Abstract
Triple negative breast cancers (TNBC), defined by the absence of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor-2 expression, account for 12-24% of all breast cancers. TNBC is associated with early recurrence of disease and poor outcome. Germline mutations in the BRCA1 and BRCA2 breast cancer susceptibility genes have been associated with up to 15% of TNBC, and TNBC accounts for 70% of breast tumors arising in BRCA1 mutation carriers and 16-23% of breast tumors in BRCA2 carriers. Whether germline mutations in other breast cancer susceptibility genes also predispose to TNBC remains to be determined. Common variation in a subset of the 72 known breast cancer susceptibility loci identified through genome wide association studies and other large-scale genotyping efforts have also been associated with risk of TNBC (TOX3, ESR1, RAD51L1, TERT, 19p13.1, 20q11, MDM4, 2p24.1, and FTO). Furthermore, variation in the 19p13.1 locus and the MDM4 locus has been associated with TNBC but not other forms of breast cancer suggesting that these are TNBC-specific loci. Thus, TNBC can be distinguished from other breast cancer subtypes by a unique pattern of common and rare germline predisposition alleles. Additional efforts to combine genetic and epidemiological data are needed to better understand the etiology of this aggressive form of breast cancer, to identify prevention and therapeutic targets, and to impact clinical practice through development of risk prediction models.
Triple negative breast cancer (TNBC): Epidemiologic and clinical characteristics
Triple negative breast cancers (TNBC) are defined by the absence of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) expression (1). Triple negative (TN) breast tumors account for 12-24% of the more than 200,000 breast cancers diagnosed each year in the United States (1, 2). Compared to other breast cancer subtypes, triple negative breast cancer is associated with a distinct set of epidemiologic risk factors, which has been reviewed in detail (1, 3). Briefly, women with TNBC are more likely to be young or premenopausal, African American or Hispanic, low socioeconomic status, and BRCA1 mutation carriers. Additional factors associated with risk of TNBC are earlier age at menarche, higher body mass index (BMI) during premenopausal years, higher parity, and a lower lifetime duration of breast feeding. Recurrence and disease progression are also relatively common for women with TNBC, with a peak risk of recurrence within the first three years after treatment (4). Poor clinical outcomes for women with TN tumors may in part be explained by intrinsically aggressive tumor pathology, including high mitotic index and nuclear pleomorphism yielding high histologic grade, high proliferation, medullary and metaplastic features, and a high frequency of TP53 mutation (1, 5).
Molecular classification of TNBC
While the three immunohistochemical (IHC) markers, ER, PR, and HER2, are routinely used in clinical practice to classify breast tumors and thereby determine potential courses of therapy, more detailed molecular characterization of breast cancers by gene expression profiling has identified at least five distinct “intrinsic” breast cancer subtypes that appear to represent distinct disease processes (6). These intrinsic subtypes include two luminal epithelial/estrogen receptor positive subgroups (A and B) differentiated by level of expression of HER2 and/or proliferation genes; a HER2 over-expressing group; a normal breast-like or unclassified group; and a basal-like group that is largely TNBC and expresses basal epithelial cell layer proteins including cytokeratins 5,6 (CK5/6) and EGFR. In addition, a claudin-low group has been identified that is also comprised largely of TN tumors (71%), characterized by lack of expression of luminal differentiation markers, enrichment for epithelial-to-mesenchymal transition markers, immune response genes and cancer stem cell-like features (7). Most recently a study of 1,992 breast tumors using gene expression arrays and copy number variation identified 10 possible subtypes of breast cancer, which differed by clinical outcome (8). The majority of basal-like tumors within that study again formed a single stable high genomic instability subgroup associated with rapid recurrence.
