Abstract
Transcription termination of RNA polymerase II between closely spaced genes is an important, though poorly understood, mechanism. This is true, in particular, in the Drosophila genome, where approximately 52% of tandem genes are separated by less than 1 kb. We show that a set of Drosophila tandem genes has a negative correlation of gene expression and display several molecular marks indicative of promoter pausing. We find that an intergenic spacing of 168 bp is sufficient for efficient transcription termination between the polo-snap tandem gene pair, by a mechanism that is independent of Pcf11 and Xrn2. In contrast, analysis of a tandem gene pair containing a longer intergenic region reveals that termination occurs farther downstream of the poly(A) signal and is, in this case, dependent on Pcf11 and Xrn2. For polo-snap, displacement of poised polymerase from the snap promoter by depletion of the initiation factor TFIIB results in an increase of polo transcriptional read-through. This suggests that poised polymerase is necessary for transcription termination. Interestingly, we observe that polo forms a TFIIB dependent gene loop between its promoter and terminator regions. Furthermore, in a plasmid containing the polo-snap locus, deletion of the polo promoter causes an increase in snap expression, as does deletion of polo poly(A) signals. Taken together, our results indicate that polo forms a gene loop and polo transcription termination occurs by an Xrn2 and Pcf11 independent mechanism that requires TFIIB.
Keywords: polo, polyadenylation, small intergenic regions, transcription termination
Introduction
The Drosophila melanogaster genome (~180 Mb) encodes over 15,000 protein coding genes that are transcribed by RNA polymerase II (RNAP II).1 Dynamic interactions of multiple factors with RNAP II promote initiation, elongation and termination of transcription. Regulation of RNAP II initiation and elongation rates has been shown to impact on gene expression2 and influence mRNA processing.3-5 Moreover, transcription termination has a role in pre-mRNA processing and may, in some circumstances, enhance protein expression.6
Transcription termination is interconnected with the other transcription steps and it can occur several kilobases after the recognition of the poly(A) signal (PAS, for review see 7, 8). Two mechanisms have been proposed for transcription termination of protein coding genes, the allosteric model9 and the torpedo model.10 However, an emerging view is that the termination mechanism more likely reflects a combination of both.11,12 It is also possible that termination occurs by more than one mechanism, depending on gene context or cell condition, as occurs for 3′ end processing.13 Studies in S. cerevisiae have shown that failure to terminate transcription of a gene affects the expression of the downstream gene.14,15 This process – called transcriptional interference – is particularly important in lower eukaryotes, where genes are often closely spaced.16 Transcriptional interference is less well characterized in higher eukaryotes, even though some mammalian genes also contain small intergenic regions, such as the human complement genes C2-Factor B.17,18
In this study, we focus on the transcription termination mechanism between two closely spaced tandem genes in Drosophila. We investigated the tandem gene pair polo-snap, which is separated by 168 nucleotides from the PAS to the transcription start site (TSS). polo encodes a kinase that acts as a major regulator of various steps of the cell cycle, including mitotic entry, centrosome organization, spindle formation, chromosome segregation and cytokinesis.19,20 We have previously shown that alternative polyadenylation in polo 3′ UTR is essential for fly viability, Polo production and histoblast proliferation, and also that RNAP II elongation rate affects polo PAS selection.4,21 snap encodes a cytosolic factor that promotes neurotransmitter release22 and has a function in vesicle fusion, in both the constitutive and regulated secretory pathways.23
Here we show that polo and snap have a negative correlation in their expression in fly tissues and in S2 cells, a feature shared with a group of genes that have a similar genomic organization. We found that the small intergenic region of 168 bp between polo and snap is sufficient for correct transcription termination by an Xrn2 and Pcf11 independent mechanism that requires TFIIB. In contrast, when we analyzed a tandem gene pair with more than 3 kb of intergenic region, transcription termination occurs further downstream of the PAS by a mechanism dependent on Xrn2 and Pcf11. In agreement with ChIP-seq studies, we observed that RNAP II is poised over the snap promoter, and displacement of RNAP II from the promoter leads to increased transcriptional read-through from polo into snap, suggesting a new role for poised RNAP II in transcription termination. We also show that polo forms a gene loop configuration, linking its promoter and terminator regions, and that this interaction is disrupted upon depletion of the transcription factor TFIIB. Moreover, polo transcription represses snap expression and polo PAS represses snap expression in a promoter dependent manner. Taken together, our results suggest that a poised RNAP II (with high levels of Ser5P) in the intergenic snap promoter regions, the presence of TFIIB (to aid the formation of a gene loop) and the polo PAS all contribute in the mechanism of transcription termination between the closely spaced genes polo and snap in D. melanogaster.
Results
Some Drosophila tandem genes display negative correlation of gene expression
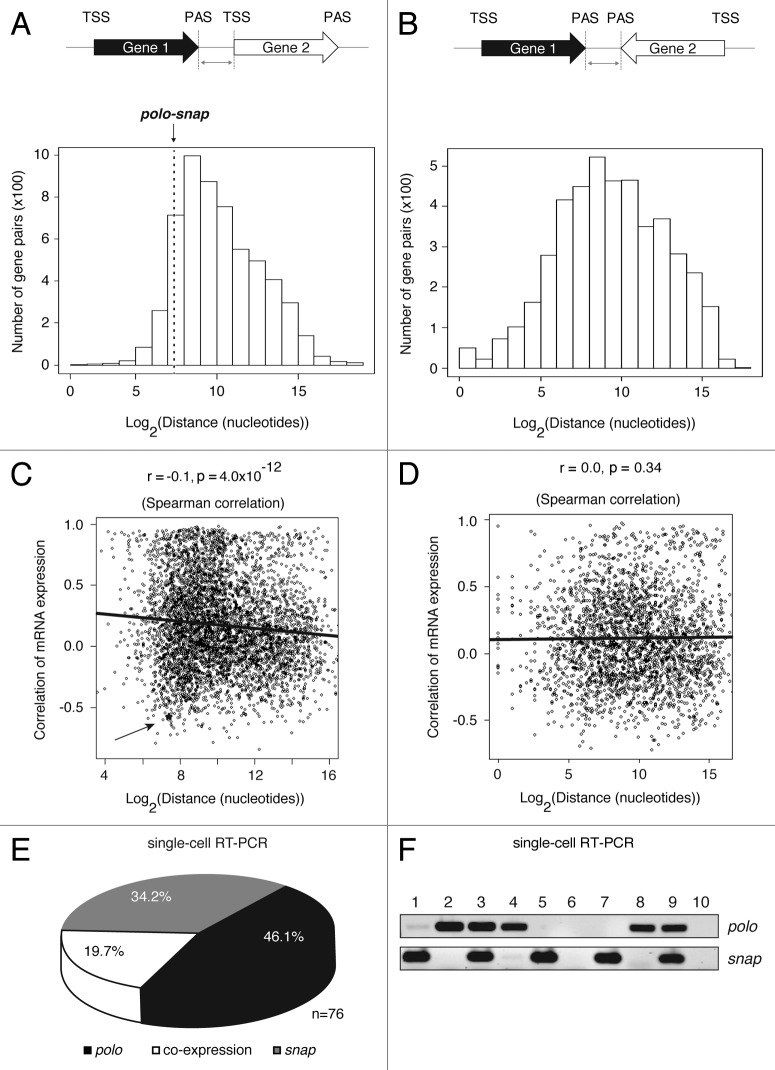
Using bioinformatics tools, we searched the genome (April 2006, BDGP release 5.12/dm3) for genes in a tandem configuration (Fig. 1A) in comparison with convergent genes with a PAS to PAS configuration (Fig. 1B) with a short intergenic region. We found that ~52% of tandem genes are separated by less than 1 kb from PAS to TSS (transcription start site), which is in contrast with higher eukaryotic genomes, where the intergenic sequence between tandem genes is on average 7 kb,24 despite the existence of sporadic overlapping genes.25 We then analyzed the pattern of gene expression in all closely spaced tandem gene pairs of Drosophila based on the data from FlyAtlas26. Correlations of gene expression between gene pairs can be classified as positive (on/on or off/off expression) or negative (on/off or off/on expression). We found that there is an overall positive correlation of expression for adjacent tandem genes with short intergenic regions – less than 1 kb from PAS to TSS. However, a set of genes including polo-snap shows a negative correlation of gene expression (asterisk and arrow in Fig. 1C and S1A). When the intergenic distance increases (> 1 kb), a negative correlation of gene expression tends to be the trend (Fig. 1C). In contrast, from the analysis of convergent genes (PAS to PAS configuration, Fig. 1B) we did not observe any correlation in gene expression (Fig. 1D and S1B).
