Abstract
Centromeres are specialized chromosomal loci that are essential for proper chromosome segregation. Recent data show that a certain level of active transcription, regulated by transcription factors Cbf1 and Ste12, makes a direct contribution to centromere function in Saccharomyces cerevisiae. Here, we discuss the requirement and function of transcription at centromeres.
Keywords: Cbf1, RNA polymerase II, Ste12, centromere transcription, topology
Introduction
Accurate chromosome segregation during mitosis and meiosis is a critical event in the transfer of genetic information to daughter cells. The centromere, the DNA element where kinetochores assemble on sister chromatids, is a key component required for faithful segregation. The centromere serves as a base for the kinetochore-microtubule attachment sites when sister chromatids are separated properly during anaphase. Loss of centromere function leads to misregulation of kinetochore architecture, which is implicated in chromosome instability and in the development of cancer and genetic disease.
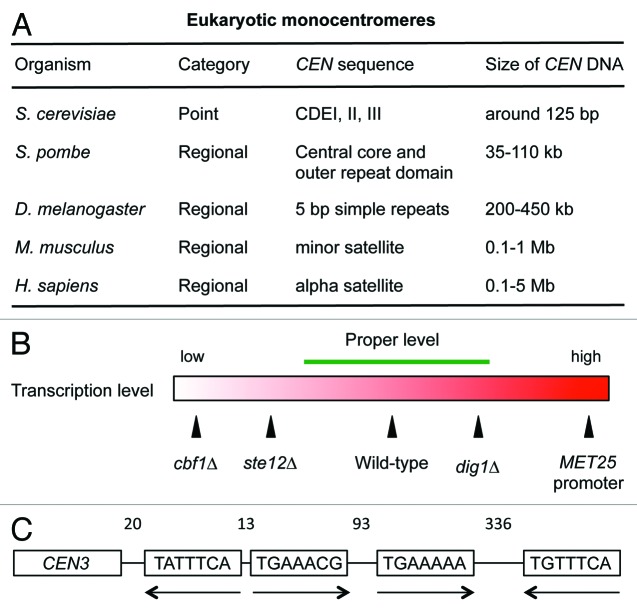
DNA sequences at the centromeres are highly variable with respect not only to the sequence itself but also to the length of the region. The centromeres found in eukaryotic cells are typically categorized into two groups: point centromeres found in the budding yeast Saccharomyces cerevisiae (S. cerevisiae) and regional centromeres found in the fission yeast Schizosaccharomyces pombe (S. pombe), plants and metazoans.1 The length of point centromeres is approximately a hundred base pairs, which are packed into a mononucleosome. Regional centromeres are large repetitive chromatin structures. In some cases, they span more than a few mega base pairs (Fig. 1A). Despite differences in centromere structure, architecture of centromeres—consisting of kinetochore, microtubules and microtubule-organizing centers (MTOCs)—and centromere function are well conserved in eukaryotes.
Figure 1. (A) Organization of centromere DNA in different eukaryotes. (B) Effects of CBF1, STE12 or DIG1 deletion and overexpression from the MET25 promoter on centromere transcription. High or low levels of centromere transcription, such as MET25 promoter and cbf1∆ or ste12∆ cells, impair centromere function. The proper, intermediate, level of transcription is indicated by the green bar. (C) Schematic representation of the organization of putative PREs in the pericentromeric region of CEN3. Numbers indicate the spacing in nucleotides between adjacent PREs.
Since the centromere DNA sequence itself is not the determinate of centromeric function, several epigenetic mechanisms are thought to regulate the centromere function. The incorporation of the centromeric histone variant, CenH3, into the centromeric nucleosome provides one important epigenetic mechanism.2,3 CenH3 is highly conserved from budding yeast to human (Cse4 in S. cerevisiae, Cnp1 in S. pombe or CENP-A in mammals).4 The presence of CenH3 dictates the characteristic single site on each sister chromatid and serves as the foundation of the kinetochore. A second epigenetic mechanism of centromere regulation involves transcriptional control through RNA interference (RNAi) and exosome pathways.3,5 In higher eukaryotes, repetitive sequences of the pericentromeric region are formed as heterochromatin by the RNAi system. We have found that a transcriptional activity is important for centromere function in S. cerevisiae.6 Although the precise role of transcription at the centromere is uncertain, it appears that transcription, per se, plays a role in generating proper centromere topology. Here, we review recent progress toward understanding transcriptional regulation and function at centromeres.