While basal-like tumors appear to have very similar molecular characteristics, it is clear that TN tumors are not synonymous with basal-like tumors. Specifically, 15-20% of TN tumors do not express basal markers and 15%-20% of non-TN tumors express basal markers. Further, since recent studies have suggested further subdivision of TNBC into immunomodulatory, mesenchymal, mesenchymal stem-like, luminal androgen receptor, and distinct basal-like subtypes (9), there are likely subtypes of TNBC that differ substantially from basal-like tumors. However, because the basal-like definition of tumors is typically available only in an experimental research setting, based on gene expression profiling, the TN phenotype is often used as a surrogate for basal-like status in clinical and observational studies. Additional work is necessary to better define TN subtypes and the epidemiologic, clinical, and prognostic characteristics of these tumors.
High risk susceptibility genes for TNBC
Genetic susceptibility to TNBC has been associated with rare, highly penetrant, germline mutations in the BRCA1 and BRCA2 breast cancer predisposition genes. Approximately 70% of breast tumors that develop in women with inherited mutations in BRCA1 exhibit low or absent expression of ER, PR and HER2 histological markers, and morphological features, recurrence patterns, and death rates (10) similar to unselected TNBC tumors (1). Consistent with these observations, several studies of unselected TN cases have shown that 9-14% overall and ∼20% of cases diagnosed under age 50 harbor germline BRCA1 mutations (11). Similarly, as many as 34% of TN cases with a family history of breast cancer and 30% of TN cases from women of Ashkenazi Jewish ancestry are associated with germline BRCA1 mutations (12, 13). To a lesser extent BRCA2 mutations are also associated with TNBC in that 16-23% of breast tumors arising in BRCA2 mutation carriers display TN properties (10). While few breast cancer susceptibility genes have been systematically evaluated for mutations in TN cases, it is already clear that up to 15% of unselected TN cases result from inherited mutations in the BRCA1 and BRCA2 high-risk susceptibility genes.
A recent report from The Cancer Genome Atlas (TCGA) Network provides further insight into the distribution of mutations in high-risk genes by breast cancer subtypes, where 49 deleterious variants in nine genes (ATM, BRCA1, BRCA2, BRIP1, CHEK2, NBN, PTEN, RAD51C, TP53) were detected in exome-sequencing data from 507 breast tumors (14). Among the 93 basal-like tumors in this group, mutations were identified in BRCA1 (9 of 13 BRCA1 mutations), BRCA2 (3 of 14 BRCA2 mutations), RAD51C (1 of 1 RAD51C mutation), and TP53 (1 of 2 TP53 mutations). This confirms the known associations with BRCA1 and BRCA2, and suggests that germline RAD51C and TP53 mutations may be found among basal-like breast cancer cases. Interestingly, no mutations in the remaining five genes (ATM, BRIP1, CHEK2, NBN, PTEN) were detected among basal tumors. Large-scale examination of the mutational spectrum of all known breast cancer susceptibility genes (BRCA1, BRCA2, CHEK2, PALB2, BRIP1, TP53, PTEN, STK11, CDH1, ATM, BARD1, RAD51C, RAD51D, NBN, XRCC2)(15) in women with TNBC, and the individual TNBC subtypes, will be necessary to fully understand the role of these genes in TN risk.
Genetic risk factors for breast cancer by ER status
Excluding BRCA1 and BRCA2, relatively little is known about the inherited genetic factors that increase risk for TNBC. The majority of information on genetic susceptibility to this aggressive form of breast cancer has come from investigation of commonly inherited variants with small effects (odds ratio (OR) <1.3) on breast cancer risk. Currently, common variants in 72 loci have been implicated in breast cancer predisposition by genome-wide association studies (GWAS) of breast cancer (16-28), candidate gene studies (29), and a large-scale custom genotyping effort from the Collaborative Oncological Gene-environment Study (COGS) (30, 31) (Supplementary Table 1).