Figure 1. Some closely spaced tandem genes, including polo-snap, have a negative correlation of gene expression. (A) Schematic representation of the analyzed tandem gene pairs across the Drosophila genome (April 2006, BDGP release 5.12/dm3). The graph represents the number of gene pairs in tandem configuration and the distribution of distances between PAS of Gene 1 and TSS of Gene 2. The distance between polo and snap is 168 nt, which is at the 10.7 percentile among all gene pairs. (B) Schematic representation of the analyzed convergent gene pairs across the Drosophila genome (dm3). The graph represents the number of gene pairs in convergent configuration and the distribution of distances between PAS of Gene 1 and Gene 2. (C) Scatter plot shows an inverse correlation between gene distance (tandem genes) and gene expression. Correlation of gene expression (r) was based on Pearson correlation of expression values from 26 fly tissues. The r and p shown in the figure are based on Spearman correlation. The gene expression correlation between polo and snap is -0.57. An asterisk and arrow mark polo-snap. (D) Scatter plot shows no correlation between gene distance (PAS to PAS only) and correlation of gene expression. Correlation of gene expression (r) was based on Pearson correlation of expression values from 26 fly tissues. The r and p shown in the figure are based on Spearman correlation. (E) S2 single cells were sorted by flow cytometry on exponential phase of growth and single- cell PCR for polo and snap was performed using specific primers. The circle graph shows that in more than 80% of the cells polo and snap are not co-expressed (n = 76). (F) A representative agarose gel showing the PCR products for polo and snap after electrophoresis is shown. Each lane corresponds to a single cell sorted by flow cytometry. Lane 6 and lane 10 represent non-template controls for PCR reactions.
As transcription termination must occur efficiently between closely spaced genes to avoid transcription interference,14 and polo is separated by only 168 nucleotides from snap, we used this gene pair as a model system to investigate the molecular mechanisms involved in transcription termination. We first analyzed the pattern of gene expression for polo and snap by single cell RT-PCR of RNA from isolated S2 cells sorted by flow cytometry into a 96-well qPCR plate (Fig. 1E) and also in several Drosophila tissues using publicly available databases26 (Fig. S1C). To prevent biased assessments and evaluate the single-cell expression pattern, different controls were performed to calculate competition, efficiency and linearity of the PCR reactions (Fig. S2A-C). As shown (Fig. 1E-F), polo and snap are separately expressed in more than 80% of cells, suggesting that polo transcription interferes with snap expression. This pattern is maintained after several cell divisions (cells were split several times between experiments), as confirmed by repetition of the single cell PCR using limiting dilution of cells with different passage numbers (Fig. S2D). It is well known that a tight connection exists between the different steps in the transcription cycle. In view of the short intergenic region between polo and snap, we hypothesized that polo transcription termination could interfere with snap transcription initiation. These considerations lead us to dissect the molecular mechanisms involved.
Stalled RNAP II accumulates in intergenic regions of closely spaced, negatively correlated tandem genes
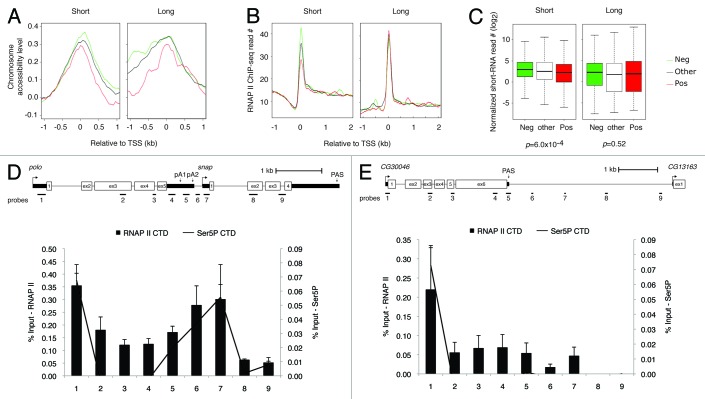
Our findings that in Drosophila ~52% of the genes are separated by less than 1 kb of intergenic DNA and that a set of closely spaced genes have a negative correlation of gene expression (named “Neg gene set” henceforth, Fig. 1C) lead us to investigate their molecular features. We first divided the Neg gene set and the set of genes that show a positive correlation of expression (Pos gene set) into “short” and “long” groups, based on the distance between PAS to TSS and PAS to PAS (see further description of gene sets and groups in Fig. 2 and Materials and Methods). We then compared several molecular features that include chromosome accessibility based on a modified DNA methylase accessibility assay (MeDIP footprint27), chromatin marks28 and RNAP II levels, based on ChIP-seq and deep sequencing of short RNAs derived from stalled RNAP II29 (data sets described in Materials and Methods), and identified different patterns of gene expression associated with distinct levels of RNAP II.
Figure 2. Molecular features around the promoter region for different sets of genes and the distinctive space requirements for transcription termination. (A) Chromosome accessibility around TSS. (B) RNAP II level based on ChIP-seq. (C) Short RNAs generated by promoter stalled RNAP II in the -50 nt to 100 nt region around TSS, normalized to the gene expression level estimated by RNA-seq. Short: gene pairs with distance < 1 kb; Long: gene pairs with distance > 1kb. Neg: negatively correlated gene pairs, corresponding to those with the lowest 20% r values in Figure 1; Pos: positively correlated gene pairs, corresponding to those with the highest 20% r values in Figure 1; Other: gene pairs not Neg or Pos. Only tandem gene pairs were used. The x-axes of all plots are distance relative to TSS with 0 being TSS. (D) Schematic representation of the polo-snap locus, where PAS and primer positions used for RT and qPCR are depicted to scale. ChIP results showing association of RNAP II (8WG16, bars) and Ser5P (4H8, line) with the different regions of polo and snap genes in Drosophila S2 cells. Numbers below each bar represent the position of qPCR primers as depicted at the top of the panel. Error bars show s.e.m. from at least three independent experiments. (E) Same as (D) but for more widely separated tandem genes CG30046-CG13163.
Knowledge of the precise location of nucleosomes in the genome is essential to understand the context in which processes such as transcription and DNA replication operate.27,30 A common theme emerging from recent genome-wide maps of nucleosome locations is a general deficiency of nucleosomes in promoter regions and an enrichment of certain histone modifications toward the 5′ end of genes.30,31 First, we analyzed chromosome accessibility and found that there is higher chromosome accessibility among Neg gene set in the intergenic region than in the Pos gene set. This suggests that this region is nucleosome depleted, and that this is independent of the intergenic distance (Fig. 2A, left panel). Considering that gene-rich regions are generally embedded in accessible chromatin, our finding was not surprising. Recent studies have shown that promoter-proximal pausing is a natural RNAP II feature, with about 20 to 30% of genes displaying 5′ end enrichment.29,32 Strikingly, this occurs for both active and inactive genes.32 Moreover, Gilchrist et al.31 have shown that paused RNAP II correlates with modified nucleosome architecture. Therefore, the correlation that we found between higher chromosome accessibility and the negative correlation of gene expression lead us to use ChIP and small RNA-seq data29 to investigate RNAP II occupancy in the Neg and Pos gene sets. As we focused on tandem genes with a short intergenic distance, the promoter of the downstream gene is included in these analyses. We observed an enrichment in RNAP II stalling (Fig. 2B, left panel), which is further supported by small RNA-seq reads for the Neg gene set (Fig. 2C, left panel). Strikingly, such a tendency was not detected for more widely spaced tandem genes (Fig. 2B-C, right panels). In addition, genes in the Neg gene set contained more insulators around the promoter, including CTCF, BEAF32 and CP190 (Fig. S3). However this feature did not appear to be related to gene distance. It might therefore be important to maintain the promoter proximal chromatin in a region that is prone to bind RNAP II.33 Importantly, all the distinct features of the Neg gene set could also be found at the snap promoter (Fig. S4), suggesting that polo-snap is a good model representing this gene set in the Drosophila genome. Taken together, these results indicate that closely spaced tandem genes with a negative correlation of expression present higher chromosome accessibility and higher RNAP II levels near the TSS. They also produce more short RNAs derived from stalled RNAP II in their promoter regions, as compared with those that have a positive correlation of expression (Fig. 2A-C). This trend is clear for closely spaced genes and indicates that gene distance is an important determining factor for such features.