Transcription at Centromeres in S. cerevisiae
The point centromeres of all 16 chromosomes in S. cerevisiae are approximately 125 bp long regions having three conserved DNA elements CDEI, CDEII and CDEIII.7 CDEI is an 8 bp sequence containing a 6 bp palindromic structure, CACRTG (R = A or G). This sequence is bound by a Cbf1 homodimer (a transcription factor and helix-loop-helix family member).8 The central element, CDEII, is 78–86 bp long, and is a highly A:T rich region. This element folds around a single nucleosome containing CenH3 (Cse4).9 The third element, CDEIII, is 25 bp in length and is an essential region for centromere function; it contains a conserved CCG motif that the CBF3 kinetochore complex binds to.10
We have used an in vitro kinetochore assembly system to search for novel proteins that associate with the centromere DNA of chromosome 3 (CEN3), and we identified the Cbf1 and Ste12 transcription factors as well as the Hst1 and Cdc14 silencing factors.6 Cbf1 binds to a CACATG motif in CDEI.11 Ste12 presumably binds a pheromone-response element [PRE; TGAAAC(A/G)] outside the CEN3 CDEIII region.6
Both Cbf1 and Ste12 contribute to CEN transcripts in an RNA Polymerase II-dependent manner. Deletion of CBF1 or STE12 influenced chromosome stability and these mutant strains were sensitive to benomyl (a microtubule-depolymerizing drug). Chromosome instability of cbf1∆ or ste12∆ cells was suppressed by transcription driven from artificial promoters, although in wild type cells strong transcription from the same artificial promoters impairs centromere function. Thus, some level of transcriptional activity at CEN DNA is required for centromere function.
Transcription Factor Cbf1
The Cbf1 transcription factor is a basic helix-loop-helix leucine zipper protein that specifically binds to the palindrome structure CACGTG in promoters of numerous genes, including genes encoding proteins of the sulfate assimilation pathway (MET genes).12 Cbf1 is likely to bind the CACGTG motif wherever it occurs in promoter-proximal regions.13 The role of Cbf1 at MET gene promoters appears to involve two separate functions. First, Cbf1 recruits the MET gene transcriptional activators MET4 and MET28.14 Second, Cbf1 plays a role in configuring the correct chromatin structure of the promoter-proximal nucleosome.15 Evidence suggests that the latter function may involve chromatin remodeling ATPase Isw1.16
Cbf1 also specifically binds to the CDEI region of centromere DNA.7 CDEI includes the consensus binding motif of Cbf1 (CACATG or CACGTG). Both consensus motifs are known to be functional in Cbf1 binding,11 although CACATG is not functional in binding of Cbf1 to promoter-proximal regions.13 We have shown that Cbf1 has a role in transcriptional control at centromeres in S. cerevisiae.6 Moreover, the chromosome missegregation phenotype of cbf1∆, which exhibits decreased levels of CEN transcripts, is suppressed by an artificial promoter for CEN transcription.
Cbf1 is conserved among point centromere species, but not in higher eukaryotic species, which have regional centromeres. It appears that Cbf1 is not a core kinetochore protein, but is a regulatory factor in CEN transcription in S. cerevisiae. It is possible that Cbf1 influences nucleosome structure at the centromere in concert with other factors, such as a transcriptional activator and a chromatin remodeling factor as in the case of MET gene regulation. We still do not know what factors mediate centromeric transcription via Cbf1, but the answer to this question is likely to shed light on the regulation of centromeric chromatin structure.