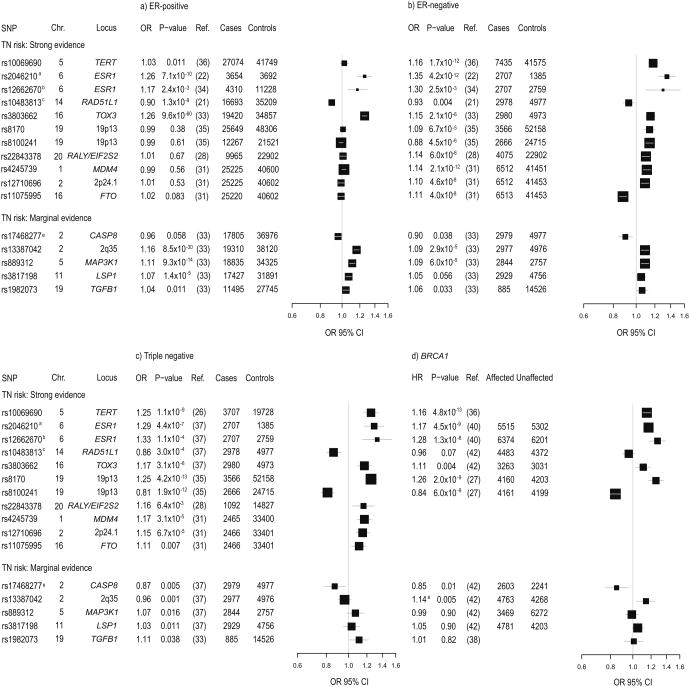
Interestingly, the incorporation of ER status into these GWAS and additional follow-up studies has shown that these risk loci are heterogeneous with respect to ER status, with only a subset associated with risk of ER-negative breast cancer. Specifically, 38 of 65 loci identified through GWAS of overall breast cancer have been associated with ER-negative breast cancer as well as ER-positive breast cancer (Figure 1a, b; Supplementary Table 1) (22, 25, 30-35). In addition, three separate meta-analyses of ER-negative breast cancer GWAS, conducted to specifically investigate genetic susceptibility to risk of ER-negative breast cancer, identified variants in seven loci associated with risk of ER-negative disease. These include TERT [rs10069690 OR=1.18, p=1.0 × 10-10] (26); 20q11 [rs22843378 OR=1.14, p= 6.0 × 10-6]; 6p14 [rs17530068 OR=1.15, p=4.1 × 10-6] (28); MDM4 (1q32.1) [rs4245739 OR=1.14, p=2.1 × 10-12]; LGR6 (1q32.1) [rs6678914 OR=1.10, p=1.4 × 10-8]; 2p24.1 [rs12710696 OR=1.10, p=4.6 × 10-8]; and FTO (16q12.2) [rs11075995 OR=1.11, p=4.0 × 10-8] (31) (Figure 1b). Further study of the TERT locus has identified three independent signals that influence the risk of different subtypes of breast and ovarian cancer (36). The rs10069690 variant and an independent variant in the TERT promoter (5-1296255 OR=0.91, p=6.15 × 10-6), accounting for two of these TERT loci, have been associated with ER-negative breast cancer. The heterogeneity observed by ER status in these studies supported the investigation of known breast cancer susceptibility loci among breast cancer subtypes defined by all three histologic markers (ER, PR, and HER2).
Figure 1. Triple negative breast cancer susceptibility loci across breast cancer subtypes.
Forest plots for thirteen TNBC susceptibility variants are shown to provide visual comparison of the strength and direction of association between each SNP and risk of a) ER-positive breast cancer, b) ER-negative breast cancer, c) triple negative breast cancer, and d) BRCA1-related breast cancers. The thirteen SNPs are stratified by strength of evidence for association with TNBC risk (strong vs. marginal). For each breast cancer subtype, estimates (odds ratios (OR) and 95% confidence intervals (CIs)) are shown for each variant from largest study of each relevant breast cancer subtype. ORs are denoted by black boxes and 95% CIs are denoted by corresponding black lines. Box heights are inversely proportional to precision of the OR estimate as influenced by sample size, such that a larger OR box indicates larger sample size and better precision. A vertical grey line is shown at the null value of 1, such that ORs to the left of the null line are <1 and ORs to the right of the null line are >1. Estimates with confidence intervals that do not overlap the null line indicate significance at p<0.05. a ER-positive and ER-negative estimates from a GWAS of Asian women; TN and BRCA1 carrier estimates from studies of women of European ancestry. b Estimate shown for rs12662670 TNBC and for rs9397435 in ER-positives, ER-negatives, and BRCA1 carriers c Previously unpublished data from TNBCC d Estimate shown for rs10483813/rs999737 in ER-negatives and ER-positives, rs999737 in TNBC, and rs1801320 (HR for GG vs CC) in BRCA1 carriers e Estimate shown for rs17468277/rs1045485 in ER-negatives and ER-positives, rs17468277 in TNBC, and rs1045485 in BRCA1 carriers.