Long and short intergenic regions require different mechanisms of RNAP II transcription termination
Our results suggest that stalled RNAP II may have other functions than solely forming a mature elongation complex or serving as an insulator.29,34 Thus, we elected to study the mechanism of transcription termination between the closely spaced tandem gene pair polo-snap. We first mapped RNAP II density using ChIP across both polo and snap transcription units. RNAP II occupancy was assayed at 9 positions (Fig. 2D) in S2 cells, with an antibody directed to the largest subunit of RNAP II. Each position corresponds to a 70 to 200 bp amplicon that was quantified by qPCR. The highest levels of RNAP II were observed on polo promoter (probe 1 in Fig. 2D), as well as over the intergenic/ snap promoter region (Fig. 2D, probes 6 and 7). To confirm these results, we used ChIP-seq data29 to analyze RNAP II occupancy in the polo-snap locus and observed that RNAP II levels are higher in the intergenic/ snap promoter region (Fig. S4). When we further characterized the phosphorylation state of the RNAP II large subunit C-terminal domain (CTD) using Ser5P specific antibody, we observed a correlation between high levels of RNAP II occupancy and high levels of Ser5P (line in graph of Fig. 2D). This suggests that stalled RNAP II is largely Ser5P modified, in agreement with recent genome-wide studies.29
We next investigated a pair of more widely separated tandem genes, containing more than 3 kb of intergenic region (CG30046-CG13163). As previously observed for some mammalian genes,35,36 we found RNAP II present in the intergenic region for more than 1 kb downstream of the PAS (up to probe 7, Fig. 2E), suggesting that termination occurs farther away from the PAS of the upstream gene. RNAP II is probably released downstream of probe 7, as RNAP II levels over probe 8 and 9 were undetectable. In contrast to polo, levels of RNAP II at the 3′ end of the upstream gene were low. The pattern of RNAP II occupancy is thus clearly distinct between these two gene pairs (polo-snap vs. CG30046-CG13163) implying distinct modes of transcription termination.
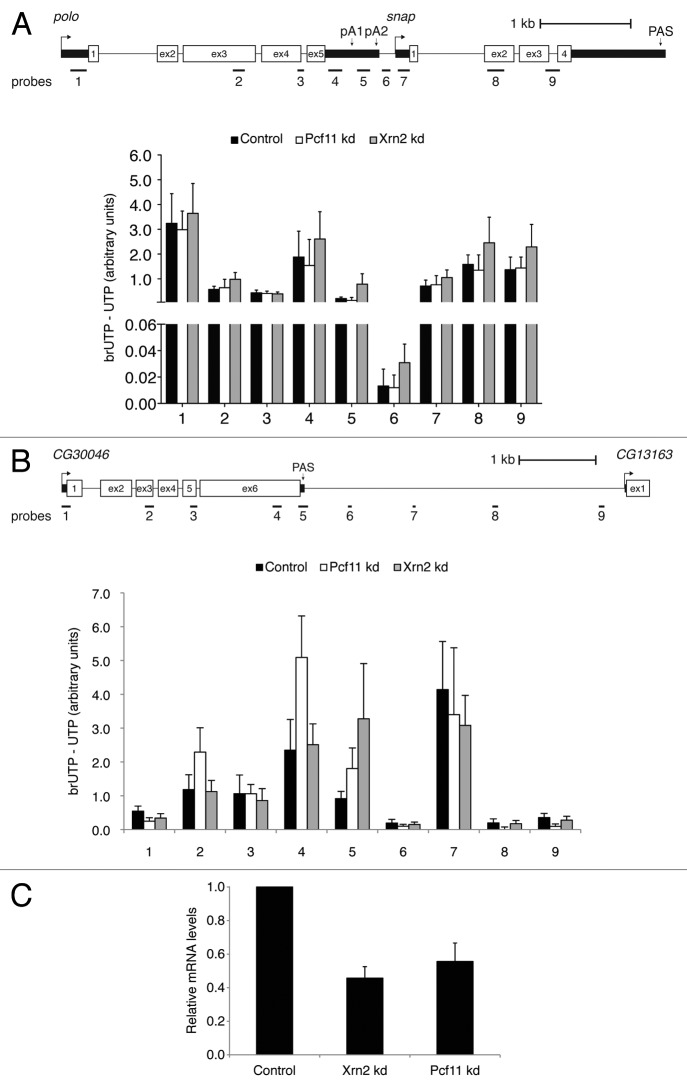
Nascent transcription was assessed across these two gene pairs by measuring the density of engaged RNA polymerases by use of a modified nuclear run-on (NRO) technique. This procedure involves incorporation of bromo-labeled UTP (BrUTP) into nascent RNAs and then immunoprecipitated using a BrUTP-specific antibody6. cDNA was synthesized with primers 1–9 and PCR amplification was performed with the same primer pairs to detect pre- RNA. In order to measure read-through unprocessed RNA at position 6 of Figure 3A, cDNA was synthesized with the reverse primer 6 and quantified by qPCR with primer pair 5. The background obtained from incorporation of UTP into nascent RNAs was subtracted. For both gene pairs we found a drop in levels of the nascent transcripts in the region downstream of the PAS, both for polo and for CG30046 (black bars in position 6, Fig. 3A and B). For polo, although there is a small decrease in the levels of unprocessed transcripts downstream of pA1, a stronger 16-fold decrease is observed between probes 5 and 6 (Fig. 3A, compare black bars in position 5 and 6). Remarkably, less than 8% of polo pre-RNAs were found to read-through into the snap-coding region. This clearly indicates that polo transcription termination occurs efficiently downstream of pA2 and that the small intergenic region is sufficient for termination to take place. For CG30046, we also observe a decrease in the levels of nascent transcripts downstream of the PAS over probe 6 (black bar, probe 6 in Fig. 3B), which occurs ~700 nt downstream of the PAS. Surprisingly, for this gene, there is an accumulation of nascent transcripts over probe 7. We predict that this may be due to the presence of an enhancer, as this region possesses a strong H3K4me mark but does not produce significant levels of polyA+ RNA.30,37 In conclusion, our BrUTP-NRO analyses allowed us to accurately map RNAP II transcription termination occurring in the 168 bp of intergenic region between polo and snap, and 700 nt downstream of the CG30046 PAS.
Figure 3. Transcription termination in the short polo-snap intergenic region by an Xrn2 and Pcf11 independent mechanism. (A) BrUTP-NRO analysis of polo and snap. Quantification shows the value obtained after subtracting the UTP control value. (B) As in (A), but for CG30046. (C) Analysis of the efficiency of Xrn2 and Pcf11 RNAi treatment by measuring mRNA levels using qRT/PCR. Error bars show s.e.m. from at least three independent experiments.
