Abstract
Rapid progress in the discovery of motor neuron disease genes in amyotrophic lateral sclerosis, the spinal muscular atrophies, hereditary motor neuropathies, and lethal congenital contracture syndromes is providing new perspectives and insights into the molecular pathogenesis of the motor neuron. Motor neuron disease genes are often expressed throughout the body with essential functions in all cells. A survey of these functions indicates that motor neurons are uniquely sensitive to perturbations in RNA processing pathways dependent upon the interaction of specific RNAs with specific RNA-binding proteins, which presumably result in aberrant formation and function of ribonucleoprotein complexes. This review provides a summary of currently recognized RNA processing defects linked to human motor neuron diseases.
Keywords: amyotrophic lateral sclerosis, spinal muscular atrophy, hereditary neuropathy, RNA processing
Introduction
The motor neuron is responsible for voluntary movement and is the target of a number of neuromuscular disorders that ultimately result in weakness. An appreciation of the selective vulnerability of the motor neuron in diseases characterized by weakness has existed for over 150 years, and the spectrum of motor neuron diseases covers a wide range of clinical severities and patterns. Spinal muscular atrophy (SMA), the most common genetic cause of infant mortality, is also the most common genetic form of motor neuron disease and is characterized by progressive proximal weakness and loss of lower motor neurons110. Clinically, progression is quite variable, and severity in SMA ranges from respiratory failure and early death in infancy to mild proximal weakness in adulthood115. Lethal congenital contracture syndrome (LCCM) and lethal arthrogryposis with anterior horn cell disease (LAAHD) are neurogenic forms of arthrogryposis congenita multiplex that are characterized pathologically by amyotrophy and a selective loss of lower motor neurons141,47. Hereditary motor neuropathies (HMNs, also known as distal SMA or Charcot-Marie-Tooth disease type 2 subtypes) are a heterogeneous group of syndromes that typically result in weakness in the distal extremities and include demyelinating and axonal forms26. Familial amyotrophic lateral sclerosis (fALS) is characterized by progressive loss of upper and lower motor neurons that occurs in adolescence or adulthood and leads to respiratory failure and death138. While the clinical spectrum and course of these diseases is broad, they all share a common pathology: primary loss of the lower motor neuron that bridges the central nervous system to the peripheral musculature. What is the mechanism of motor neuron cell death in these diseases? What is the basis of motor neuron selectivity in these diseases? What explanations exist to account for the large number of genes that specifically affect the motor neuron?
The molecular genetics of motor neuron diseases is providing welcome new perspectives and insights into the molecular basis of motor neuron death and dysfunction by providing pieces to the puzzle of motor neuron-selective vulnerability. To date, there are 18 distinct genes that are mutated in primary diseases of the motor neuron (Table 1) (also see http://www.molgen.ua.ac.be/CMTMutations/Home/IPN.cfm and http://neuromuscular.wustl.edu/synmot.html). Some generalities may be gleaned from this list. For example, genes essential for axonal transport (DCTN1, NEFL) and oxidative cell stress response (SOD1) point to these pathways as critical for motor neuron cell survival.
Table 1.
Motor neuron disease genes. Mutations in these genes result in motor neuron disease syndromes. SBMA=spinal and bulbar muscular atrophy, ALS=amyotrophic lateral sclerosis, SMA=spinal muscular atrophy, CMT=Charcot-Marie-Tooth disease, SMARD=spinal muscular atrophy with respiratory distress, HMN=hereditary motor neuropathy, dSMA=distal spinal muscular atrophy, LCCS=lethal congenital contracture syndrome.
| Gene | Symbol | Syndrome | Year | Refs |
|---|---|---|---|---|
| Androgen Receptor | AR | SBMA (X-Linked) | 1991 | 73 |
| Superoxide dismutase 1 | SOD1 | ALS1 | 1993 | 113 |
| Survival Motor Neuron | SMN | SMA | 1995 | 76 |
| Neurofilament Protein, Light Polypeptide | NEFL | CMT2E | 2000 | 88 |
| Immunoglobulin Mu Binding Protein 2 | IGHMBP2 | SMARD1/HMN6 | 2001 | 39 |
| Dynactin 1 | DCTN1 | HMN7B | 2003 | 111 |
| ALS susceptibility | 2004 | 95 | ||
| αHeat Shock Protein 27-kD | HSPB1 | CMT1F | 2004 | 29 |
| Glycyl tRNA Synthetase | GARS | CMT2D, dSMA5 | 2003 | 6 |
| Vesicle-Associated Membrane Protein-Associated Protein B | VAPB | ALS8 | 2004 | 101 |
| Heat Shock Protein 22-kD | HSPB8 | CMT-2L/ HMN2A | 2004 | 134 |
| CMT-2F/ HMN2B | 2005 | 54 | ||
| Seipin | BSCL2 | Silver Spastic Paraplegia/HMN5 | 2004 | 147 |
| Senataxin | SETX | ALS4 | 2004 | 18 |
| Tyrosyl tRNA Synthetase | YARS | CMT dominant/intermediate type C | 2006 | 57 |
| Pleckstrin Homology Domain-Containing Protein, Family G, Member 4 | PLEKHG5 | dSMA4 (Lower Motor Neuron Syndrome) | 2007 | 85 |
| Tyrosine Kinase-Type Cell Surface Receptor HER3 | HER3 | LCCS2 | 2007 | 99 |
| Phosphatidyl Inositol-4-Phosphate 5-Kinase, Type I, Gamma | PIP5K1C | LCCS3 | 2007 | 98 |
| TAR DNA Binding Protein, 43-kD | TDP-43 | ALS10 | 2008 | 124 |
| GLE1 | GLE1 | LCCS1 | 2008 | 102 |
| Fused in Sarcoma | FUS | ALS6 | 2009 | 72,139 |
This review will focus on motor neuron disease genes that appear to play a role in RNA processing pathways (Table 2). While there is no one cellular pathway that is exclusively responsible for these syndromes, it is instructive to place the ultimate molecular pathology responsible for motor neuron loss in this class of disease, including sporadic ALS, into the context of ubiquitous RNA processing events. We will begin with a brief review of RNA processing pathways.
Table 2.
Motor Neuron Disease Genes Involved in RNP formation
| Gene | RNA processing pathway |
|---|---|
| SMN | UsnRNP assembly, RNP assembly |
| SETX | Transcription, processing |
| TDP-43 | Splicing |
| FUS | Splicing |
| GLE1 | mRNA transport |
| IGHMBP2 | Translation |
| GARS/YARS | Amino-acyl tRNA assembly |
| HSPB1 | AU-rich element mediated mRNA decay |
Transcription to Translation
RNA is never naked within the cell. From the moment it is transcribed until it is degraded, RNA is bound by protein and assembled into ribonucleoprotein (RNP) complexes. The RNA-associated proteins in these complexes insure that each RNA transcript is properly spliced, processed, transported from the nucleus to the cytoplasm and translated by the protein synthesis machinery of the cell. The stages of this intricately choreographed process are complex and incompletely understood, although much has been learned over the past decade about the major pathways involved.
RNA processing begins while RNA polymerase II is in the process of transcribing each protein coding gene. The first step is the addition of an inverted guanosine to the 5’ end and its methylation to create a ‘cap’ that marks the beginning of the mRNA. Proteins binding to the cap structure coordinate subsequent steps in pre-mRNA processing, export, translation and decay. In pre-mRNA, the protein-coding sequences (exons) are interrupted by numerous non-coding sequences (introns) that are removed in the nucleus by pre-mRNA splicing. Several hundred proteins and some noncoding RNAs participate in the overall splicing reaction. Moreover, the inclusion or exclusion of selected exonic sequences in specific cells at specific developmental stages and in response to specific extracellular stimuli generates multiple mRNA transcripts from the same pre-mRNA sequence via alternative-splicing pathways. Perhaps nowhere is this more prevalent than in neurons, and this accounts for much of the remarkable variability in gene expression seen in the nervous system. The role of RNA splicing in neurological diseases is well recognized36,78.