Common susceptibility loci for TNBC
Several large-scale follow-up studies with extensive pathology data have investigated the relevance of common breast cancer risk loci to breast tumor subtypes defined by ER, PR, and HER2.. The first of these studies, from the Breast Cancer Association Consortium (BCAC) (33), found that five of the 12 variants investigated were associated with TNBC [TOX3 OR=1.21, p=3.1 × 10-6; 2q35 OR=1.12, p=0.001; MAP3K1 OR=1.11, p=0.016; LSP1 OR=1.11, p=0.011; TGFB1 OR=1.11, p=0.038] (Figure 1c). However, a much larger study by the Triple Negative Breast Cancer Consortium (TNBCC) involving nearly 3,000 TN cases, only confirmed an association for the TOX3 locus [OR=1.17, p=3.7 × 10-5] (37) (Figure 1c). In addition, TNBCC found that SNPs from more recently identified risk loci including rs2046210 and rs12662670 in the ESR1 locus [rs2046210 OR=1.29, p=4.4 × 10-7; rs12662670 OR=1.33, p=1.1 × 10-4], and rs10483813 in the RAD51L1 locus [OR=0.86, p=3.0 × 10-4], were strongly associated with TNBC. These RAD51L1 findings were consistent with evidence from a recent BCAC study of TNBC [OR=0.89, p=0.02] (32) (Figure 1c). In contrast an association between CASP8 and TNBC risk was observed by TNBCC [OR=0.87, p=0.005], but not by BCAC (OR=0.92, p=0.15) (33). Thus, SNPs in the TOX3, ESR1, and RAD51L1 loci, and possibly in 2q35, MAP3K1, LSP1, TGFB1, and CASP8, that influence the risk of both ER-positive and ER-negative breast cancer (Figure 1a,b), have also been identified as genetic risk factors for TNBC (Figure 1c).
Several of the loci identified in ER-negative breast cancer studies also appear to influence risk of TNBC. The ER-negative TERT variant, rs10069690, has been associated with an increased risk of TNBC [rs10069690 OR=1.25, 95% CI 1.16 - 1.34, p=1.1 × 10-9] (Figure 1c) (26). In addition, separation of HER2-positive ER/PR-negative cases [n=376, OR=1.03, p=0.71] from HER2-negative cases [n=3,707, OR=1.25, p=1.1 × 10-9] has suggested that this variant may be uniquely associated with TNBC (Figure 1c) (26). However, given the complexity of the associations between variation in this locus and breast cancer risk (36), further work must be done to evaluate the relevance of the three TERT signals to TNBC. The 20q11 locus from the Siddiq et al meta-analysis of ER-negative breast cancer was also shown to be strongly associated with TNBC [rs22843378 OR=1.16, 95% CI 1.04 - 1.29, p=6.4 × 10-3] (Figure 1c) (28). Furthermore, of the four loci identified by Garcia-Closas, et al., MDM4 [OR=1.17, 95% CI 1.09 - 1.26, p=3.1 × 10-5], 2p24.1 [OR=1.15, 95% CI 1.07 - 1.23, p=6.7 × 10-5], and FTO [OR=1.11, 95% CI 1.03 - 1.20, p=0.007] were associated with TNBC in subtype analyses (Figure 1c) (31). Of these, the MDM4 locus may have a specific association with TNBC [OR=1.17, p=3.1 × 10-5], since no significant association has been seen with non-TN ER-negative breast cancer [OR=1.02, 95% CI 0.92 - 1.12, p=0.711] (pHet=0.005).