To dissect the molecular mechanism of polo transcription termination, we next investigated the role of Xrn2 and Pcf11.8,10,38 Xrn2 is a 5′-3′ exoribonuclease necessary for termination,10 while Pcf11 (cleavage and polyadenylation factor subunit) is a subunit of CFI, that has been shown to have a function both in mRNA 3′ end formation and in transcription termination.7,38,39 After successful RNAi mediated-depletion of each factor (Fig. 3C), endogenous unprocessed transcripts of polo were measured using BrUTP-NRO. RNA was isolated and reverse transcribed with primers 1–9, and cDNA was amplified by qRT-PCR using the same strategy as above. Surprisingly, no increase in polo transcriptional read-through was observed upon depletion of Xrn2 or Pcf11 (Fig. 3A, compare black with white and gray bars - Pcf11 and Xrn2 knockdown respectively - for probes 5 and 6). The same results were obtained using a read-through assay: RNA was isolated and reverse transcribed with primers 3R and 6R and cDNA was amplified by qRT-PCR using primers 3F/3R and 6F/6R, to detect RNA beyond polo pA2 (see Fig. S5A and Materials and Methods for primers description). Upon normalization of the read-through RNA to polo pre-RNA and comparison to the control condition, no increase in transcriptional read-through was observed upon depletion of Xrn2 and Pcf11 (Fig. S5A). In agreement with these results, Xrn2 and Pcf11 depletion did not affect the level of polo mRNA (Fig. S5B). This data indicates that the mechanism of polo transcription termination is Xrn2 and Pcf11 independent. We followed the same approach to measure transcriptional read-through of CG30046 following RNAi depletion of Pcf11 and Xrn2. With Pcf11 depletion levels of nascent transcripts over probes 2, 4 and 5 increased (Fig. 3B, white bars). Moreover, depletion of Xrn2 leads to increased levels of nascent transcripts over probe 5 (Fig. 3B, gray bars). These results indicate that Xrn2 and Pcf11 are necessary for CG30046 transcription termination suggesting that genes with short and long intergenic regions require different termination factors.
TFIIB is enriched among negative gene set and its depletion leads to increased polo read-through
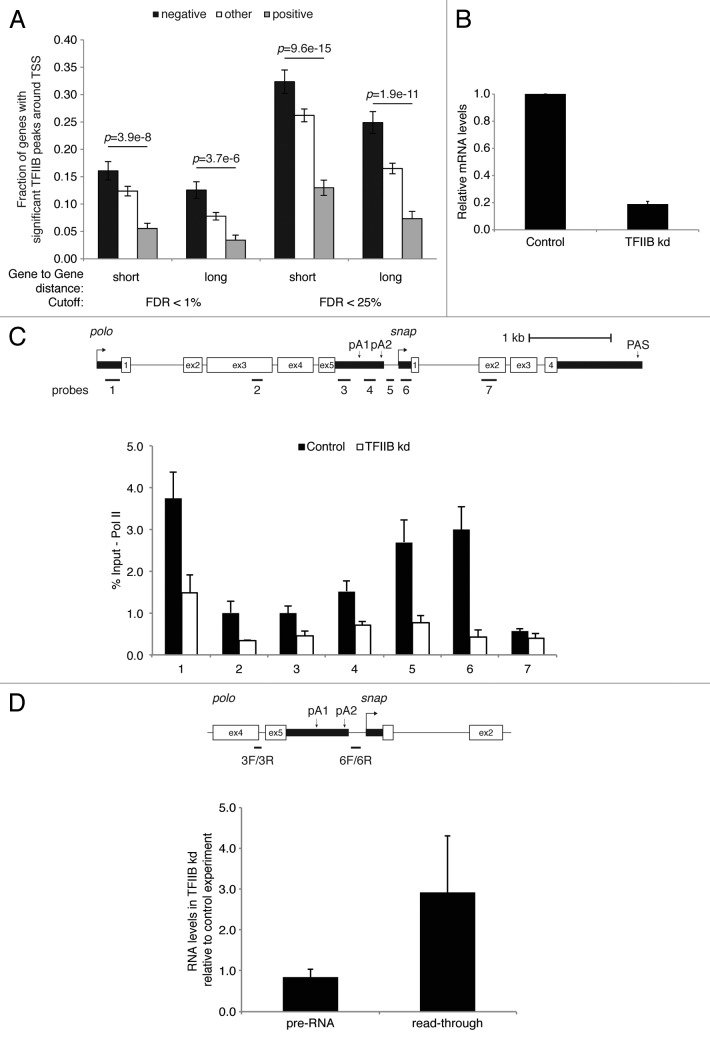
TFIIB is known to promote interactions between the promoter and terminator.40 It interacts with yeast and human cleavage and polyadenylation factor (yCPF and hCPSF) and CstF complexes41,42 and this interaction contributes to the juxtaposition of promoter and terminator DNA at active gene loci enabling RNAP II recycling and rapid re-initiation. Moreover, the interaction of TFIIB with 3′ end processing complexes is regulated by the phosphorylation of TFIIB at Ser65. This helps productive transcription initiation and most likely facilitates gene looping through its interaction with CstF components.41 As TFIIB has been previously reported to bind to polo and snap promoters,43 we investigated its role in our Neg and Pos gene sets and in polo transcription termination. We observed a TFIIB enrichment among the Neg gene set in comparison with the Pos gene set (Fig. 4A, compare black with gray bars). As TFIIB was shown to engage RNAP II complexes that are involved in mRNA 3′ end processing and transcription termination,41,44 we hypothesized that TFIIB depletion could lead to a transcription termination defect. Upon successful TFIIB RNAi- mediated depletion (Fig. 4B), we observed a substantial reduction in RNAP II levels across polo-snap, most pronounced in the intergenic/ snap promoter region (Fig. 4C, compare black and white bars in probe 5 and 6) as measured by ChIP against the largest RNAP II subunit. Furthermore, RNAP II displacement was accompanied by a 3-fold increase of transcriptional read-through measured by primer pair 6F/6R in comparison with nascent pre-RNA in TFIIB depleted samples, using the previously described qRT-PCR assay that recapitulates NRO analysis12 (Fig. 4D). These results indicate that TFIIB is involved in polo transcription termination and further supports a connection between the transcription initiation and termination processes.
Figure 4. Depletion of TFIIB leads to increased read-through of polo transcripts. (A) TFIIB peaks in the promoter regions of different groups of genes. The different groups of genes are described in Figure 2A-C. (B) Analysis of the efficiency of TFIIB RNAi treatment, as in Figure 3C. (C) Schematic representation of the polo and snap locus and ChIP showing association of RNAP II (8WG16) with the different regions of polo and snap genes in Drosophila Kc cells following TFIIB depletion. Numbers below each bar represent the position of qPCR primers as depicted at the top of the panel. (D) RT-qPCR of read-through transcription. The diagram shows the primer positions for reverse transcription and qPCR analysis. The graph shows RT-PCR quantitation of endogenous pre-RNA (nascent) and read-through RNA in Kc cells depleted for TFIIB and compared with control depletion. Primers used for reverse transcription (3R and 6R) and qPCR (3F/3R and 6F/6R) are shown in the diagram. Error bars show s.e.m. from at least three independent experiments.