The pre-mRNA splicing process is essential for eukaryotic gene expression and is carried out by the nuclear spliceosome. The major components of the spliceosome are the U1, U2, U5, and U4/U6 small nuclear ribonucleoproteins (snRNPs), each of which is composed of one snRNA molecule, a set of seven common proteins, and several proteins that are specific to individual snRNAs81,59,146. A critical aspect of snRNP biogenesis is that their assembly is highly regulated and requires both ATP hydrolysis and dedicated assembly factors87,108. With the exception of U6, which never leaves the nucleus, the newly transcribed U snRNAs are initially exported to the cytoplasm, where the major assembly process of the snRNPs occurs. The common proteins, called Sm proteins, B/B’, D1, D2, D3, E, F, and G are arranged into a stable heptameric ring, the Sm core, on a highly conserved, uridine-rich sequence motif, the Sm site, of the snRNAs59,1,125. The 5’ ends of snRNAs begin with the same 7-methyl guanosine cap structure as mRNA; however, once assembled, it undergoes additional methylation to a 2,2,7-trimethyl guanosine cap, which signals the re-import of the properly assembled and modified snRNPs into the nucleus where additional snRNP-specific proteins associate to form fully functional snRNPs83,30,46,31,84,146. Mature snRNPs then carry out the process of pre-mRNA splicing. Splicosomal U snRNP biogenesis occurs in the cytoplasm and is mediated by the SMN (survival motor neuron) complex. The SMN complex directly recognizes and binds to both the protein and the RNA components of the RNP and facilitates their interaction, thereby ensuring a strict specificity of the snRNP assembly process (reviewed in150,68).
Transcribing RNA polymerase continues past the end of the gene, and downstream sequences are removed by a precise processing event that involves assembly of a large protein complex guided by the conserved AAUAAA sequence and GU-rich sequences downstream of the processing site. This multiprotein complex contains an endonuclease that cleaves the pre-mRNA to create the 3’ end and poly(A) polymerase, which adds the poly(A) tail, a homopolymer of ~250 adenosine residues. These latter steps are coordinated by interaction between a splicing factor bound to the last intron of the pre-mRNA and poly(A) polymerase.
At this point the capped, spliced and polyadenylated mRNA is assembled into an mRNP complex that is competent for export from the nucleus to the cytoplasm. As with snRNP assembly, the export process is a highly coordinated event involving the formation and disruption of specific RNA-protein complexes and intermediaries. The objective in this case is to move mRNPs through nuclear pore complexes (NPCs), large protein assemblies that offer the only channel between the nucleus and cytoplasm93,20,127. This transport requires that another protein complex, the Mex67:Mtr2 heterodimer, binds to the mRNP prior to transport127. Once transported to the cytoplasmic side of the NPC, Mex67:Mtr2 is removed, which prevents retrograde transport back into the nucleus. This removal is catalyzed by the DEAD-box RNA helicase Dbp5. Dbp5 shuttles between the nucleus and cytoplasm, but it is on the cytoplasmic side of the nuclear pore complex, where it binds to and is activated by Gle1. The associated loss of Mex67:Mtr2 from the mRNP allows this to then complete transit through the NPC into the cytoplasm. DEAD-box helicases use the energy in ATP to unwind RNP complexes, and maximum ATPase activity of Dbp5 is achieved when it associates with InsP6, a molecule which facilitates and regulates the interaction between Gle1 and Dbp5151.
mRNA faces numerous potential fates after it arrives in the cytoplasm. It is currently thought that coincident with export, the mRNP undergoes a surveillance process where it is scanned by a ‘pioneer round’ of translation that determines whether the transcript will encode a full-length protein. This is an important, evolutionarily conserved process that protects the cell from the accumulation of C-terminally truncated proteins with potential dominant negative function that may arise from alternative splicing, processing errors or certain inherited mutations, all of which generate mRNAs with a termination codon upstream of the correct position. Splicing deposits a complex of proteins at each exon junction (the exon junction complex, EJC) that accompany the newly processed mRNP into the cytoplasm, and these provide positional information for the location of a stop codon. On a correct mRNA, ribosomes will scan through these. The ribosomes remove each of the complexes in the process until they reach the normal stop codon and are released. However, if the mRNA carries a termination codon that is upstream of an EJC, the ribosome will pause at that position, and this activates the degradation of that mRNA through a process termed ‘nonsense-mediated mRNA decay’ or NMD. NMD is ultimately responsible for the loss of protein product in many genetic disorders that result from the inheritance of one or more genes that carry a premature termination codon.
Once through the surveillance process, the mRNA can be bound by initiation factors and translated, silenced (for example by binding of microRNAs), stored or degraded. Each of these processes in itself is highly regulated, and the fate of each mRNA is determined by cis-acting sequence elements within the mRNA and by proteins (and microRNAs) that bind to these elements. A case in point is the large family of mRNAs that carry adenosine plus uridine-rich sequence elements (AU-rich elements, or AREs) in their 3’ untranslated regions. There are over 3000 mRNAs with AREs8, and their differential binding by a number of ARE-binding proteins (including HSPB1121) determine their overall accumulation, translation and decay. The role of mRNA stability in neurons is increasingly recognized and important in neuronal development and regeneration12.
mRNA translation is mediated by the ribosome and is conventionally divided into the initiation, elongation and termination stages. During initiation, the ribosome and the tRNA for the first amino acid residue, methionine, recognize and bind to the first codon, AUG, of the mRNA transcript. The polypeptide encoded by the mRNA is assembled during the elongation phase, and termination occurs when the ribosome encounters a ‘stop’ codon that results in release of the completed polypeptide and dissociation of the ribosomal subunits. The entire process is, once again, mediated by the complex association of a host of specific RNP complexes, non-coding RNAs, and proteins.
If one considers the ribosome to be a protein-synthesizing machine, the raw materials are the mRNA template and the individual aminoacyl-tRNAs. Aminoacyl-tRNAs are covalently linked RNA-protein complexes that are produced by a number of aminoacyl-tRNA synthetases (ARSs) that work to catalyze the esterification of specific amino acids to their respective tRNAs. A number of additional functions for particular ARSs have recently come to light, including roles in transcription, splicing, apoptosis, inflammation and angiogenesis (reviewed in67,104,105).
Ultimately all mRNAs are degraded, and their rate of decay is a controlling factor in the timing, quantity and perhaps even location of their encoded proteins. In general, mRNA decay begins with shortening of the poly(A) tail, followed by removal of the 5’ cap and degradation by exonucleases from both the 5’ and 3’ ends of the mRNA96. This is a highly regulated process that involves the interplay between cis-acting sequences and the proteins that bind to them. Both translation and decay are targets of numerous signal transduction pathways, and we are just beginning to understand how they are controlled.
Motor neuron disease genes
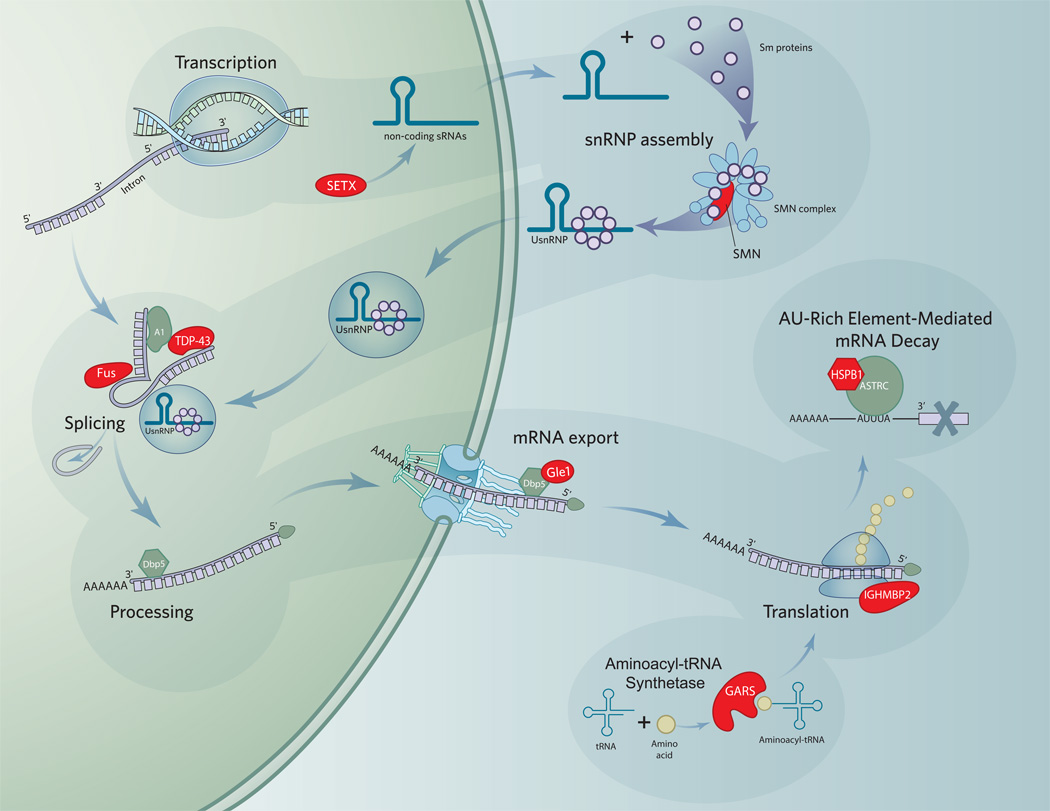
A large number of known motor neuron disease genes are directly involved in RNA processing (Figure 1). While many of these genes theoretically impact upon all mRNAs as they are processed, this is not proven. In neurons, there may be unique susceptibility for distinct groups of mRNAs to mutations in the following genes (Table 2).
