Abstract
Scaffolded DNA origami is a widely used technology for self-assembling precisely structured nanoscale objects that contain a large number of addressable features. Typical scaffolds are long, single strands of DNA (ssDNA) that are folded into distinct shapes through the action of many, short ssDNA staples that are complementary to several different domains of the scaffold. However, sources of long single stranded DNA are scarce, limiting the size and complexity of structures that can be assembled. Here we demonstrated that dsDNA scaffolds can be directly used to fabricate integrated DNA origami structures that incorporate both of the constituent ssDNA molecules. Two basic principles were employed in the design of scaffold folding paths – folding path asymmetry and periodic convergence of the two ssDNA scaffold strands. Asymmetry in the folding path minimizes unwanted complementarity between staples, and incorporating an offset between the folding paths of each ssDNA scaffold strand reduces the number of times that complementary portions of the strands are brought into close proximity with one another, both of which decrease the likelihood of dsDNA scaffold recovery. Meanwhile, the folding paths of the two ssDNA scaffold strands were designed to periodically converge to promote the assembly of a single, unified structure rather than two individual ones. Our results reveal that this basic strategy can be used to reliably assemble integrated DNA nanostructures from dsDNA scaffolds.
Keywords: DNA origami, double stranded DNA scaffold, self-assembly, DNA nanotechnology, scale up
DNA origami1 technology is a commonly used technique to assemble precise nanoscale structures with custom geometries and fully addressable surfaces. The technique involves folding long, single stranded DNA (ssDNA) scaffolds into arbitrarily designed shapes through the action of many short, synthetic DNA oligonucleotides (staples). The most commonly used scaffold is M13mp18, a single stranded, 7249 nucleotide, and circular DNA genome derived from bacteria phage M13. M13mp18 scaffolds have been used to build a wide variety of nanoscale structures ranging from simple 2-dimensional (2D)1-2 and 3-dimensional (3D)3-5 lattices to complex 3D shapes,6-9 with and without surface curvature. However, the complexity, size, and diversity of structures that can be created with M13mp18 or similar scaffolds are limited by the length of the ssDNA templates. For example, with 7249 nucleotides (nts), the maximum area and volume that an M13mp18 DNA origami structure can occupy is 78 × 78 nm2 and 24.7 × 24.7 × 24.7 nm3 in 2D and 3D, respectively. Identifying reliable methods to scale up the assembly of DNA origami structures is a key challenge.
Several groups have reported different approaches to address this problem. Woolley and co-workers used ssDNAs (756 to 4808 nts) amplified from polymerase chain reaction (PCR) as templates to fabricate several origami “letters”.10 More recently, longer single stranded PCR products (26 kilobases) have been achieved and folded into 108 nm × 238 nm rectangular structures by Fan and co-workers.11 Beyond PCR, Zhao et al. used a nested scaffold strategy in which a second pre-folded ssDNA template (from phiX 174 phage) was used to organize individual M13mp18 based origami unit tiles into larger patterns.12 Several groups also attempted to create DNA origami super-structures by linking individual origami unit tiles. Li et al. used short linking strands to connect rectangular origami tiles into zigzag arrays, twisted ribbons and even tube structures.13 Meanwhile, Seeman and coworkers achieved a 2D origami crystal based on a “cross” shape origami unit tile assisted by linking strands.14 Later, Rothemund engaged geometric compatibility and double helical base-stacking interactions to assemble higher-order DNA origami architectures from discrete origami tiles.15 The challenges with these hierarchically assembled ‘super-origami’ structures are the lack of intrinsic super-structure stability at inter-unit connection points, and absence of practical purification strategies.
Alternatively, Shih, Simmel and co-workers developed a protocol to use double stranded DNA (dsDNA) as a source of scaffold.16 This is important because the supply of natural, double stranded genomic DNA is virtually endless, and the methods for producing and manipulating long dsDNA molecules are far more developed than those for ssDNA. They used a denaturing dialysis method to separate a dsDNA molecule (derived from m13mp18) into two ssDNA scaffolds, and used two unique sets of staple strands to fold each individual ssDNA into two independent nanoscale objects.