Juxtaposition of polo initiation and termination regions depends on TFIIB
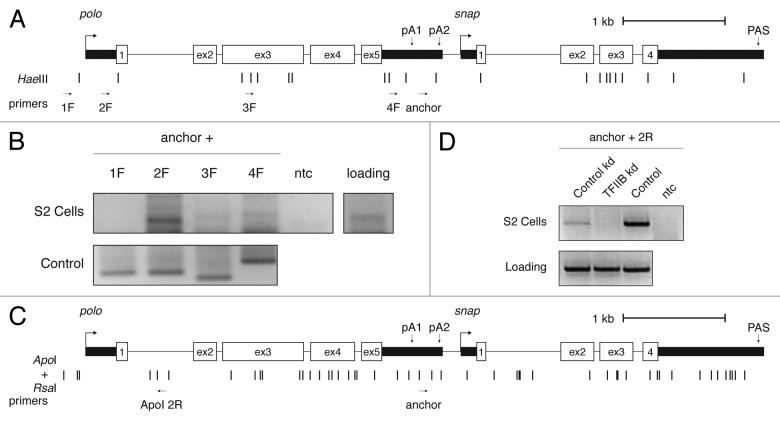
A tight connection between initiation and termination machinery through gene loop formation has been demonstrated in specific genes of yeast and mammals.40,41,45,46 These gene loops were shown to be dependent both on TFIIB40 and a functional PAS.45 As we observed an effect of TFIIB in polo transcription termination and an accumulation of RNAP II in polo initiation and termination regions, we went on to investigate the existence of a gene loop structure in the polo locus by chromosome conformation capture (3C) analyses. Using primers throughout the polo locus (Fig. 5A), we found a selective interaction between the polo promoter region and the 3′ end of the gene (Fig. 5B, lane 2F). As we also find that TFIIB is necessary for polo transcription termination, and this is one well-described factor known to be responsible in the maintenance of gene loop conformation (reviewed in 42), we asked whether TFIIB was necessary for polo loop formation. After TFIIB RNAi depletion we used primers in the same region where we observed an interaction (Fig. 5A and C). However, we used different restriction enzymes not only to complement the initial result, but also to increase the resolution of the assay (see Fig. 5C). After this modification to the protocol, we detect loss of the gene loop configuration in TFIIB depleted cells (Fig. 5D). These results indicate that polo forms a TFIIB dependent gene loop. Additionally, in combination with increased read-through of polo transcripts upon depletion of this factor, we infer that the interaction between the 5′ and 3′ end of polo may have a role in the transcription termination mechanism of this gene.
Figure 5.polo initiation and terminator regions interact through a gene loop dependent of TFIIB. (A) Schematic representation of polo and snap locus with restriction enzyme sites and primers for HaeIII 3C analysis. Arrows indicate primer direction and name. polo PAS are indicated. (B) polo 3C analysis. An agarose gel is shown. Positive lanes are internal polo PCR products on S2 gDNA (control panel) and chromatin (for cells). Common PCR primer (anchor) is shown above the figure, with the second primer shown above each lane (1F-4F). (C) Schematic representation of polo and snap locus with restriction enzyme sites and primers for ApoI/ RsaI 3C analysis. Arrows indicate primer direction and name. polo PAS are indicated. (D) polo 3C analysis upon TFIIB depletion. A representative agarose gel is shown for control kd, TFIIB depletion and control PCR panel (control) is shown. Positive lanes are internal polo PCR products on S2 gDNA (control panel) and chromatin (for cells). anchor and ApoI 2R PCR primers were used.
Transcription of polo represses snap while polo PAS deletion increases snap mRNA levels in a promoter-dependent manner
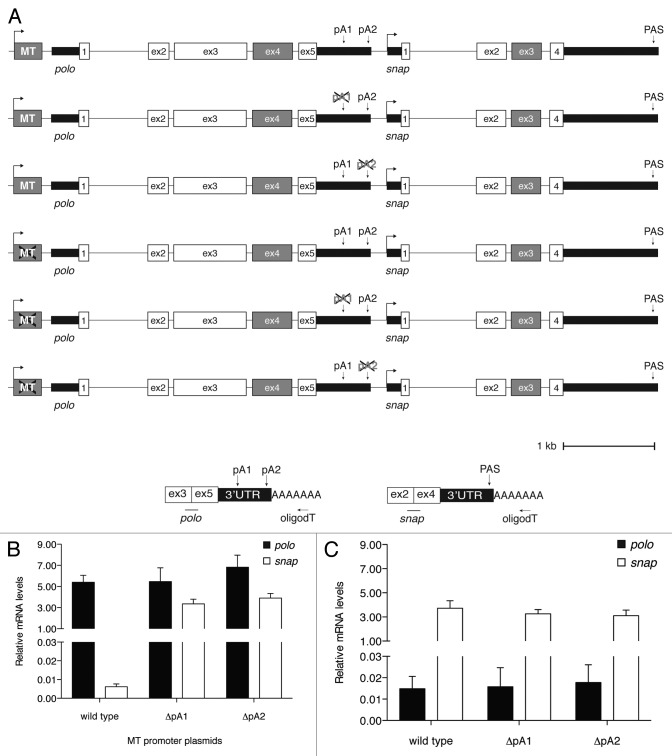
We have previously shown that polo auto-regulates its own protein levels by alternative polyadenylation (APA) and that polo pA2 is necessary to produce high levels of Polo protein necessary for histoblast proliferation.4 As polo PAS are very close to the snap transcription initiation site, we investigated the role of the two polo PAS in snap transcription. For this purpose we constructed a plasmid (named wild type) containing the polo-snap locus with the exception of polo exon 4 and snap exon 3 that were removed to distinguish between plasmid-driven transcripts from the endogenous polo and snap mRNAs (Fig. 6A). Using the wild type plasmid as a template, each polo PAS was then deleted separately, producing plasmids, ΔpA1 and ΔpA2. The three plasmids were transiently transfected into S2 cells and the resultant mRNAs were quantified by qRT-PCR. As shown, snap mRNA levels produced by the wild type plasmid are decreased in comparison with polo mRNA levels (compare black with white bars in wild type, Fig. 6B). This is in agreement with our single cell PCR result that showed that in most cells polo is transcribed when snap is not expressed (Fig. 1E, S1C and S2D). Surprisingly, when we deleted either polo PAS (ΔpA1 and ΔpA2), snap expression is increased (ΔpA1 and ΔpA2 in Fig. 6B). In light of previous results showing that deletion of PAS interfere with transcription at a downstream promoter,14 this result was intriguing. We cannot discard the possibility that, due to the close proximity between polo PAS and snap TSS, deletion of polo PAS renders snap transcription initiation site available/free for recruitment of snap activators. Therefore, we further investigated that effect. To rule out the possibility that transcripts could read-through the PAS into snap and further determine whether the promoter contributes for the effect observed, we deleted the promoter from these plasmids, producing and testing three promoter-less plasmids: wild type, ΔpA1 and ΔpA2 (Fig. 6A). As expected, mutation of the promoter inactivates polo transcription (Fig. 6B). Remarkably, this is accompanied by an increase in snap transcription. These results clearly show that inhibition of polo transcription enhances snap transcription indicating that transcription of polo represses snap expression (wild type in Fig. 6B-C). They are in agreement with the negative correlation of gene expression we observed in the single-cell PCR and FlyAtlas microarray data. Moreover, when polo transcription is abolished by using the promoter-less plasmids, deletion of either polo PAS does not affect snap expression (ΔpA1 and ΔpA2 in Fig. 6C). This implies that the increase in snap mRNA levels observed when polo PAS were deleted (Fig. 6B) is not due to an increase in read-through transcripts derived from the upstream promoter and eliminates the possibility of the PAS, per se, acting as repressors of snap.
Figure 6. Deletion of polo PAS affect snap expression. (A) Detailed schematic representation of different polo-snap containing plasmids. Note the absence of polo exon 4 and snap exon 3 (gray boxes) for differentiation between endogenous and plasmid derived transcripts. The crosses over either the promoter (MT) or the PAS (pA1 or pA2) represent the deletion of these structures in the respective plasmids. (B) Quantification of polo and snap mRNA levels using the plasmids where polo is expressed under the control of the metallothionein promoter. Polyadenylated transcripts from the plasmid were quantified as depicted in the scheme on the right and normalized to a co-transfected plasmid expressing eGFP. Oligod(T) was used for the RT reaction and primers on exons 3 and 5, and 2 and 4, on polo and snap, respectively, were used on the qPCR. (C) Same as (B) but for quantification of polo and snap mRNA levels using plasmids where the metallothionein promoter cassette was deleted. Polyadenylated transcripts were quantified as in (A).
