 2003;22:2360–9. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, et al. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24:1479–84. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne ACC, Vogel JL, Ortega N, et al. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J Biol Chem. 2010;285:13364–71. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–85. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–88. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luciano RL, Wilson AC. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J Biol Chem. 2003;278:51116–24. doi: 10.1074/jbc.M303470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Smith ER, Cayrou C, Huang R, Lane WS, Côté J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–88. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng H, Nogueira ML, Vogel JL, Kristie TM. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci U S A. 2010;107:2461–6. doi: 10.1073/pnas.0911128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–25. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–19. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 24.Knez J, Piluso D, Bilan P, Capone JP. Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol Cell Biochem. 2006;288:79–90. doi: 10.1007/s11010-006-9122-x. [DOI] [PubMed] [Google Scholar]
- 25.Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–47. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mani K, Fay DS. A mechanistic basis for the coordinated regulation of pharyngeal morphogenesis in Caenorhabditis elegans by LIN-35/Rb and UBC-18-ARI-1. PLoS Genet. 2009;5:e1000510. doi: 10.1371/journal.pgen.1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piluso D, Bilan P, Capone JP. Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J Biol Chem. 2002;277:46799–808. doi: 10.1074/jbc.M206226200. [DOI] [PubMed] [Google Scholar]
- 28.Deléhouzée S, Yoshikawa T, Sawa C, Sawada J, Ito T, Omori M, et al. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells. 2005;10:717–31. doi: 10.1111/j.1365-2443.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- 29.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–51. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Binné UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr., et al. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–32. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–12. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–50. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 33.Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–28. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 34.Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–88. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol Cell Biol. 2010;30:5260–72. doi: 10.1128/MCB.00484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao AR, Rabinovich R, Xu M, Xu X, Jin VX, Farnham PJ. Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome. J Biol Chem. 2011;286:11985–96. doi: 10.1074/jbc.M110.217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin SM, Gall AL, Zheng N, Pavletich NP. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093–106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 38.El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103:13351–6. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–71. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang SE, McMahon SB, Cole MD, Hearing P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem. 2001;276:32627–34. doi: 10.1074/jbc.M102067200. [DOI] [PubMed] [Google Scholar]
- 41.Hsu SI, Yang CM, Sim KG, Hentschel DM, O’Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J. 2001;20:2273–85. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]