Herein we describe the use of dsDNA scaffolds to fabricate integrated DNA origami structures that incorporate both of the constituent ssDNA molecules. Unlike the previously described report in which a dsDNA molecule is completely separated into two individual ssDNA scaffolds that can be sequestered separately by excess staples, our design requires a certain level of cooperation between the two ssDNA components to form the integrated structure. This is particularly challenging because there is an increased possibility that the complementary ssDNA molecules will recombine to form the initial dsDNA due to their spatial proximity. We placed special emphasis on developing general design strategies to address this issue and evaluating how various experimental conditions influence the assembly process, such as annealing temperature and DNA strand concentration.
Results and discussion
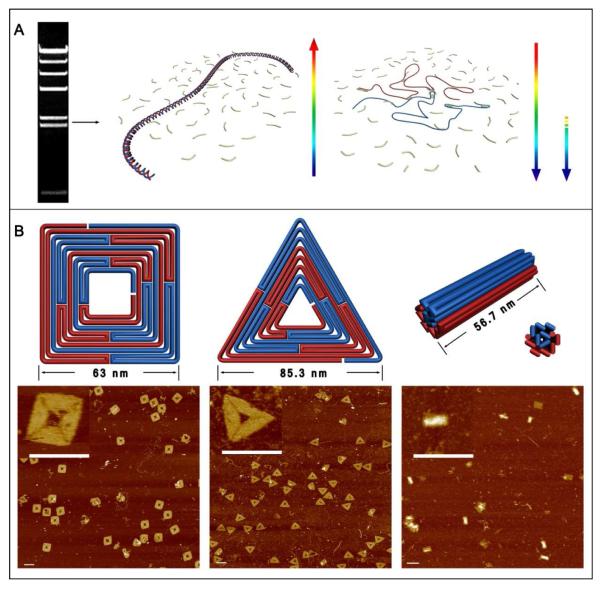
Figure 1A illustrates the overall assembly process of the square, triangle, and 24-helix bundle structures that were initially constructed. The double stranded scaffolds were first prepared by digesting λDNA into smaller fragments using the restriction enzyme Hind III. The fragments were separated by gel electrophoresis and λ5 and λ6 (2322 bps and 2027 bps, respectively) fragments were isolated. Fragment λ5 served as the scaffold for the square and bundle structures, while λ6 was used for the triangle. For our folding path designs (schematic diagrams in Fig. 1B), both of the ssDNA components of the λDNA fragments (red and blue) are required for the assembly of a complete structure. Here it is convenient to consider the two ssDNAs as a long (twice as long as the individual ssDNA components) ssDNA loop with the two strands aligned head to tail (with two nick points). A reliable folding path should promote effective hybridization of correct staples while simultaneously suppressing the recovery of the initial dsDNA template.
Figure 1.
Assembling integrated DNA origami structures from both strands of a dsDNA scaffold. A. The assembly process: A double stranded DNA template is isolated by gel electrophoresis and denatured by heating at 90°C for 15 minutes. The scaffold strands (blue and red) are subsequently folded by a collection of short oligonucleotides staples present in large excess with rapid cooling and re-annealing from a moderate temperature (See SI for details). The arrows indicate the temperature changes. B. Top row: Schematic diagrams of a square, triangle, and 24-helix bundle structure, assembled using λ5 or λ6 (2322 bps and 2027 bps, respectively) dsDNA as the scaffold. B. Bottom row: AFM images of the assembled structures. Scale bars are 100 nm. The Inset in each image panel shows a zoom in image.
The two basic principles that were applied to the folding path design are: folding path asymmetry and periodic convergence of the two ssDNA scaffold strands. Relative asymmetry between the folding paths of each ssDNA scaffold component is essential; without it, there would be partial or even full complementarity between staples. Interactions among staples should be minimized to reduce the likelihood that the ssDNA scaffold strands recombine to form the original dsDNA molecule. In addition, an offset between the folding paths of each ssDNA strand reduces the number of times that complementary portions of the strands are brought into close proximity with one another, further decreasing the likelihood of dsDNA scaffold recombination. On the other hand, periodic convergence of the folding paths of the two ssDNA scaffold strands promotes the assembly of a single, fully integrated structure rather than two individual ones. As shown in Fig. 1B, the folding paths of each ssDNA scaffold strand are partially intertwined to facilitate their cooperative assembly. The atomic force microscope (AFM) images shown in Fig. 1B confirm that our basic strategy can be used to reliably assemble integrated DNA nanostructure from the dsDNA molecules. It should be noted that the 24-helix bundle structure is an asymmetric structure without extensive strand convergence. The interface between the 12 helices depicted in red in Fig. 1B and those depicted in blue is quite broad, as 40% of the staples participate in linkages between the two domains to form a solid bundle. Despite this, a number of imperfectly assembly products are visible in the AFM images.