Figure 1.
RNA processing pathways involved in motor neuron disease.
Abbreviations: snRNA, small nuclear ribonucleic acid; ATP, adenosine triphosphosphate; mRNP, messenger ribonucleoprotein; InsP6, inositol hexakisphosphate 6; UsnRNP, uridine-rich small nuclear ribonucleoprotein; UPF1, Up Frameshift 1; hnRNP, heteronuclear ribonucleoprotein;
Survival Motor Neuron (SMN)
SMA is an autosomal recessive neurodegenerative disease that is the leading inherited cause of infant mortality. It affects 1 in 10,000 live births and has a carrier frequency of ~1 in 40107,22. The hallmark features of SMA are progressive weakness, dysphagia, dyspnea and death and are the result of a selective loss of motor neurons at an early age28,51,131. Clinically, SMA is classified based on age of onset and severity defined by major motor milestones. Children with the most severe type I (Werdnig-Hoffman) are never able to sit unassisted and usually die of respiratory insufficiency by age 2 years. Children with the intermediate type II are never able to stand unassisted and have a variable survival. The mild type III (Kugelberg-Welander) is characterized by onset of weakness after the age of 18 months, and survival is prolonged well into adulthood. However these patients often become wheelchair bound during youth or adulthood. There is currently no treatment for SMA.
SMA is caused by mutation of both alleles of the survival of motor neurons 1 (SMN1) gene76, however all patients retain at least one copy of the SMN2 gene. The SMN1 gene produces full-length SMN mRNA, whereas the SMN2 gene produces full-length mRNA and mRNA lacking exon 7 (SMNΔ7), as well as small amounts of mRNA lacking exon 5 or exon 3 or combinations thereof80,92. SMNΔ7 mRNA encodes an unstable, truncated protein79. SMA is postulated to result from insufficient expression levels of full-length SMN protein in motor neurons, perhaps during a critical stage of motor neuron development, however, the exact mechanism by which SMN mutations result in SMA is unknown86,114,34. Several studies have shown that SMN mRNA and protein are reduced in cell lines and tissues derived from type I SMA patients compared to controls21,77,37,64,123.
The capacity of cells to assemble small nuclear ribonucleoproteins (snRNPs) is reduced when the expression of the SMN protein is reduced in cell culture, SMA patient cells and in SMA mouse model tissues150,143,34,68,153. Thus, it appears that defects in the formation of specific RNPs can result in motor neuron disease109. The consequences of decreased snRNP assembly include alterations of the relative expression of spliceosomal U snRNAs that subsequently alter specific mRNA splicing events34,153. The precise alterations in the splicing of mRNAs that occur in motor neurons in SMA are not yet known. It is also unclear whether these alterations cause motor neuron death in SMA, and why the motor neuron is so vulnerable to these changes. One can imagine that nearly all cellular processes are vulnerable to splicing changes. For example, there is evidence that structural abnormalities at the neuromuscular junction precede the loss of motor neuron cell bodies in a mouse model of SMA62. One can speculate that genes involved in neuromuscular junction integrity or formation may be improperly spliced or otherwise processed as a result of reduced SMN levels.
Senataxin (SETX)
Several missense mutations in the senataxin gene (SETX) (L389S, R2136H, and T3I) cause a rare autosomal dominant form of juvenile ALS (ALS4) characterized by distal muscle weakness and atrophy with pyramidal signs, in which sensation remains unaffected18. Other mutations in SETX (L1976R, L1977F, N603D–Q653K) have been shown to cause oculomotor apraxia 2 (AOA2), an autosomal recessive cerebellar ataxia characterized by cerebellar atrophy, oculomotor apraxia, early loss of reflexes, late peripheral neuropathy, and slow progression leading to severe disability94,32,10.
The senataxin protein is a member of the UPF1-like helicase within the DNA/RNA helicase superfamily. It contains 7 helicase motifs and is thought to unwind both DNA and RNA substrates18,45. While little is known about the function of senataxin in motor neurons, Sen1p, the senataxin ortholog in Saccharomyces cerevisiae, has been well studied. Mutations in the helicase domain of Sen1p alter the cellular abundance of many diverse RNA species including intron-containing tRNA precursors, ribosomal RNA precursors and certain small nucleolar RNAs and snRNAs148,136,112,126. In yeast, Sen1p plays a role in the normal formation of the U5 spliceosomal snRNA137. It will be interesting to compare the changes in U5 snRNA abundance that result from SETX mutations to the U5 snRNA abundance changes that are now being reported as a result of reduced SMN expression.
TAR DNA binding protein (TDP-43)
TDP-43 was initially shown to be the ubiquitinated protein that comprises the ubiquitin-positive inclusions found in pathologic specimens from individuals with sporadic ALS and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U)100. In one study, immunohistochemistry performed on postmortem spinal cord from individuals with sporadic ALS and familial ALS with and without the SOD1 mutation revealed TDP-43 positive inclusions in all of 59 sporadic and 11 SOD1-negative familial ALS specimens82. TDP-43 positive inclusions were NOT detected in 15 SOD1-positive familial ALS specimens82. A causal role of TDP-43 in ALS pathogenesis has been demonstrated by recent reports of TDP-43 mutations in familial ALS patients58,69,116,124,38.
TDP-43 is an RNA-binding protein that contains two RNA recognition motifs (RRMs) that are similar to those found in well-characterized heteronuclear RNPs such as hnRNP A1 and poly A binding protein (PABP)103,14,15. TDP-43 binds to (UG)n-repeated sequences near the 3’ splice site of the cystic fibrosis transmembrane conductance regulator (CFTR) exon 9 and, and its binding inhibits the inclusion of this exon14. TDP-43 also binds to UG dinucleotide repeats at the 3’ splice site of apolipoprotein A-II intron 3 and intron 2 of the human cardiac Na+-Ca2+ exchanger35,7. The C-terminal domain of TDP-43 binds to several splicing inhibitors including hnRNP A1 and hnRNP A2/B116. Thus, TDP-43 has splicing inhibitor activity and appears to form a molecular anchor to recruit hnRNP proteins at specific pre-mRNA dinucleotide sequences in order to effect splicing at that site.
Most TDP-43 mutations described in fALS (a mutation in RRM1) are found in the C-terminal domain of the protein. They are predicted to disrupt hnRNP binding, which would confer a functional consequence of reduced splicing inhibitory activity58,124. fALS-causing TDP-43 mutations alter the subcellular distribution of the protein from the nucleus to the cytoplasm, and this may be a result of protein aggregation of the mutant protein that precludes import into the cytoplasm124. Thus, the mutations would interfere with TDP-43 mediated splicing activity by directly decreasing the available amount of active protein. While these mutations are rare, the global presence of TDP-43 in sporadic ALS inclusions suggests that, in the case of sporadic ALS, TDP-43 mediated splicing inhibitory activity may be reduced secondarily via excessive ubiquitination and sequestration.