Common susceptibility loci for BRCA1 mutation carriers
Because TNBC and breast cancer in BRCA1 mutation carriers are phenotypically similar, studies of genetic modifiers of breast cancer risk in BRCA1 mutation carriers have provided further insight into genetic risk factors for TNBC. Specifically, SNPs in the 19p13.1 locus that displayed genome wide significant associations with breast cancer in a GWAS of BRCA1 mutation carriers [rs8170 Hazard Ratio (HR)=1.26, p=2.3 × 10-9; rs8100241 HR=0.84, p=3.9 × 10-9] (Figure 1d) (27), have also been associated with TNBC risk in the general population [rs8170 OR=1.27, p=2.3 × 10-8; rs8100241 OR=0.84, p=8.7 × 10-7] (Figure 1c) (37). In BCAC and TNBCC combined, the 19p13.1 locus was associated with TNBC risk [rs8170 OR=1.25, p=4.2 × 10-13; rs8100241 OR=0.81, p=1.9 × 10-12] but was not associated with risk of ER-positive or ER-negative, non-TN breast cancer (Figure 1c) (35). Furthermore, these variants appeared to be specifically associated with tumors that were positive for the basal markers CK5/6 or EGFR [OR=1.27, 95% CI 1.07 - 1.50, p=0.0069], indicating specificity for the basal subtype. In addition, variants in ESR1, PTHLH, TOX3, CASP8, and TERT are also associated with both TNBC and risk of breast cancer in BRCA1 carriers (Figure 1c, d) (36, 38-42). Of the known TNBC risk factors, only RAD51L1 was not found to be a modifier of BRCA1-related breast cancer risk (Figure 1d). Recent data also demonstrate that BRCA2 ER-negative tumors have pathologic characteristics similar to BRCA1 ER-negative tumors (10). Thus, further studies of BRCA1 and BRCA2 breast tumors, stratified by ER or TN tumor status, may provide additional valuable insight into genetic susceptibility to TNBC.
Together with studies of overall breast cancer risk loci by ER, PR, and HER2 subtypes, these findings suggest that TNBC and the other subtypes of breast cancer may have distinct genetic risk factor profiles. Although the 19p13.1 locus and MDM4 appear to be specific to TNBC, it is important to note that these loci were also significantly associated with overall ER-negative breast cancer (Figure 1b). Whether this overall association is driven by the inclusion of TN or basal tumors or reflects meaningful associations with other non-TNBC ER-negative tumors remains to be determined. On this basis, it will be important to continue to evaluate new breast cancer loci as candidate risk factors for TNBC and other subtypes of breast cancer if a comprehensive understanding of genetic predisposition to breast cancer is to be attained.
Future directions and implications of understanding TNBC genetics
The exact biological mechanisms underlying TNBC genetic risk loci are currently unknown, and additional fine-mapping, re-sequencing, and functional studies are necessary to determine whether single or multiple variants at these loci affect TN risk through the disregulation of nearby genes or though long-range genetic effects. One hypothesis is that causal variants at these loci directly initiate and promote development of a TN tumor through pathways that are specific to this hormone-receptor negative subtype. Interestingly, three loci specific to TNBC contain genes (TERT, c19orf62, and MDM4) encode proteins involved in DNA repair and the preservation of genomic stability. The TERT gene encodes the catalytic subunit of telomerase, which controls telomere maintenance, and has been associated with genomic instability and linked to tumorigenesis (43). MDM4 is a repressor of TP53 and TP73 transcription and is important for cell cycle regulation and apoptosis in response to DNA damage (44). C19orf62 encodes the MERIT40 protein, which is integral to the localization of the BRCA1-A complex during DNA double-strand break (DSB) repair, through the recruitment and retention of the BRCA1-BARD1 ubiquitin ligase and the BRCC36 deubiquitination enzyme (45). Telomere maintenance, DSB repair, and DNA damage checkpoints have been linked as coordinating factors in genomic integrity, and the disruption of this pathway, resulting in genomic instability, has been implicated in cancer (46, 47). Indeed, one proposed mechanism of spontaneous telomere loss in cancer cells is a deficiency in DSB repair combined with oncogene-mediated DNA replication stress (46). Additionally, evidence suggests that DNA damage checkpoint and DNA repair proteins have an essential role in telomere maintenance, by controlling the processing of telomeric DNA and through other mechanisms that have yet to be delineated (47). This highlights a potential common biologic pathway that may be specifically associated with development of TNBC. Focusing on these pathways involved in DNA repair and the preservation of genomic stability, as highlighted by the associations of TNBC with genetic variation in MERIT40, MDM4, and TERT, may lead to the development of targeted prevention and/or therapeutic agents for TNBC patients, analogous to PARP inhibitors and the homologous recombination DNA repair pathway in BRCA1 and BRCA2-deficient carriers (48).