In addition to designing the folding path, energetic factors play an important role in the assembly process. Theoretically, a DNA origami structure (with hundreds of staple-scaffold interactions) assembled from a dsDNA scaffold will contain twice as many base pair interactions as the linear dsDNA template, making its assembly enthalpically more favorable, but entropically less favorable. The melting temperature of the linear dsDNA of this size is higher than that of the origami structure due to longer stretches of uninterrupted base-pair interactions. At temperatures higher than the melting temperature (Tm) of the target origami structure (but lower than that of the dsDNA template),17-19 entropy will dominate and the linear dsDNA template will be the prevailing structure. Conventional annealing programs that consist of a gradual, uniform cooling step will result in recombination of the ssDNA strands to form the lowest energy structure, the dsDNA scaffold. Here, we employed a non-conventional DNA origami assembly protocol to discourage the formation of the linear dsDNA. Initially, a mixture of the dsDNA scaffold and a 50 fold excess of staple strands was heated to 90°C (for 15 min) and rapidly cooled down to 25°C (in < 1 minute) to denature the dsDNA and prevent the system from equilibrating to its lowest energy state. We expect that the separated ssDNA strands are immediately surrounded and sequestered by the large excess of staple strands (rather than recombining to form the dsDNA), especially because of the faster diffusion kinetics of the smaller staple strands. Next, the mixture was heated to 45°C and slowly cooled to 4°C, providing the staples with enough thermal kinetic energy to fold the scaffold into the desired arrangement. 45°C was identified as the optimal starting temperature for this step through a series of control experiments that are shown in Fig. S3. Higher temperatures (50°C) result in dsDNA recombination and lower temperatures (40°C) yield incomplete structures. The overall goal of this annealing protocol is to kinetically trap the DNA in a metastable state (assembled origami structures), rather than allowing the system to gain enough thermal energy to reach the lowest free energy state (dsDNA scaffold and free staples), by providing just enough thermal energy and time for the system to equilibrate. As evidenced by gel electrophoresis and AFM characterization, our method successfully converted the dsDNA scaffolds into discrete DNA origami structures with about 85% yield (See additional images in Fig.S2A).
We emphasize that the absolute concentrations of the DNA components, both the dsDNA template and the ssDNA staples, are also important factors in the assembly process. Control experiments revealed that dsDNA templates with 5 nM concentrations yielded significantly more partially formed and cross-linked structures than those with 1 nM concentration (Fig. S4). We also found that the molar ratio of staples to dsDNA scaffold should be higher than 50:1 to effectively sequester each ssDNA strand that are separated from the initial dsDNA scaffold. A summary of the optimized experimental conditions is included in Supporting Information.
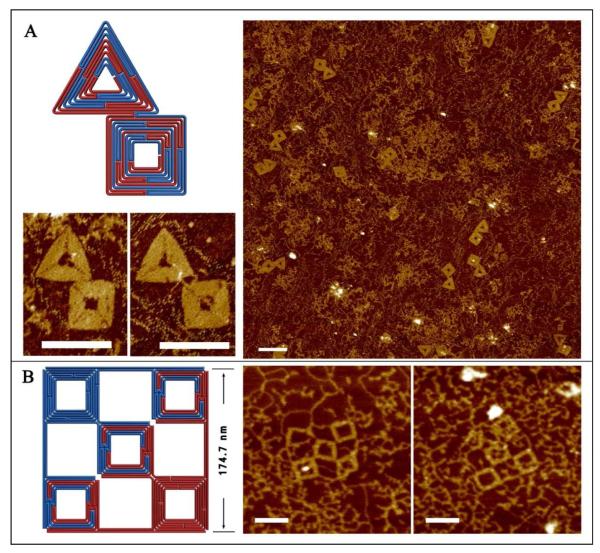
After optimizing our design and assembly conditions, we sought to translate the smaller square and triangle structures into a more complex arrangement using an internal segment of the full-length λ phage DNA genome, i.e. λ5 and λ6 together as the scaffold without excising the extra dsDNA. The design is expected to grow into an inter-connected square and triangle shaped structure with long dsDNA tails extending from each of the shapes. The staples corresponding to both shapes were mixed with full-length λDNA and subjected to the annealing program described above. As confirmed by the AFM images shown in Fig. 2A, the DNA origami structures were successfully assembled within a very large, dsDNA genome. However, several questions remained: what is the largest structure that can be assembled from a dsDNA scaffold, and what are the factors that influence the upper limit?