Fused in Sarcoma (FUS)
Mutations in FUS have been discovered in three families with fALS72,139. In at least one of the reported families, the onset of weakness was in the proximal upper extremities with no bulbar involvement72. The FUS gene encodes a protein that contains three RGG repeat domains, an RNA-recognition motif (RRM) and a zinc-finger domain52. FUS associates with hnRNP A1 and C1/C2, suggesting a role in RNA splicing154. FUS also binds to the actin-associated molecular motors KIF5 and Myosin Va, and colocalizes with RNA-transport granules60,152.
A molecular consequence of fALS-causing FUS mutations is that FUS protein, which is predominantly localized in the nucleus, is redistributed and found within inclusions in the cytoplasm72,139. Thus, FUS and TDP-43 share many of the same molecular properties (interaction with RNA and splicing factors) and are similarly altered when they are mutated (nuclear to cytoplasmic redistribution), resulting in fALS. The molecular overlap of these proteins in motor neuron disease may be further demonstrated should the FUS protein be found to co-localize with TDP-43 positive inclusions found in sporadic ALS patients.
GLE1
Mutations in GLE1 can lead to forms of lethal congenital contracture syndrome (LCCS), an autosomal recessive motor neuropathy characterized by early fetal hydrops, akinesia, agnathia, loss of anterior horn neurons, and various other features, which result in prenatal or immediately postnatal death102. LCCS has been divided into types 1, 2, and 3 based upon specific clinical criteria, and they are caused by mutations in GLE1, HER3, and PIP5K1C, respectively98,99,102. Remarkably, the latter two proteins are required in the phosphatidyl inositol pathway. Both are involved in the synthesis of inositol hexakisphosphate (InsP6), which binds directly to the yeast homolog of GLE1.
GLE1 encodes the protein Gle1 which is a required mediator of mRNA export. Gle1 is found with the protein Dbp5 on the cytoplasmic side of nuclear pore complexes (NPCs) along nuclear pore filaments5,145, where it activates DExD/H-box protein 5 (Dbp5) to affect mRNA export from the nucleus. Dbp5 is an ATP-dependent RNA helicase, although it is too inefficient alone to be considered active under biological conditions. However, binding of activated Gle1 (via the InsP6 pathway) increases the catalytic efficiency of Dbp5 by 27-fold145.
Immunoglobulin µ-binding protein 2 (IGHMBP2)
Mutations in the gene that encodes immunoglobulin µ-binding protein 2 (IGHMBP2) have been demonstrated to cause distal spinal muscular atrophy with respiratory distress type 1 (SMARD1)39,42. This severe form of SMA results in diaphragmatic paralysis with weakness in the upper limbs and distal muscles. The pattern of phenotypic features is somewhat variable with some involvement of sensory and autonomic nerves in some confirmed cases40,110.
IGHMBP2 shares a 42% sequence homology with SETX and contains DExxQ-type helicase/ATPase domains that are conserved among DNA replication, repair, and recombination proteins, and a nuclear localization signal in the protein’s C-terminal region89,91. IGHMBP2 immunoreactivity occurs in the cytosol and in axons of neuronal cells41. Recombinant IGHMBP2 directly interacts with ribosomes. It hydrolyzes ATP and is capable of unwinding duplex RNA or DNA in vitro43. The functional consequences of each currently known disease-causing mutation are a loss of ATPase activity with resultant helicase function reduction or direct helicase impairment43. One of the IGHMBP2 mutations, T493I, does not affect ATPase or helicase activity in vitro, however this mutation may lead to decreased stability of the protein. It produces a lower expression level, as has been demonstrated in DSMA1 patient cell lines44.
Aminoacyl-tRNA synthetase family
Aminoacyl-tRNA synthetases attach amino acids to their cognate tRNAs. Mutations in a number of these enzymes result in neurological diseases, including motor neuron disease. Glycyl-tRNA synthetase (GARS) mutations result in CMT2D, an axonal form of CMT with upper limb predominance, and dHMNV, a very similar disease that lacks the distal sensory loss of CMT2D6,122,23,55,118,25. Tyrosyl-tRNA synthetase (YARS) mutations cause CMT-dominant/intermediate type C, including G41R in a North American family, E196K in a Bulgarian family, and a 12-bp in-frame deletion (153–156delVKQV) in a Bulgarian individual57.
Aminoacyl-tRNA synthetases are comprised of 3 domains: a WHEP-TRS domain that conjugates with other ARSs, a core catalytic domain for ligation, and an anticodon-binding domain to recognize the correct tRNAs33. To become active, the GARS protein (GlyRS) forms a homodimer.149. Mutations that affect the strength of the GlyRS dimer interaction also lead to GlyRS mislocalization within neurons97,149. Building upon the findings that the related tyrosil ARS (TyrRS) localizes to sprouting neurites57, researchers found that GlyRS also localizes to neurites, but mutant forms do so poorly97. These dimer-localization studies suggest a potential role for a subset of tRNA synthetases in neuron development. In support of this hypothesis, a recent study in Drosophila has shown that the GARS ortholog (gars) is necessary for proper neuron outgrowth19. Additionally, mutant flies could be rescued with transgenic human GARS, but outgrowth improved to lesser degrees when flies were given two different human CMT2D mutant forms of GARS.
Aminoacyl-tRNA synthetases may also play a role in splicing. The fungal protein homologous to TyrRS in the mitochondria of Neurospora crassa (mt TryRS, encoded by cyt-18) is required not only for tyrosyl-tRNA synthesis, but also for efficient splicing of group I introns in vitro and in vivo4,117,90,106. TyrRS may also be secreted and have cytokine properties under conditions of cell stress142.
Mutations in this family of enzymes are recognized in many human diseases of the nervous system, autoimmune disease and cancer105. For example, mitochondrial aspartyl-tRNA synthetase mutation resulting in leukoencephalopathy with brain stem and spinal cord involvement119, and a missense mutation in the alanyl-ARS of mouse leads to a late onset neurological disease characterized by patchy fur and mild tremors progressing to ataxia and Purkinje cell loss (esp. in the rostral cerebellum)75. Lastly, splicing errors in tRNA have been shown to cause pontocerebellar hypoplasia13.
α-crystallin type heat shock protein family
The α-crystallin type heat shock protein family (sHSPs) is comprised of ten multifunctional molecular chaperones that are induced in response to a wide variety of physiological and environmental factors and are numbered HSPB1 – HSPB1061. All sHSPs share an α-crystallin domain which is thought to mediate protein-protein interactions. Substitution mutations in three of the ten sHSP genes result in neuromuscular disease, including two (HSPB1 and HSPB8) which result in distal hereditary motor neuropathy11,135,53,54. Mutations in HSPB1 have been shown in dominant, recessive and sporadic cases of hereditary motor neuropathy29,133,65,24,50,56. In nearly every case, these mutations target positively charged arginines or lysines that are critical for the structural and functional integrity of the α-crystallin domain contained in each of these proteins and appear to alter sHSP chaperone-like activity29,54,66. The functional consequences of these mutations as they relate to the motor neuron are not known, however, HSPB1 appears to be neuroprotective and have roles in the maintenance and regulation of the neuronal cytoskeleton and axonal transport128,2,74.
HSPB1 plays a role in AU-rich element mediated mRNA decay121. As noted above, approximately 3000 mRNAs contain AU-rich elements in the 3’-UTR. As a group these mRNAs are inherently unstable, and it is this property that cells exploit to effect rapid changes in protein expression. Induction of an unstable mRNA results in a more rapid acquisition of a new steady state than a similar induction of a stable mRNA, and withdrawing the stimulus for this induction results in similarly rapid decrease in steady state, with matching decrease in protein production. While inflammatory cytokines are the mRNAs that are most commonly studied, they represent only a fraction of the mRNAs that contain AU-rich elements. The turnover of many of these mRNAs is controlled by a complex of RNA-binding proteins, many of which bind selectively to the different AU-rich elements. One of these, AUF1 (hnRNP D) is noteworthy, because differentially spliced isoforms act either to stabilize or destabilize these mRNAs. Recently, HSPB1 was identified as a constituent of a destabilizing complex with AUF1, Hsp70 and Hsc70121. HSPB1 binds to AU-rich elements, and its knockdown stabilizes a reporter mRNA that contains this destabilizing sequence. These findings raise the intriguing possibility that mutations in HSPB1 (and perhaps other members of this protein family) might result in motor neuron-specific changes in the stability of specific mRNAs.