Discussion
The fine-tuning of gene expression involves the cooperation of multiple molecular processes. These include the acquisition of specific chromatin marks as well as nucleosome positioning and transcription factor binding. Also, RNAP II levels and modification to its CTD, as well as the association of several proteins with RNA polymerase II, all combine to facilitate the initiation, elongation and termination stages of transcription.47-49 Genome wide studies have provided invaluable insight into the characterization of these features.1,30,50,51 Nevertheless, global analyses do not generally provide mechanistic information within a biological context. Only a few examples have been described where the mechanisms involved in transcription termination in higher eukaryotes were molecularly characterized (for reviews see 7, 8, 52). Transcription termination is intimately correlated with mRNA 3′ end cleavage/polyadenylation and has a strong impact on gene expression.52,53 In higher eukaryotes, transcription termination usually occurs several kilobases downstream of the end of the transcriptional unit.35,54 However, this process must occur in a different manner when genes are closely spaced within the genome to avoid interference with the downstream gene.
There are very few described examples of mechanisms that mediate transcription termination between closely spaced tandem genes in eukaryotes. A study on the transcriptional interactions between the GAL10 and GAL7 tandem genes of S. cerevisiae showed the importance of correct transcription termination, as deletion of the GAL10 PAS completely inactivated the use of the GAL7 promoter, resulting in transcriptional interference.14 Another example describing transcription termination between two closely spaced genes was the human C2 complement, which is separated by only 160 bp downstream from the gene, factor B. In this case, the MAZ protein binds downstream of the C2 complement PAS and has a role in the termination process.17 More recently, another termination mechanism was described in S. pombe, occurring in the G2 phase of the cell cycle, where cohesin is concentrated into the intergenic regions of several convergent genes, blocking the passage of elongating RNAP II and forcing gene-proximal abrupt termination.55 How RNAP II terminates transcription in genes that undergo alternative polyadenylation and in short intergenic regions remains largely unknown. We have previously shown that, in Drosophila, transcription of polo, which is only 168 bp downstream from the tandem gene snap, generates two APA-mediated mRNAs that differ at their 3′UTR4.
Here we investigated how transcription termination occurs within the 168 nucleotides that separate polo and snap. This is an important general question because, as we show, 50% of genes in Drosophila contain short intergenic regions; therefore, transcription must efficiently terminate in a very short space so that RNAP II does not read-through into the downstream gene. Using genomic tools we analyzed pairs of tandem genes with short intergenic regions in Drosophila and showed that a set of short tandem genes have a negative correlation of gene expression, i.e., when one gene is transcribed the other one is inactive. polo and snap are an example of closely spaced tandem genes included in this set. In contrast, when genes exist in a convergent arrangement, no correlation between intergenic distance and gene expression was observed. polo codes for a key cell cycle kinase and is expressed in dividing cells.20 snap promotes synaptic vesicle fusion and neurotransmitter release and is mainly expressed in the central nervous system.22 Thus, in vivo, polo and snap are rarely co-expressed in the same cell. We determined polo and snap expression by single-cell PCR analysis and found that in ~80% of the cells polo and snap are not co-expressed. This finding was corroborated using the publicly available microarray data and using plasmids transfected into Schneider cells, where we showed that when polo is being actively transcribed, snap is repressed. These results indicate that transcription of one gene interferes with transcription of the other.
Transcription termination mechanisms are viewed as the coordination of several events where the PAS, nascent pre-mRNA cleavage, 5′-3′ exonucleolytic RNA degradation and protein factors all play a role (reviewed in 7, 8). Two of the most important protein factors involved in the transcription termination process are Xrn2 and Pcf11. Xrn2 was described as the 5′-3′ exonuclease responsible for degrading the unprotected 5′ end exposed upon cleavage at the PAS or at the site of transcription termination.10 Pcf11 had been described as a cleavage and polyadenylation factor.56 Moreover, it was also shown that it interacts with RNAP II and dismantles the elongation complex by a CTD-dependent mechanism where high Ser2 phosphorylation density provides a signal to regulate Pcf11 recruitment at the transcription termination region.38,39 Hence, it was surprising that we found no effect in polo transcription termination upon RNAi depletion of either factor. In contrast, CG30046 transcription termination was affected both by Xrn2 and Pcf11 knockdown, causing RNAP II to read-through. It is possible that the short intergenic region between polo and snap forces certain constraints on the mechanism of transcription termination or that the protein factors bound to this region present a block to the passage of the elongating RNAP II, forcing abrupt termination to occur.
One of the key events for proper transcription termination is the recognition of a functional PAS,12 which relies on the presence of the canonical hexamer AAUAAA and additional auxiliary sequence elements, or just a potent DSE and A-rich upstream sequence.57 Moreover, mutation of the hexamer leads to a decrease in transcription initiation of the same gene44 and of the downstream gene by a phenomenon known as promoter occlusion.14 This may be related to a higher-order chromatin structure, such as gene loops (reviewed in 42), where the initiation region is associated with the termination region.45,46 High levels of CTD Ser5 phosphorylation are generally found at the 5′ end of actively transcribed genes and decline downstream on the body of these genes.58,59 Usually, genes that form loops display a Ser5P RNAP II accumulation at the 3′ end as RNAP II “bridges” the promoter and terminator regions.45,59 Interestingly, polo displays increased levels of Ser5P at the 3′ end and, by 3C analysis, we showed that polo forms a gene loop between its promoter and terminator region. Some proteins have been linked to gene loop conformations such as TFIIB.40,60 In agreement with this, we also show an enrichment of TFIIB in closely spaced genes that display a negative correlation of gene expression. Moreover, we show that TFIIB depletion increases RNAP II read-through and loop disruption, indicating that TFIIB is part of the polo gene loop structure.
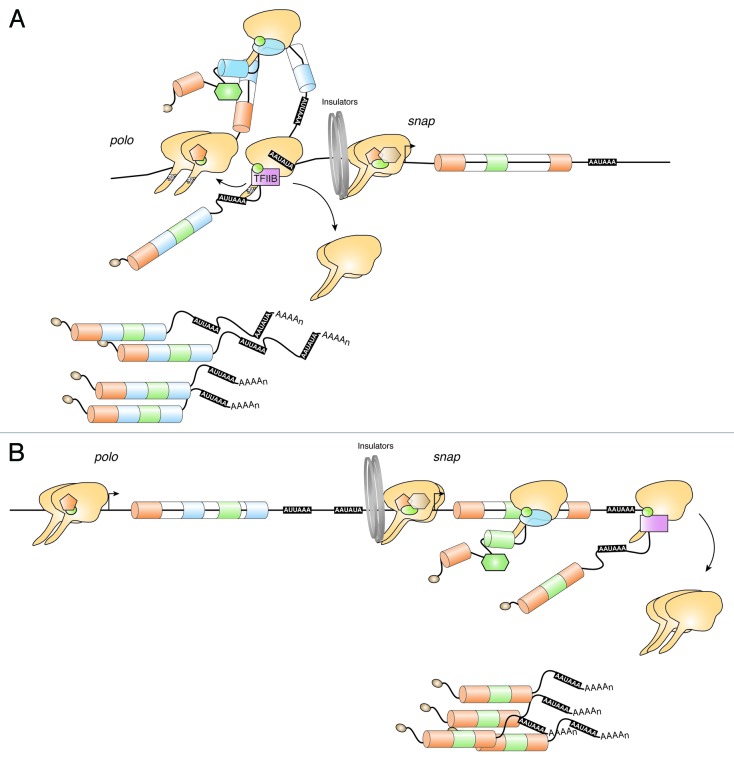
Our working model for polo-snap transcription termination (Fig. 7A) shows a physical interaction between the 5′ and 3′ end of polo that involves RNAP II and TFIIB. In agreement with this model, we observed high levels of RNAP II occupancy, concomitant with increased levels of Ser5 CTD phosphorylation at the 5′ and 3′ end of polo. It is then tempting to suggest that polo gene looping aids in the coordination of gene expression: when a gene loop is formed, polo is transcribed and snap expression is inhibited because its promoter is hidden by protein complexes involved in loop formation. As it has been shown that maintenance of a gene loop is transcription dependent,42,45 it is possible that when polo transcription is decreased polo gene loop is dismantled and snap can start to be transcribed (Fig. 7B). Such an effect is seen using the wild-type promoter-less plasmid, where snap mRNA levels increase in the absence of polo expression. Surprisingly, we found an increased expression of snap when either polo PAS was deleted from a plasmid containing the polo-snap locus, transfected into Schneider cells. Interestingly, this effect seems to be dependent on the upstream promoter, as promoter-less plasmids from which PAS were deleted do not show any alteration in snap expression. This discards the possibility that polo PAS acts as snap repressor and further implies that interactions between the promoter transcriptional machinery and the PAS are important for snap repression, consistent with our proposed working model.