An alternative hypothesis is that variants or even combinations of variants in the TNBC associated risk loci may act to change existing malignant breast lesions to a TN phenotype during the formation of the tumor. This is particularly intriguing considering the relevance of the ESR1 locus, which is directly involved in estrogen signaling, to ER-negative and TN breast cancer in addition to ER-positive breast cancer. While a particular locus such as ESR1 may predispose breast epithelium to cancer in general, others such as TERT and 19p13.1 may act further downstream after tumorigenesis has begun to direct tumors towards the TN phenotype. Thus, the identification of these TNBC genetic loci offer exciting opportunities to better understand how TN tumors arise.
Beyond gaining insight into TNBC etiology, accurately defining the spectrum of high-risk mutations in breast cancer susceptibility genes among women with TNBC has the potential to modify clinical practice. A recent study from the United Kingdom showed that up to a third of TNBCs found to carry BRCA1 mutations would not have been clinically tested for these mutations based on traditional risk profiling (11) and would not have benefitted from the modified clinical care associated with known cancer predisposing mutations. By characterizing the association between mutations in all high-risk susceptibility genes and family history of cancer, age of onset of cancer, and other epidemiologic risk factors, improved breast cancer risk prediction models can be developed that more accurately identify women at risk for TNBC.
Similarly, identification of additional common genetic variants associated with risk of TNBC will likely have utility for breast cancer risk prediction. The expectation is that inclusion of all 72 common breast cancer risk loci in the Gail model, in addition to clinical and epidemiologic risk factors, will improve risk model performance (49), and the effectiveness of these models will likely be further improved by tailoring them to specific subtypes of breast cancer, including TNBC. Based on the specificity of 19p13.1, TERT, and MDM4 for this subtype, TN-specific risk models may be feasible. Accurate risk prediction models for TNBC that incorporate genetic information from both rare, high-risk and common, low-risk susceptibility loci would argue for screening women for cancer at a younger age and may assist in identifying high-risk women earlier in life.
Although we have made progress in understanding TNBC genetics, it is clear that there is much to learn about genetic susceptibility to TNBC and that there is a scientific and clinical need to continue this line of work. We must continue to combine high quality genetic, phenotypic, and pathologic data from large breast cancer studies and consortia in order to better define genetic susceptibility to TNBC, particularly considering that TNBC is a relatively rare subtype of breast cancer. Analyses that attempt to explain the complex biology of human cancers are necessary to make progress in understanding the etiology of TNBC and to impact disease prevention and clinical care.