Discussion/Convergence
The clustering of recognized motor neuron disease genes in various RNA processing pathways suggests that much more needs to be done to understand the mechanisms of RNA processing and metabolism as they relate to motor neurons (and associated cell types). The riddle of motor neuron selectivity, when one of a handful of RNA processing genes are mutated, suggests that a key to understanding sporadic ALS may be to focus on a characterization of the unique sets of mRNA, splicing isoforms, and the expansive repertoire of non-coding RNAs that a normal motor neuron is expected to produce throughout its lifetime. Defects in RNA processing may be subtle and have consequences that relate to other, relevant pathogenic pathways. For example, defects in the RNA editing pathway of GluR2 pre-mRNA have been described in the spinal cords of sporadic ALS patients which results in altered calcium permeability of the resultant AMPA receptor, presumably resulting in increased susceptibility to glutamate excitotoxicity129,63,71,130. Ultimately, a key concept going forward is that motor neurons (and motor neuron-specific RNA expression patterns) appear to be susceptible to errant ribonucleoprotein formations.
The critical function of axonal transport mechanisms in motor neuron disease and neuropathies has been well-established49,27. Mutations in DCTN1, which encodes dynactin, the principle protein responsible for dynein-mediated movement of organelles along microtubules, have been associated with a form of fALS111,95. Two isoforms of dynactin exist in neuronal tissue, created by alternative splicing of the mRNA. While both isoforms bind dynein in the cytoplasm, only the p150 isoform binds microtubules, suggesting that the p135 isoform is functionally distinct48. It is conceivable that primary errors in the expression of an RNA processing gene or of a non-coding RNA could produce a similar motor neuron disease by secondarily altering the alternative splicing of dynactin.
Dynactin may also be considered an important protein in the processing of RNAs in neuronal processes. Dynactin directly mediates the transport of RNAs along microtubules, in turn increasing the overall minus-end motility of RNA transport140. There is also evidence that the seipin protein, which is encoded by the BSCL2 gene and mutated in yet another HMN, may be involved in RNA transport3,147. Thus, a distinction between incorrect processing of RNAs destined to encode proteins essential for axonal transport and the incorrect transport of RNAs destined to travel along the axon may be a “chicken-or-egg” riddle that is not mutually exclusive in motor neuron disease.
It is well known that mutations in SOD1 result in fALS113. Oxidative damage is an established contributor to motor neuron pathology in ALS9. Remarkably, direct mRNA oxidation is also a common feature in ALS that may occur early in disease pathogenesis17. Moreover, mRNAs that encode genes linked to other motor neuron disease genes, such as SOD1 and DCTN1, were highly oxidized relative to other genes17. Since the oxidation of mRNA clearly interferes with subsequent processing steps120,132, a convergence of errors in oxidative stress buffering capacity and RNA processing errors must also be considered in our riddle. For example, mutant SOD1 but not wild-type SOD1 interacts with lysyl-tRNA synthetase and translocation-associated protein δ, suggesting that SOD1 mutations have the ability to affect RNA processing by binding to and altering the normal function of key proteins70. And it is, perhaps, no coincidence that oxidative stress inhibits the assembly of spliceosomal UsnRNPs by decreasing the activity of the SMN complex144.
The production of mature proteins from the human genome is an extremely complex and wonderfully organized process that, at every step, presents an opportunity for error. Given the added complexity of gene expression required within the nervous system, which must respond to connectivity and neuronal activity cues, it is not surprising that primary disruptions of RNA processing might prove deleterious to specific populations of neurons. How RNA processing defects result in the selective loss of motor neurons is an important area of focus to gain another foothold in our understanding of the pathogenesis of motor neuron disease, including sporadic ALS. The emerging importance of RNA processing in motor neuron disease calls for: 1) the identification of alternative splicing events that are pathogenic in MND, 2) characterization of essential non-coding RNAs in the cells of the motor unit (motor neuron, glial cells and muscle), 3) characterization of the full complement of mature mRNAs and RNPs in the motor unit, 4) an understanding of RNA processing events in the axoplasm, 5) an inclusive approach to motor neuron disease gene pathogenesis with an appreciation for molecular pathway convergence and 6) facilitated collaboration between RNA biologists and motor neuron disease investigators.
Acknowledgements
We are grateful to John T. Kissel for helpful comments and suggestions. SJK is supported by startup funds from The Ohio State University Medical Center. DRS is supported by PHS grant GM38277 and GM079707 from the National Institute of General Medical Science.
Bibliography
- 1.Achsel T, Stark H, Luhrmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerley S, James PA, Kalli A, French S, Davies KE, Talbot K. A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes. Human molecular genetics. 2006;15:347–354. doi: 10.1093/hmg/ddi452. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal AK, Garg A. Seipin: a mysterious protein. Trends Mol Med. 2004;10:440–444. doi: 10.1016/j.molmed.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Akins RA, Lambowitz AM. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987;50:331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- 5.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nature cell biology. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 6.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, Sivakumar K, Ionasescu V, Funalot B, Vance JM, Goldfarb LG, Fischbeck KH, Green ED. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. American journal of human genetics. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrisi-Mercado P, Romano M, Muro AF, Baralle FE. An exonic splicing enhancer offsets the atypical GU-rich 3' splice site of human apolipoprotein A-II exon 3. The Journal of biological chemistry. 2004;279:39331–39339. doi: 10.1074/jbc.M405566200. [DOI] [PubMed] [Google Scholar]
- 8.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic acids research. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber SC, Mead RJ, Shaw PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochimica et biophysica acta. 2006;1762:1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Bassuk AG, Chen YZ, Batish SD, Nagan N, Opal P, Chance PF, Bennett CL. In cis autosomal dominant mutation of Senataxin associated with tremor/ataxia syndrome. Neurogenetics. 2007;8:45–49. doi: 10.1007/s10048-006-0067-8. [DOI] [PubMed] [Google Scholar]
- 11.Benndorf R, Welsh MJ. Shocking degeneration. Nature genetics. 2004;36:547–548. doi: 10.1038/ng0604-547. [DOI] [PubMed] [Google Scholar]
- 12.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 13.Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, E TTB, Aronica E, van der Knaap MS, Hohne W, Toliat MR, Crow YJ, Steinlin M, Voit T, Roelens F, Brussel W, Brockmann K, Kyllerman M, Boltshauser E, Hammersen G, Willemsen M, Basel-Vanagaite L, Krageloh-Mann I, de Vries LS, Sztriha L, Muntoni F, Ferrie CD, Battini R, Hennekam RC, Grillo E, Beemer FA, Stoets LM, Wollnik B, Nurnberg P, Baas F. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nature genetics. 2008 doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 14.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. The Journal of biological chemistry. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 15.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. The EMBO journal. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. The Journal of biological chemistry. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 17.Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR, Wong PC, Lin CL. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS ONE. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) American journal of human genetics. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nature neuroscience. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- 20.Cole CN, Scarcelli JJ. Unravelling mRNA export. Nature cell biology. 2006;8:645–647. doi: 10.1038/ncb0706-645. [DOI] [PubMed] [Google Scholar]