Figure 7. Proposed working model: mechanism of transcription termination in the short intergenic region between the tandem genes polo-snap. (A) A Ser5P RNAP II is localized in polo-snap intergenic region. Stalled RNAP II together with TFIIB are necessary for polo transcription termination to occur in the 168 nt region between polo and snap. polo forms a TFIIB-dependent gene loop. As polo and snap are rarely co-expressed in the same cell, we suggest that this is due to the polo loop components that occupy the intergenic/snap-promoter region inhibiting the pre-initiation complex to form in the snap promoter. (B) In 80% of the cells, snap is transcribed when polo is not. As RNAP II and TFIIB in the polo terminator region physically interact with polo initiation region (A) we suggest that snap transcription initiation can only occur when the loop is disassembled, allowing initiation of snap transcription.
Our results reinforce the view, that in spite of RNAP II being poised within the promoter region of the downstream gene, it only moves into productive elongation when the required signals are received,61 in addition to a correct chromatin environment being present.31
Taken together, our results suggest that the presence of a gene loop, stalled RNAP II, TFIIB, specific chromatin marks and polo PAS combine to terminate transcription in the short intergenic region between polo and snap, two closely spaced tandem genes in Drosophila.
Materials and Methods
BromoUTP Nuclear Run On (BrUTP-NRO)
The BrUTP-NRO protocol was performed as previously described in.36 Briefly, cells were harvested by centrifugation (5min at 500xg), washed with PBS, and resuspended in HLB (10mM Tris [pH 7.5], 10mm NaCl, 2.5mM MgCl2) + 0.5% NP-40. After incubation on ice (5min), nuclei were pelleted through a cushion of HLB + 0.5% NP-40 + 10% sucrose. Nuclear pellets were resuspended in transcription buffer (40mM Tris [pH 7.9], 300mM KCl, 10mM MgCl2, 40% glycerol, 2mM DTT) and 10mM mix of ATP, CTP, GTP, and Br-UTP or UTP (in the control samples). The NRO reaction was performed at 20°C for 15min. Total RNA was isolated using TRIzol reagent (Invitrogen) according to manufacturer’s instructions and treated with RNase-free DNase I (Roche) for 1h at 37°C. 2μl of anti-BrU antibody (Sigma) were pre-incubated with 30μl of Protein G Dynabeads (Upstate) and 10μg tRNA per sample for 1h at 4°C. The beads were washed three times with ice-cold RSB-100 buffer (10mM Tris [pH 7.4], 100mM NaCl, 2.5mM MgCl2, 0.4% Triton X-100) and resuspended in 150μl RSB-100 with 40U RNase-OUT (Invitrogen) and 5μg of glycogen. Total RNA was added to beads and incubated for an additional hour at 4°C. Then beads were washed three times with RSB-100 buffer and RNA-bound to beads was extracted with TRIzol reagent. The reverse transcription reaction was performed using SuperScript III Reverse Transcriptase (Invitrogen) and gene-specific primers following the manufacturer’s instructions. Real-time quantitative PCR was performed using a Rotor-Gene Q machine (Qiagen). The PCR mixture contained SensiMix™ SYBR No-ROX master mix (Bioline), 2μl of template cDNA (1:5 dilution), and 200nM of each primer. Cycling parameters were 95°C for 10min, followed by 50 cycles of 95°C for 10s, 58°C for 10s, and 72°C for 10s. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer. The data was presented as the enrichment of BrU-RNA over the U-RNA produced over a specific probe.
Chromatin Immunoprecipitation (ChIP)
Aliquots of 5x107 cells were lysed after resuspension in 300μl of cell lysis buffer (5mM PIPES/KOH [pH 8.0], 85mM KCl, 0.5% NP-40, supplemented with 0.5mM PMSF, 1μg/ml pepstatin and 1μg/ml leupeptin) for 10min on ice. Nuclei were pelleted by centrifugation for 5min at 4°C and 500xg in a tabletop centrifuge, and lysed in 400μl of nuclei lysis buffer (5mM EDTA [pH 8.0], 50mM Tris [pH 8.0], 1% SDS, supplemented with 0.5mM PMSF, 1μg/ml pepstatin and 1μg/ml leupeptin) on ice for additional 10min. Chromatin was sonicated using a Bioruptor® sonicator (Diagenode) for 15min (30 sec on, 30 sec off, medium amplitude) with the tubes submerged in ice-cold water. Supernatants were collected through centrifugation at 15.000xg and 4°C for 10min and diluted to a final volume of 3mL in IP dilution buffer (1.5mM EDTA [pH 8.0], 20mM Tris [pH 8.0], 200mM NaCl, 0.012% SDS, and 1.3% Triton X-100). 300μl were taken to serve as input controls and saved at -20°C. Sonicated samples were pre-cleared for 1.5h with 50μl of 50% A/G bead slurry (Santa Cruz Biotechnology), then immunoprecipitated overnight at 4°C with 1μg of 8WG16 antibody (Abcam) and 3μg of 4H8 antibody (Abcam). Antigen-antibody complexes were immunoprecipitated with 30μl of 50% A/G bead slurry, washed once with low salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA [pH 8.0], 20mM Tris [pH 8.0] and 150mM NaCl), once with high salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA [pH 8.0], 20mM Tris [pH 8.0] and 500mM NaCl), once with lithium chloride buffer (250mM LiCl, 1mM EDTA [pH 8.0], 10mM Tris [pH 8.0], 1% NP-40, 1% sodium deoxycholate), and finally three times with Tris-EDTA (10mM Tris [pH 8.0] and 1mM EDTA [pH 8.0]). Samples were eluted twice in IP elution buffer (1% SDS and 100mM NaHCO3), reverse crosslinked and chromatin DNA was then purified using the QIAquick PCR purification kit (Qiagen). Eluted chromatin was quantified by qPCR. Each value for ChIP experiments was derived from IPs of at least two independent Drosophila S2 cell culture samples. EDTA-free protease inhibitors (Roche, France) were added to all washing buffers to a final concentration of 1x together with 0.5mM PMSF and 1μg/ml pepstatin and 1μg/ml leupeptin.
Chromosome Conformation Capture (3C)
Cells were fixed with 1% formaldehyde for 10min at room temperature (22°C) and fixation was stopped with glycine (0.125 M). 107 (HaeIII digestion) or 0.5x106 (ApoI + RsaI digestion) cells per sample were lysed (10mM Tris [pH 8], 10mM NaCl, 5mM MgCl2, 0.2% NP-40) for 30min on ice. The nuclei were pelleted and resuspended in 0.5 ml 1x digestion buffer (NEB4, New England Biolabs) and permeabilized with SDS (0.5% final concentration) for 1h (HaeIII) or 10min (ApoI + RsaI) at 37°C, shaking at 800 r.p.m. and 3.3% Triton X-100 were added for an additional 1h at 37°C. 2,000 U HaeIII or RsaI (New England Biolabs) were added before incubation overnight at 37°C (800 r.p.m.) and inactivated with or without SDS (1.5%, 65°C, 30min). A further digestion by ApoI at 50°C was performed overnight before heat inactivated at 80°C. The reaction was diluted in 6.2 ml 1.1x T4 ligase buffer (New England Biolabs) and incubated at 37°C for 1h after addition of 1% Triton X-100. 800 U T4 DNA ligase (New England Biolabs) was added for 4h at 16°C, crosslinking was reversed by incubation at 65°C with 300μg proteinase K for 16 h. 300μg RNase A was added for 1h at 37°C. DNA was column purified (QIAGEN). Two microliters of input was used in a standardized PCR reaction of 35 cycles of 95°C for 45s, 58°C for 45s, 72°C for 1min, and final extension for 5min at 72°C. polo internal PCR controls were obtained by PCR amplification using 1μg S2 cells genomic DNA as above, column purified, and quantified to supply a random pool of religated products. Equimolar amounts of PCR products were digested 4 h at 37°C and ligated overnight at 4°C, and a 1/500 dilution was used for agarose gel analysis.