Transcription Factor Ste12
S. cerevisiae Ste12 is a transcription factor that controls two different developmental programs, mating and invasive/pseudohyphal growth.17 Ste12 is regulated by the Fus3 and Kss1 mitogen-activated protein kinase (MAPK) pathways. In response to mating pheromones, Ste12 activates mating-specific gene expression via the Fus3 MAPK cascade. In response to pseudohyphal signals, Ste12 activates genes required for pseudohyphal growth via the Kss1 MAPK pathway. In both pathways, Ste12 activity is inhibited by two functionally redundant inhibitors, Dig1 and Dig2.18
Dig1 was shown to repress Ste12-dependent transcription at centromeres.6 Interestingly, the fidelity of chromosome segregation in dig1∆ cells is higher than in wild-type cells. This is important because expression levels at centromeres correlate closely with the fidelity of chromosome segregation. There is less segregation fidelity when transcription levels are low, as in cbf1∆ and ste12∆ cells, but higher fidelity at medium transcription levels, as in wild-type cells, and high fidelity at high transcription levels as in dig1∆ cells, although strong expression disrupts centromere function (Fig. 1B). Therefore, we conclude that some level of transcription, per se, is important for centromere function. The dig1∆dig2∆ double mutant showed increased fidelity compared with dig1∆ single mutant cells (unpublished data). Thus, Dig1 and Dig2 appear to be functionally redundant for centromere activity as well as for MAPK pathways.
The consensus sequence of the Ste12 PRE binding site is present in the pericentromeric region of CEN3. A recent study has shown that many pheromone-responsive genes appear to possess multiple PREs in their promoter regions.19 In vitro data indicate that Ste12 likely binds to a single PRE as a monomer, therefore a minimum of two PREs would be necessary for Ste12 multimerization required to activate the pheromone response in vivo. There are multiple PREs located within a 500 nucleotide sequence to the right side of CEN3 (Fig. 1C). Multiple PREs are found in the pericentromeric regions of other CENs; thus, regulation by Ste12 in pericentromeric regions is likely similar to that in promoter regions of pheromone-responsive genes, which are regulated by MAPK pathways. It is not known if MAPK pathways are involved in kinetochore regulation or transcriptional control at centromeres. But it is possible that there exist cell signaling cascades, which are regulated by activators and inhibitors, to convey the information of environmental signals to the centromeres. Further investigation will be required to determine if the signaling cascades such as Fus3 and Kss1 MAPK pathways play a role in kinetochore regulation or CEN transcription.
Transcription at Regional Centromeres
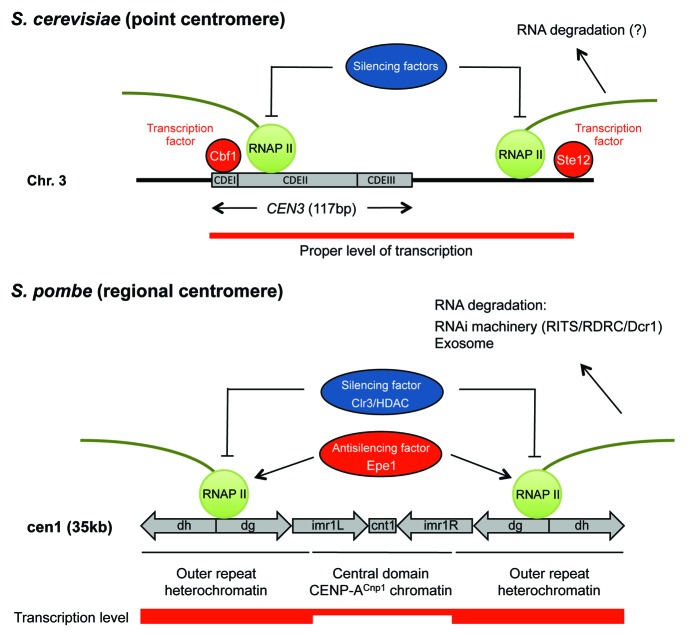
Our findings suggest that there are similarities between point and regional centromeres regarding the role of transcription at centromeres (Fig. 2). First, centromeric regions are transcribed, and the transcription at centromeres and pericentromeres plays an important role in centromere function. In S. cerevisiae, which is known to have point centromeres, some level, albeit poorly defined, of transcriptional activity is required for centromere function.6 In S. pombe, which has regional centromeres, centromeres are transcribed, and the transcription is required for the assembly and maintenance of centromeric heterochromatin.20 The transcription level at pericentromeres appears to be much higher than the level at the central domain of CenH3/CENP-A chromatin in S. pombe.21 TRNA (tRNA) genes are thought to mediate a boundary function separating central and outer repeat domains.