Supplementary Material
Footnotes
Conflict of Interest: None
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Breast Cancer Facts & Figures, 2011-2012. Atlanta, GA: American Cancer Society; p. 2012. [Google Scholar]
- 3.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–39. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nofech-Mozes S, Trudeau M, Kahn HK, Dent R, Rawlinson E, Sun P, et al. Patterns of recurrence in the basal and non-basal subtypes of triple-negative breast cancers. Breast Cancer Res Treat. 2009;118:131–7. doi: 10.1007/s10549-008-0295-8. [DOI] [PubMed] [Google Scholar]
- 5.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012 doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21:134–47. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson L, Hanson H, Seal S, Warren-Perry M, Hughes D, Howell I, et al. BRCA1 testing should be offered to individuals with triple-negative breast cancer diagnosed below 50 years. Br J Cancer. 2012 doi: 10.1038/bjc.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26:4282–8. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comen E, Davids M, Kirchhoff T, Hudis C, Offit K, Robson M. Relative contributions of BRCA1 and BRCA2 mutations to “triple-negative” breast cancer in Ashkenazi Women. Breast Cancer Res Treat. 2011;129:185–90. doi: 10.1007/s10549-011-1433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumors. Nature. 2012 doi: 10.1038/nature11412. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–33. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 19.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, et al. Novel Breast Cancer Susceptibility Locus at 9q31.2: Results of a Genome-Wide Association Study. J Natl Cancer Inst. 2011 doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 25.Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44:312–8. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43:1210–4. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 30.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne R, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013 doi: 10.1038/ng.2563. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Closas M, Couch F, Lindstrom S, Michailidou K, Schmidt MK, Brook M, et al. Genome-wide association studies identify four ER-negative specific breast cancer risk loci. Nat Genet. 2013 doi: 10.1038/ng.2561. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueroa JD, Garcia-Closas M, Humphreys M, Platte R, Hopper JL, Southey MC, et al. Associations of common variants at 1p11.2 and 14q24.1 (RAD51L1) with breast cancer risk and heterogeneity by tumor subtype: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:4693–706. doi: 10.1093/hmg/ddr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:3289–303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stacey SN, Sulem P, Zanon C, Gudjonsson SA, Thorleifsson G, Helgason A, et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, Lindblom A, et al. 19p13.1 is a triple negative-specific breast cancer susceptibility locus. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bojesen S, Pooley K, Johnatty SE, Beesley J, Michailidou K, Tyrer J, et al. Multiple independent TERT variants associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013 doi: 10.1038/ng.2566. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens KN, Vachon CM, Lee AM, Slager S, Lesnick T, Olswold C, et al. Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer Res. 2011;71:6240–9. doi: 10.1158/0008-5472.CAN-11-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebbeck TR, Antoniou AC, Llopis TC, Nevanlinna H, Aittomaki K, Simard J, et al. No association of TGFB1 L10P genotypes and breast cancer risk in BRCA1 and BRCA2 mutation carriers: a multi-center cohort study. Breast Cancer Res Treat. 2009;115:185–92. doi: 10.1007/s10549-008-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–54. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoniou AC, Kartsonaki C, Sinilnikova OM, Soucy P, McGuffog L, Healey S, et al. Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2011;20:3304–21. doi: 10.1093/hmg/ddr226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antoniou AC, Kuchenbaecker KB, Soucy P, Beesley J, Chen X, McGuffog L, et al. Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res. 2012;14:R33. doi: 10.1186/bcr3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne RL, Antoniou AC. Genetic modifiers of cancer risk for BRCA1 and BRCA2 mutation carriers. Ann Oncol. 2011;22(1):i11–7. doi: 10.1093/annonc/mdq660. [DOI] [PubMed] [Google Scholar]
- 43.Daniel M, Peek GW, Tollefsbol TO. Regulation of the human catalytic subunit of telomerase (hTERT) Gene. 2012;498:135–46. doi: 10.1016/j.gene.2012.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mancini F, Di Conza G, Monti O, Macchiarulo A, Pellicciari R, Pontecorvi A, et al. Puzzling over MDM4-p53 network. Int J Biochem Cell Biol. 2010;42:1080–3. doi: 10.1016/j.biocel.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–28. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murnane JP. Telomere dysfunction and chromosome instability. Mutat Res. 2012;730:28–36. doi: 10.1016/j.mrfmmm.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian L, Nakamura TM. To fuse or not to fuse: how do checkpoint and DNA repair proteins maintain telomeres? Front Biosci. 2010;15:1105–18. doi: 10.2741/3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–87. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 49.Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–93. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.