Plasmids and Transfections
pMT polo/snap: polo and snap locus was amplified from genomic DNA with primers containing HindIII and SpeI restriction sites and cloned into the same restriction sites present in the pMT-CIDeGFP plasmid.
Deletion of the metallothionein promoter and the PAS were done with divergent primers for circular PCR.
pAc5.1 eGFP: eGFP sequence was removed from peGFP-N2 (Clontech) with EcoRI and NotI restriction enzymes and inserted in pAc5.1-V5 HisA (Invitrogen) into the same restriction sites.
All enzymes used were from New England Biolabs® and all plasmids were sequenced. For PCRs Phusion© High-Fidelity DNA polymerase (Finnzymes) was used.
1μg of DNA from plasmids pMTpolosnap wt, ΔpA1 or ΔpA2 was co-transfected with 500ng of pAc-5.1eGFP into Drosophila S2 cells with Fugene HD (Roche) following manufacturer’s instructions. Cells were induced for 18h with 500μM of CuSO4 prior to the harvest 48h after transfection.
RNA Functional Assays
RNA was extracted with TRIzol (Invitrogen) and reverse transcribed with the primers specified in the figures with SuperScript III following manufacturer’s instructions (Invitrogen). For oligod(T) reverse transcription phased oligos were used. For quantification cDNAs were diluted 3–5 times and 2μl of the diluted cDNA was analyzed by qPCR (200nM of each oligo, 2μl of cDNA, 10μl of iQ™ Sybr® Green Supermix (Biorad), and water to a final volume of 20μl). Experiments were quantified after subtraction of values obtained from minus RT samples, and normalized to respective housekeeping gene (rp49 or 7SL RNA) and Control experiment.
RNA interference
In vitro transcripts with 500–700bp of size were produced with MEGAscript® RNAi kit (Ambion) following manufacturer’s instructions. After annealing, 20μg of dsRNA was added for 96h to 2x106 Drosophila S2 cells cultivated in Schneider’s Insect Medium (Sigma) supplemented with 10% FCS (Gibco) in a 6-well plate. For Pcf11 and Xrn2 depletions a second boost was done at 48h and cells were starved at 96h. For TFIIB depletion, only one boost of dsRNA was done for 72h.
Single Cell PCR
Cells were separated into a 96-well PCR plate by a FACSAria Cell Sorter (BD Biosciences) and promptly frozen in liquid nitrogen. The following protocol was performed as described in.62
Correlation of Gene Expression between Adjacent Gene Pairs
The gene expression data were obtained from FlyAtlas.26 The information on adjacent gene pairs was based on annotation of RefSeq genes. The Pearson correlation value was calculated based on gene expression in 26 tissues. Genes were grouped based on correlation coefficient (r). The negatively correlated gene pairs were those with the lowest 20% r values, and positively correlated gene pairs were those with the highest 20% r values. Other gene pairs were put into the ‘other’ group.
Analysis of ChIP-seq, ChIP-chip and short RNA-seq data
We separated tandem genes into two groups based on the distance between gene pairs. The gene pairs were considered as short distance gene pairs if the distance is < 1kb, and were otherwise long distance gene pairs. Pol II level around TSS was based on the number of ChIP-seq reads mapped to a genomic position. Gene expression levels in S2 cells were calculated by the RPKM (reads per kilobase of exon per million mapped reads) method. Short RNA-seq reads were analyzed as described in.29 Only genes expressed in S2 cells (RPKM > 3) were used in the analysis. Chromosome accessibility and insulator levels were based on the mean of ChIP-chip signals for a genomic position.
The data sets and oligonucleotides list used in this study are shown in Table S1 and S2 respectively, in Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to J. Lis (Cornell University, USA), D. Gilmour (Pennsylvania State University, USA) and H. Maiato (IBMC, Porto) for antibodies and reagents. We also thank K. Adelman (NIEHS, USA), J. Lis, D. Price (University of California, USA), M. Carmo-Fonseca (IMM, Lisbon), C. Sunkel (IBMC, Porto), and S. Murphy (University of Oxford) for helpful suggestions and critical insight throughout this work. We thank J. Saraiva and G. Rodrigues for technical support and all the members of the laboratories for suggestions on the work.
TH was funded by FCT-POCTI PhD fellowship (SFRH/BD/30916/2006) and APBRF and EMBO (ASTF 393.00–2010) short-term fellowships. AM laboratory is funded by FEDER through COMPETE-Programa Operacional Fatores de Competitividade and by National Funds through FCT - Fundação para a Ciência e a Tecnologia FCOMP-01–0124-FEDER-022718 (PEst-C / SAU / LA0002 / 2011) and PTDC / SAU – GMG / 116621/2010.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21967
References
- 1.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juven-Gershon T, Hsu J-Y, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–9. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–44. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–18. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West S, Proudfoot NJ. Transcriptional termination enhances protein expression in human cells. Mol Cell. 2009;33:354–64. doi: 10.1016/j.molcel.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–69. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–94. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logan J, Falck-Pedersen E, Darnell JE, Jr., Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A. 1987;84:8306–10. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West S, Gromak N, Proudfoot NJ. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–5. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–89. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–10. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 14.Greger IH, Proudfoot NJ. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17:4771–9. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci U S A. 2002;99:8796–801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–45. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashfield R, Patel AJ, Bossone SA, Brown H, Campbell RD, Marcu KB, et al. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 1994;13:5656–67. doi: 10.1002/j.1460-2075.1994.tb06904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira A, Wollerton M, Monks J, Proudfoot NJ. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–19. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, et al. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5(12A):2153–65. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 20.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–75. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 21.Moreira A. Integrating transcription kinetics with alternative polyadenylation and cell cycle control. Nucleus. 2011;2:556–61. doi: 10.4161/nucl.2.6.18064. [DOI] [PubMed] [Google Scholar]
- 22.Babcock M, Macleod GT, Leither J, Pallanck L. Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles. J Neurosci. 2004;24:3964–73. doi: 10.1523/JNEUROSCI.5259-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Chin LS. The molecular machinery of synaptic vesicle exocytosis. Cell Mol Life Sci. 2003;60:942–60. doi: 10.1007/s00018-003-2240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 25.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 27.Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, et al. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- 28.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, et al. modENCODE Consortium Unlocking the secrets of the genome. Nature. 2009;459:927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–8. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–51. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–2. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–8. doi: 10.1016/S0959-437X(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 34.Chopra VS, Cande J, Hong J-W, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–9. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–96. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, et al. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2010;17:1195–201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–16. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Fairley JA, Roberts SGE. Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol. 2010;20:548–53. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hampsey M, Singh BN, Ansari A, Lainé JP, Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul. 2011;51:118–25. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, et al. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell. 2010;40:410–22. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105:5160–5. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–8. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muñoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–8. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 51.Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–5. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 52.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–82. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Almeida SF, Carmo-Fonseca M. Cotranscriptional RNA checkpoints. Epigenomics. 2010;2:449–55. doi: 10.2217/epi.10.21. [DOI] [PubMed] [Google Scholar]
- 54.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–8. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–95. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 56.Sadowski M, Dichtl B, Hübner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–77. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA. 2012;3:385–96. doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–60. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grosso AR, de Almeida SF, Braga J, Carmo-Fonseca M. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res. 2012;22:1447–56. doi: 10.1101/gr.138057.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14(10A):1938–47. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, et al. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11:R42. doi: 10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.