- 21.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Human molecular genetics. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 22.Czeizel A, Hamula J. A hungarian study on Werdnig-Hoffmann disease. J Med Genet. 1989;26:761–763. doi: 10.1136/jmg.26.12.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Bo R, Locatelli F, Corti S, Scarlato M, Ghezzi S, Prelle A, Fagiolari G, Moggio M, Carpo M, Bresolin N, Comi GP. Coexistence of CMT-2D and distal SMA-V phenotypes in an Italian family with a GARS gene mutation. Neurology. 2006;66:752–754. doi: 10.1212/01.wnl.0000201275.18875.ac. [DOI] [PubMed] [Google Scholar]
- 24.Dierick I, Baets J, Irobi J, Jacobs A, De Vriendt E, Deconinck T, Merlini L, Van den Bergh P, Rasic VM, Robberecht W, Fischer D, Morales RJ, Mitrovic Z, Seeman P, Mazanec R, Kochanski A, Jordanova A, Auer-Grumbach M, Helderman-van den Enden AT, Wokke JH, Nelis E, De Jonghe P, Timmerman V. Relative contribution of mutations in genes for autosomal dominant distal hereditary motor neuropathies: a genotype-phenotype correlation study. Brain. 2008;131:1217–1227. doi: 10.1093/brain/awn029. [DOI] [PubMed] [Google Scholar]
- 25.Dubourg O, Azzedine H, Yaou RB, Pouget J, Barois A, Meininger V, Bouteiller D, Ruberg M, Brice A, LeGuern E. The G526R glycyl-tRNA synthetase gene mutation in distal hereditary motor neuropathy type V. Neurology. 2006;66:1721–1726. doi: 10.1212/01.wnl.0000218304.02715.04. [DOI] [PubMed] [Google Scholar]
- 26.Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. II. Neurologic, genetic, and electrophysiologic findings in various neuronal degenerations. Archives of neurology. 1968;18:619–625. doi: 10.1001/archneur.1968.00470360041003. [DOI] [PubMed] [Google Scholar]
- 27.El-Kadi AM, Soura V, Hafezparast M. Defective axonal transport in motor neuron disease. J Neurosci Res. 2007;85:2557–2566. doi: 10.1002/jnr.21188. [DOI] [PubMed] [Google Scholar]
- 28.Eng GD, Binder H, Koch B. Spinal muscular atrophy: experience in diagnosis and rehabilitation management of 60 patients. Arch Phys Med Rehabil. 1984;65:549–553. [PubMed] [Google Scholar]
- 29.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, Van Impe K, Fedotov V, Dadali E, Auer-Grumbach M, Windpassinger C, Wagner K, Mitrovic Z, Hilton-Jones D, Talbot K, Martin JJ, Vasserman N, Tverskaya S, Polyakov A, Liem RK, Gettemans J, Robberecht W, De Jonghe P, Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nature genetics. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 30.Fischer U, Luhrmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science (New York, NY. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 31.Fischer U, Sumpter V, Sekine M, Satoh T, Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. The EMBO journal. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fogel BL, Perlman S. Novel mutations in the senataxin DNA/RNA helicase domain in ataxia with oculomotor apraxia 2. Neurology. 2006;67:2083–2084. doi: 10.1212/01.wnl.0000247661.19601.28. [DOI] [PubMed] [Google Scholar]
- 33.Freist W, Logan DT, Gauss DH. Glycyl-tRNA synthetase. Biological chemistry Hoppe-Seyler. 1996;377:343–356. [PubMed] [Google Scholar]
- 34.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein Assembly Defects Correlate with Spinal Muscular Atrophy Severity and Preferentially Affect a Subset of Spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabellini N. A polymorphic GT repeat from the human cardiac Na+Ca2+ exchanger intron 2 activates splicing. European journal of biochemistry / FEBS. 2001;268:1076–1083. doi: 10.1046/j.1432-1327.2001.01974.x. [DOI] [PubMed] [Google Scholar]
- 36.Gallo JM, Jin P, Thornton CA, Lin H, Robertson J, D'Souza I, Schlaepfer WW. The role of RNA and RNA processing in neurodegeneration. J Neurosci. 2005;25:10372–10375. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH. Differential SMN2 expression associated with SMA severity. Nature genetics. 1998;20:230–231. doi: 10.1038/3030. [DOI] [PubMed] [Google Scholar]
- 38.Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, Baker M, Al-Lozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Annals of neurology. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C, Bertini E, Leonhardt-Horti H, Muntoni F, Ouvrier R, Pfeufer A, Rossi R, Van Maldergem L, Wilmshurst JM, Wienker TF, Sendtner M, Rudnik-Schoneborn S, Zerres K, Hubner C. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nature genetics. 2001;29:75–77. doi: 10.1038/ng703. [DOI] [PubMed] [Google Scholar]
- 40.Grohmann K, Varon R, Stolz P, Schuelke M, Janetzki C, Bertini E, Bushby K, Muntoni F, Ouvrier R, Van Maldergem L, Goemans NM, Lochmuller H, Eichholz S, Adams C, Bosch F, Grattan-Smith P, Navarro C, Neitzel H, Polster T, Topaloglu H, Steglich C, Guenther UP, Zerres K, Rudnik-Schoneborn S, Hubner C. Infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1) Annals of neurology. 2003;54:719–724. doi: 10.1002/ana.10755. [DOI] [PubMed] [Google Scholar]
- 41.Grohmann K, Rossoll W, Kobsar I, Holtmann B, Jablonka S, Wessig C, Stoltenburg-Didinger G, Fischer U, Hubner C, Martini R, Sendtner M. Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1) Human molecular genetics. 2004;13:2031–2042. doi: 10.1093/hmg/ddh222. [DOI] [PubMed] [Google Scholar]
- 42.Guenther UP, Schuelke M, Bertini E, D'Amico A, Goemans N, Grohmann K, Hubner C, Varon R. Genomic rearrangements at the IGHMBP2 gene locus in two patients with SMARD1. Human genetics. 2004;115:319–326. doi: 10.1007/s00439-004-1156-0. [DOI] [PubMed] [Google Scholar]
- 43.Guenther UP, Handoko L, Laggerbauer B, Jablonka S, Chari A, Alzheimer M, Ohmer J, Plottner O, Gehring N, Sickmann A, von Au K, Schuelke M, Fischer U. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1) Human molecular genetics. 2009 doi: 10.1093/hmg/ddp028. [DOI] [PubMed] [Google Scholar]
- 44.Guenther UP, Handoko L, Varon R, Stephani U, Tsao CY, Mendell JR, Lutzkendorf S, Hubner C, von Au K, Jablonka S, Dittmar G, Heinemann U, Schuetz A, Schuelke M. Clinical variability in distal spinal muscular atrophy type 1 (DSMA1): determination of steady-state IGHMBP2 protein levels in five patients with infantile and juvenile disease. Journal of molecular medicine (Berlin, Germany) 2009;87:31–41. doi: 10.1007/s00109-008-0402-7. [DOI] [PubMed] [Google Scholar]
- 45.Hadjigeorgiou GM, Kim SH, Fischbeck KH, Andreu AL, Berry GT, Bingham P, Shanske S, Bonilla E, DiMauro S. A new mitochondrial DNA mutation (A3288G) in the tRNA(Leu(UUR)) gene associated with familial myopathy. Journal of the neurological sciences. 1999;164:153–157. doi: 10.1016/s0022-510x(99)00062-3. [DOI] [PubMed] [Google Scholar]
- 46.Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 47.Herva R, Leisti J, Kirkinen P, Seppanen U. A lethal autosomal recessive syndrome of multiple congenital contractures. American journal of medical genetics. 1985;20:431–439. doi: 10.1002/ajmg.1320200303. [DOI] [PubMed] [Google Scholar]
- 48.Holzbaur EL, Tokito MK. Localization of the DCTN1 gene encoding p150Glued to human chromosome 2p13 by fluorescence in situ hybridization. Genomics. 1996;31:398–399. doi: 10.1006/geno.1996.0068. [DOI] [PubMed] [Google Scholar]
- 49.Holzbaur EL. Motor neurons rely on motor proteins. Trends in cell biology. 2004;14:233–240. doi: 10.1016/j.tcb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Houlden H, Laura M, Wavrant-De Vrieze F, Blake J, Wood N, Reilly MM. Mutations in the HSP27 (HSPB1) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology. 2008;71:1660–1668. doi: 10.1212/01.wnl.0000319696.14225.67. [DOI] [PubMed] [Google Scholar]
- 51.Iannaccone ST. Spinal muscular atrophy. Seminars in neurology. 1998;18:19–26. doi: 10.1055/s-2008-1040858. [DOI] [PubMed] [Google Scholar]
- 52.Iko Y, Kodama TS, Kasai N, Oyama T, Morita EH, Muto T, Okumura M, Fujii R, Takumi T, Tate S, Morikawa K. Domain architectures and characterization of an RNA-binding protein, TLS. The Journal of biological chemistry. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- 53.Irobi J, Tissir F, De Jonghe P, De Vriendt E, Van Broeckhoven C, Timmerman V, Beuten J. A clone contig of 12q24.3 encompassing the distal hereditary motor neuropathy type II gene. Genomics. 2000;65:34–43. doi: 10.1006/geno.2000.6149. [DOI] [PubMed] [Google Scholar]
- 54.Irobi J, Van Impe K, Seeman P, Jordanova A, Dierick I, Verpoorten N, Michalik A, De Vriendt E, Jacobs A, Van Gerwen V, Vennekens K, Mazanec R, Tournev I, Hilton-Jones D, Talbot K, Kremensky I, Van Den Bosch L, Robberecht W, Van Vandekerckhove J, Van Broeckhoven C, Gettemans J, De Jonghe P, Timmerman V. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nature genetics. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- 55.James PA, Cader MZ, Muntoni F, Childs AM, Crow YJ, Talbot K. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology. 2006;67:1710–1712. doi: 10.1212/01.wnl.0000242619.52335.bc. [DOI] [PubMed] [Google Scholar]