Figure 2. Model illustrating transcription at point and regional centromeres. In S. cerevisiae, the length of the CEN3 sequence is only 117 bp and contains three DNA elements, CDEI, CDEII and CDEIII. The transcription factor Cbf1 binds the CEN3 CDEI site, and Ste12 binds a site pericentromeric of CEN3. RNA Polymerase II (RNAP II) is required for centromeric transcription regulated by Cbf1 and Ste12. Antagonists such as silencing factors might contribute to transcriptional regulation. In S. pombe, the length of the cen1 sequence is 35 kb, and includes a central core (cnt) of non-repetitive sequence and inner (imr) and outer (comprising dg and dh repeats) repeats. RNA Pol II is required for pericentromeric transcription regulated by opposing activities of Clr3 (histone deacetylase, HDAC) and Epe1 (transcriptional activator). The balance of the antagonists determines the functional status at the pericentromeric heterochromatin. Centromeric transcripts are degraded by RNAi machinery and exosomes. The red bar represents the proposed transcriptional levels at repetitive and non-repetitive sequences of the centromere.
Second, RNA Polymerase II is responsible for transcription of both point and regional centromeres. This implies that centromere transcription is evolutionary conserved. Third, the balance of antagonists (e.g., transcriptional activators vs. silencers or RNAi) determines the transcription level and/or status at centromeres. We have identified Cbf1 and Ste12 as transcription factors, and Hst1 and Cdc14 as silencing factors.6 Sir1 (budding yeast silencing protein) and Cdc14 are functional components of centromeric chromatin in S. cerevisiae.22,23 On the other hand, the anti-silencing factor Epe1 and silencing factor Clr3 together regulate the transition between heterochromatin and euchromatin at the outer repeats of centromeres in S. pombe.24
The RNAi pathway has been lost in S. cerevisiae, although some budding yeast species such as Saccharomyces castellii, Kluyveromyces polysporus and Candida albicans (C. albicans) have retained the RNAi system.25 RNAi of S. pombe corresponds to the outer repeats of the centromeres and contributes to heterochromatin formation and maintenance.20 The point centromeres of S. cerevisiae are only around 125 bp, and no repetitive sequences exist in the core and pericentromeric regions. In C. albicans, each centromere is a 3–4.5 kb sequence, which represents an intermediate size between point and regional centromeres.26 Repetitive sequences such as inverted repeats and long-terminal repeats are present in pericentromeric regions of some C. albicans centromeres.26 Therefore, it is possible that there are two mechanisms involved in transcription at centromeres. The first may be to direct heterochromatin formation at repetitive regions of centromeres using the RNAi pathway. The second may be to incorporate CenH3 into the non-repetitive core region sequences of centromeres. In S. pombe, transcripts at the centromere core region might occur as a result of remodeling events that promote CenH3/CENP-A deposition and the transcripts are degraded by the exosome pathway. An increased level of RNA, apparently derived from CEN3, was detected in an exosome mutant of S. cerevisiae.27 The exosome pathway in S. cerevisiae might also regulate the centromeric transcription. Thus, there might be a link between CenH3 incorporation, which is an initial step in the architecture of kinetochore, and transcriptional regulation at the centromere non-repetitive core region.
Interestingly, functional CenH3 nucleosomes induce positive supercoils in eukaryotic cells, although H3 nucleosomes induce negative supercoils.28,29 Dynamic topological changes of DNA occur during the transcription process.30 Perhaps transcription at CenH3 nucleosomes is responsible for regulating not only CenH3 incorporation but also proper topology formation.