- 56.James PA, Rankin J, Talbot K. Asymmetrical late onset motor neuropathy associated with a novel mutation in the small heat shock protein HSPB1 (HSP27) Journal of neurology, neurosurgery, and psychiatry. 2008;79:461–463. doi: 10.1136/jnnp.2007.125179. [DOI] [PubMed] [Google Scholar]
- 57.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, Rao CV, Tournev I, Gondim FA, D'Hooghe M, Van Gerwen V, Callaerts P, Van Den Bosch L, Timmermans JP, Robberecht W, Gettemans J, Thevelein JM, De Jonghe P, Kremensky I, Timmerman V. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nature genetics. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 58.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nature genetics. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 59.Kambach C, Walke S, Nagai K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Current opinion in structural biology. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- 60.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 61.Kappe G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1-10. Cell stress & chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Human molecular genetics. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 64.Kernochan LE, Russo ML, Woodling NS, Huynh TN, Avila AM, Fischbeck KH, Sumner CJ. The role of histone acetylation in SMN gene expression. Human molecular genetics. 2005 doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 65.Kijima K, Numakura C, Goto T, Takahashi T, Otagiri T, Umetsu K, Hayasaka K. Small heat shock protein 27 mutation in a Japanese patient with distal hereditary motor neuropathy. J Hum Genet. 2005;50:473–476. doi: 10.1007/s10038-005-0280-6. [DOI] [PubMed] [Google Scholar]
- 66.Kim MV, Kasakov AS, Seit-Nebi AS, Marston SB, Gusev NB. Structure and properties of K141E mutant of small heat shock protein HSP22 (HspB8, H11) that is expressed in human neuromuscular disorders. Archives of biochemistry and biophysics. 2006;454:32–41. doi: 10.1016/j.abb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 67.Ko YG, Park H, Kim S. Novel regulatory interactions and activities of mammalian tRNA synthetases. Proteomics. 2002;2:1304–1310. doi: 10.1002/1615-9861(200209)2:9<1304::AID-PROT1304>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 68.Kolb SJ, Battle DJ, Dreyfuss G. Molecular Functions of the SMN Complex. Journal of child neurology. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 69.Kuhnlein P, Sperfeld AD, Vanmassenhove B, Van Deerlin V, Lee VM, Trojanowski JQ, Kretzschmar HA, Ludolph AC, Neumann M. Two German kindreds with familial amyotrophic lateral sclerosis due to TARDBP mutations. Archives of neurology. 2008;65:1185–1189. doi: 10.1001/archneur.65.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunst CB, Mezey E, Brownstein MJ, Patterson D. Mutations in SOD1 associated with amyotrophic lateral sclerosis cause novel protein interactions. Nature genetics. 1997;15:91–94. doi: 10.1038/ng0197-91. [DOI] [PubMed] [Google Scholar]
- 71.Kwak S, Kawahara Y. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. Journal of molecular medicine (Berlin, Germany) 2005;83:110–120. doi: 10.1007/s00109-004-0599-z. [DOI] [PubMed] [Google Scholar]
- 72.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science (New York, NY. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 73.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 74.Latchman DS. HSP27 and cell survival in neurones. Int J Hyperthermia. 2005;21:393–402. doi: 10.1080/02656730400023664. [DOI] [PubMed] [Google Scholar]
- 75.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 76.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 77.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nature genetics. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 78.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 79.Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nature genetics. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 80.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luhrmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochimica et biophysica acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 82.Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, Eisen A, McClusky L, Kretzschmar HA, Monoranu CM, Highley JR, Kirby J, Siddique T, Shaw PJ, Lee VM, Trojanowski JQ. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Annals of neurology. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 83.Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 84.Mattaj IW, Boelens W, Izaurralde E, Jarmolowski A, Kambach C. Nucleocytoplasmic transport and snRNP assembly. Mol Biol Rep. 1993;18:79–83. doi: 10.1007/BF00986760. [DOI] [PubMed] [Google Scholar]
- 85.Maystadt I, Rezsohazy R, Barkats M, Duque S, Vannuffel P, Remacle S, Lambert B, Najimi M, Sokal E, Munnich A, Viollet L, Verellen-Dumoulin C. The nuclear factor kappaB-activator gene PLEKHG5 is mutated in a form of autosomal recessive lower motor neuron disease with childhood onset. American journal of human genetics. 2007;81:67–76. doi: 10.1086/518900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. The Journal of cell biology. 2003;162:919–932. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nature cell biology. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 88.Mersiyanova IV, Perepelov AV, Polyakov AV, Sitnikov VF, Dadali EL, Oparin RB, Petrin AN, Evgrafov OV. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. American journal of human genetics. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizuta TR, Fukita Y, Miyoshi T, Shimizu A, Honjo T. Isolation of cDNA encoding a binding protein specific to 5'-phosphorylated single-stranded DNA with G-rich sequences. Nucleic acids research. 1993;21:1761–1766. doi: 10.1093/nar/21.8.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mohr G, Rennard R, Cherniack AD, Stryker J, Lambowitz AM. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. Journal of molecular biology. 2001;307:75–92. doi: 10.1006/jmbi.2000.4460. [DOI] [PubMed] [Google Scholar]
- 91.Molnar GM, Crozat A, Kraeft SK, Dou QP, Chen LB, Pardee AB. Association of the mammalian helicase MAH with the pre-mRNA splicing complex. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7831–7836. doi: 10.1073/pnas.94.15.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Human molecular genetics. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 93.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science (New York, NY. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 94.Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, Pandolfo M, Schulz JB, Pouget J, Calvas P, Shizuka-Ikeda M, Shoji M, Tanaka M, Izatt L, Shaw CE, M'Zahem A, Dunne E, Bomont P, Benhassine T, Bouslam N, Stevanin G, Brice A, Guimaraes J, Mendonca P, Barbot C, Coutinho P, Sequeiros J, Durr A, Warter JM, Koenig M. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nature genetics. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 95.Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, Ludolph AC. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- 96.Murray EL, Schoenberg DR. A+U-rich instability elements differentially activate 5'-3' and 3'–5' mRNA decay. Molecular and cellular biology. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nangle LA, Zhang W, Xie W, Yang XL, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Narkis G, Ofir R, Landau D, Manor E, Volokita M, Hershkowitz R, Elbedour K, Birk OS. Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKI gamma of the phophatidylinsitol pathway. American journal of human genetics. 2007;81:530–539. doi: 10.1086/520771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Narkis G, Ofir R, Manor E, Landau D, Elbedour K, Birk OS. Lethal congenital contractural syndrome type 2 (LCCS2) is caused by a mutation in ERBB3 (Her3), a modulator of the phosphatidylinositol-3-kinase/Akt pathway. American journal of human genetics. 2007;81:589–595. doi: 10.1086/520770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science (New York, NY. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 101.Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. American journal of human genetics. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nature genetics. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. Journal of virology. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends in biochemical sciences. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 105.Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paukstelis PJ, Coon R, Madabusi L, Nowakowski J, Monzingo A, Robertus J, Lambowitz AM. A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface. Molecular cell. 2005;17:417–428. doi: 10.1016/j.molcel.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 107.Pearn JH, Hudgson P, Walton JN. A clinical and genetic study of spinal muscular atrophy of adult onset: the autosomal recessive form as a discrete disease entity. Brain. 1978;101:591–606. doi: 10.1093/brain/101.4.591. [DOI] [PubMed] [Google Scholar]
- 108.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science (New York, NY. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 109.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pitt M, Houlden H, Jacobs J, Mok Q, Harding B, Reilly M, Surtees R. Severe infantile neuropathy with diaphragmatic weakness and its relationship to SMARD1. Brain. 2003;126:2682–2692. doi: 10.1093/brain/awg278. [DOI] [PubMed] [Google Scholar]
- 111.Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nature genetics. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 112.Rasmussen TP, Culbertson MR. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Molecular and cellular biology. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 114.Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. The Journal of cell biology. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Russman BS. Spinal muscular atrophy: clinical classification and disease heterogeneity. Journal of child neurology. 2007;22:946–951. doi: 10.1177/0883073807305673. [DOI] [PubMed] [Google Scholar]
- 116.Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJ, Sreedharan J, Vance C, Sorenson E, Lippa C, Bigio EH, Geschwind DH, Knopman DS, Mitsumoto H, Petersen RC, Cashman NR, Hutton M, Shaw CE, Boylan KB, Boeve B, Graff-Radford NR, Wszolek ZK, Caselli RJ, Dickson DW, Mackenzie IR, Petrucelli L, Rademakers R. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS genetics. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saldanha RJ, Patel SS, Surendran R, Lee JC, Lambowitz AM. Involvement of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing. A new method for purifying the protein and characterization of physical and enzymatic properties pertinent to splicing. Biochemistry. 1995;34:1275–1287. doi: 10.1021/bi00004a022. [DOI] [PubMed] [Google Scholar]
- 118.Sambuughin N, Sivakumar K, Selenge B, Lee HS, Friedlich D, Baasanjav D, Dalakas MC, Goldfarb LG. Autosomal dominant distal spinal muscular atrophy type V (dSMA-V) and Charcot-Marie-Tooth disease type 2D (CMT2D) segregate within a single large kindred and map to a refined region on chromosome 7p15. Journal of the neurological sciences. 1998;161:23–28. doi: 10.1016/s0022-510x(98)00264-0. [DOI] [PubMed] [Google Scholar]
- 119.Scheper GC, van der Klok T, van Andel RJ, van Berkel CG, Sissler M, Smet J, Muravina TI, Serkov SV, Uziel G, Bugiani M, Schiffmann R, Krageloh-Mann I, Smeitink JA, Florentz C, Van Coster R, Pronk JC, van der Knaap MS. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nature genetics. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 120.Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. Faseb J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 121.Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, Maher LR, Scrudato S, Rivera YM, Gupta S, Turrin DK, De La Cruz MP, Pestka S, Brewer G. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Molecular and cellular biology. 2008;28:5223–5237. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, Sambuughin N, Christodoulou K, Beggs JL, Zamba-Papanicolaou E, Ionasescu V, Dalakas MC, Green ED, Fischbeck KH, Goldfarb LG. Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain. 2005;128:2304–2314. doi: 10.1093/brain/awh590. [DOI] [PubMed] [Google Scholar]
- 123.Soler-Botija C, Cusco I, Caselles L, Lopez E, Baiget M, Tizzano EF. Implication of fetal SMN2 expression in type I SMA pathogenesis: protection or pathological gain of function? Journal of neuropathology and experimental neurology. 2005;64:215–223. doi: 10.1093/jnen/64.3.215. [DOI] [PubMed] [Google Scholar]
- 124.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science (New York, NY. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 126.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3'-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 127.Stewart M. Ratcheting mRNA out of the nucleus. Molecular cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 128.Sun Y, MacRae TH. The small heat shock proteins and their role in human disease. The FEBS journal. 2005;272:2613–2627. doi: 10.1111/j.1742-4658.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- 129.Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takuma H, Kwak S, Yoshizawa T, Kanazawa I. Reduction of GluR2 RNA editing, a molecular change that increases calcium influx through AMPA receptors, selective in the spinal ventral gray of patients with amyotrophic lateral sclerosis. Annals of neurology. 1999;46:806–815. doi: 10.1002/1531-8249(199912)46:6<806::aid-ana2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 131.Talbot K. Spinal muscular atrophy. J Inherit Metab Dis. 1999;22:545–554. doi: 10.1023/a:1005516625866. [DOI] [PubMed] [Google Scholar]
- 132.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tang B, Liu X, Zhao G, Luo W, Xia K, Pan Q, Cai F, Hu Z, Zhang C, Chen B, Zhang F, Shen L, Zhang R, Jiang H. Mutation analysis of the small heat shock protein 27 gene in chinese patients with Charcot-Marie-Tooth disease. Archives of neurology. 2005;62:1201–1207. doi: 10.1001/archneur.62.8.1201. [DOI] [PubMed] [Google Scholar]
- 134.Tang BS, Zhao GH, Luo W, Xia K, Cai F, Pan Q, Zhang RX, Zhang FF, Liu XM, Chen B, Zhang C, Shen L, Jiang H, Long ZG, Dai HP. Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Human genetics. 2005;116:222–224. doi: 10.1007/s00439-004-1218-3. [DOI] [PubMed] [Google Scholar]
- 135.Timmerman V, De Jonghe P, Spoelders P, Simokovic S, Lofgren A, Nelis E, Vance J, Martin JJ, Van Broeckhoven C. Linkage and mutation analysis of Charcot-Marie-Tooth neuropathy type 2 families with chromosomes 1p35-p36 and Xq13. Neurology. 1996;46:1311–1318. doi: 10.1212/wnl.46.5.1311. [DOI] [PubMed] [Google Scholar]
- 136.Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic acids research. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ursic D, Chinchilla K, Finkel JS, Culbertson MR. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic acids research. 2004;32:2441–2452. doi: 10.1093/nar/gkh561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Valdmanis PN, Rouleau GA. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–152. doi: 10.1212/01.wnl.0000296811.19811.db. [DOI] [PubMed] [Google Scholar]
- 139.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science (New York, NY. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vendra G, Hamilton RS, Davis I. Dynactin suppresses the retrograde movement of apically localized mRNA in Drosophila blastoderm embryos. RNA (New York, NY. 2007;13:1860–1867. doi: 10.1261/rna.509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vuopala K, Ignatius J, Herva R. Lethal arthrogryposis with anterior horn cell disease. Human pathology. 1995;26:12–19. doi: 10.1016/0046-8177(95)90109-4. [DOI] [PubMed] [Google Scholar]
- 142.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science (New York, NY. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 143.Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The Survival of Motor Neurons Protein Determines the Capacity for snRNP Assembly: Biochemical Deficiency in Spinal Muscular Atrophy. Molecular and cellular biology. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wan L, Ottinger E, Cho S, Dreyfuss G. Inactivation of the SMN complex by oxidative stress. Molecular cell. 2008;31:244–254. doi: 10.1016/j.molcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nature cell biology. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 146.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Current opinion in cell biology. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 147.Windpassinger C, Auer-Grumbach M, Irobi J, Patel H, Petek E, Horl G, Malli R, Reed JA, Dierick I, Verpoorten N, Warner TT, Proukakis C, Van den Bergh P, Verellen C, Van Maldergem L, Merlini L, De Jonghe P, Timmerman V, Crosby AH, Wagner K. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nature genetics. 2004;36:271–276. doi: 10.1038/ng1313. [DOI] [PubMed] [Google Scholar]
- 148.Winey M, Culbertson MR. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988;118:609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xie W, Nangle LA, Zhang W, Schimmel P, Yang XL. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9976–9981. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends in cell biology. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 151.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science (New York, NY. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 152.Yoshimura A, Fujii R, Watanabe Y, Okabe S, Fukui K, Takumi T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 153.Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes & development. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]