Conclusions
Some level of transcriptional activity at point centromeres is required for centromere function in S. cerevisiae. This is a reminiscent of the requirement for transcription at regional centromeres in higher eukaryotes. Our phenotypic and biochemical analyses revealed that transcripts at centromeres are regulated by the transcription factors Cbf1 and Ste12 in an RNA Polymerase II-dependent manner. Opposing factors, including silencing factors, might contribute to transcriptional regulation. This regulation seems to be similar to that occurring in promoter regions. We propose that the balance of antagonists is important for maintaining the appropriate transcriptional status at centromeres, and that transcription generates proper topology formation at CenH3 nucleosomes.
Glossary
Abbreviations:
- S. cerevisiae
Saccharomyces cerevisiae
- S. pombe
Schizosaccharomyces pombe
- MTOC
microtubule-organizing center
- RNAi
RNA interference
- PRE
pheromone-response element
- MAPK
mitogen-activated protein kinase
- tRNA
transfer RNA
- C. albicans
Candida albicans
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/20884
References
- 1.Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–8. doi: 10.1016/S0959-437X(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 2.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–37. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 4.Torras-Llort M, Moreno-Moreno O, Azorín F. Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J. 2009;28:2337–48. doi: 10.1038/emboj.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:239–46. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Ohkuni K, Kitagawa K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol. 2011;21:1695–703. doi: 10.1016/j.cub.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegemann JH, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–60. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 8.Mellor J, Jiang W, Funk M, Rathjen J, Barnes CA, Hinz T, et al. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–26. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–13. doi: 10.1016/S0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 10.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–25. doi: 10.1016/0092-8674(91)90501-O. [DOI] [PubMed] [Google Scholar]
- 11.Wilmen A, Pick H, Niedenthal RK, Sen-Gupta M, Hegemann JH. The yeast centromere CDEI/Cpf1 complex: differences between in vitro binding and in vivo function. Nucleic Acids Res. 1994;22:2791–800. doi: 10.1093/nar/22.14.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–29. [PMC free article] [PubMed] [Google Scholar]
- 13.Kent NA, Eibert SM, Mellor J. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J Biol Chem. 2004;279:27116–23. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- 14.Kuras L, Barbey R, Thomas D. Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 1997;16:2441–51. doi: 10.1093/emboj/16.9.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent NA, Tsang JS, Crowther DJ, Mellor J. Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor 1. Mol Cell Biol. 1994;14:5229–41. doi: 10.1128/mcb.14.8.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreau JL, Lee M, Mahachi N, Vary J, Mellor J, Tsukiyama T, et al. Regulated displacement of TBP from the PHO8 promoter in vivo requires Cbf1 and the Isw1 chromatin remodeling complex. Mol Cell. 2003;11:1609–20. doi: 10.1016/S1097-2765(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 17.Wong Sak Hoi J, Dumas B. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot Cell. 2010;9:480–5. doi: 10.1128/EC.00333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:4794–805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su TC, Tamarkina E, Sadowski I. Organizational constraints on Ste12 cis-elements for a pheromone response in Saccharomyces cerevisiae. FEBS J. 2010;277:3235–48. doi: 10.1111/j.1742-4658.2010.07728.x. [DOI] [PubMed] [Google Scholar]
- 20.Partridge JF. Centromeric chromatin in fission yeast. Front Biosci. 2008;13:3896–905. doi: 10.2741/2977. [DOI] [PubMed] [Google Scholar]
- 21.Choi ES, Strålfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem. 2011;286:23600–7. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp JA, Krawitz DC, Gardner KA, Fox CA, Kaufman PD. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 2003;17:2356–61. doi: 10.1101/gad.1131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–4. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 24.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 25.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, et al. RNAi in budding yeast. Science. 2009;326:544–50. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–65. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- 27.Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–13. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proceedings of the National Academy of Sciences of the United States of America 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–40. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]