Figure 2.
Assembling more complex DNA origami arrangements using a long segment within full-length λDNA. Left panel – schematic diagrams. Right panel – corresponding AFM images of the assembled products. A. A combined triangle and square DNA origami structure is assembled using λ5 and λ6 fragments within full length λDNA. B. A five-unit square lattice structure is assembled using ~ 1/5 of the λDNA as scaffold. Scale bars are 100 nm.
To address these questions, we designed the 5 unit square lattice structure shown in Fig.2B. This structure encompasses approximately one fifth (9,280 bps) of the entire λDNA genome (48,502 bps). Unlike the smaller square and triangle structures, the termini of the folding paths of the ssDNA scaffold strands are completely separated into opposite corners, and 3 of the square frames are formed from both ssDNA scaffolds while 2 are formed using only a single scaffold. Although the pattern appears symmetric in Fig. 2B, a close examination of the folding paths and sequences of the ssDNA fragments reveals an asymmetric arrangement. Nevertheless, this structure formed with poor yield. Most of the observed structures formed only partially (usually 4 out of the 5 square units) suggesting that at least one of the squares is not stable. The reasons for this are likely very complicated, especially from an energetic perspective. However, a few fully assembled 175×175 nm2 lattices were observed, which prompted us to try even larger structures.
Towards this end, we designed a 500×500 nm2 square lattice, consisting of 3×3 squares using the entire double stranded λDNA genome (shown in Fig.S5). The structure was designed and assembled using the same principles and strategy as described above. Unfortunately, the fabrication of this structure was a failure, despite the multitude of experimental conditions that were adjusted. We only observe partial formation of the structure, with evidence of significant aggregation. The most likely reasons for this include: (1) the scaffold strand in the outermost layers of the structure is relatively long with reduced rotational and translational dynamics, making it more difficult for the scaffold to traverse the large area required to join distant domains; (2) the structure is too flexible at the connection points between the squares, and, selected sides are only composed of 2 helices which may not be rigid enough to support cooperative assembly. Therefore, the individual localized sections are relatively free to move independently in all three dimensions and do not cooperatively settle into the ‘correct’ positions within the overall two dimensional plane; (3) the unique sequence limitation. The shortest binding domain within each staple is 8 nts for a total of 48 (65,536) possible sequence combinations. Within the 96,988 nts of the scaffold, (quantitatively greater than 65536 unique domains), it is very likely that the staple strands will be complementary to multiple positions along the scaffold.
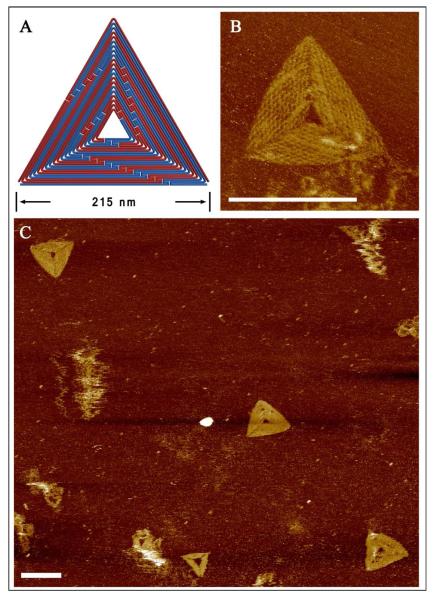
In an effort to address the challenges of scaling up DNA origami with dsDNA, we carefully designed a more rigid triangular structure (215 nm edges) that utilized 1/4 of the ds λDNA scaffold. We attempted to overcome the problem of inadequate sequence space by changing the crossover design to 26 bps between crossovers, rather than 16 bps, so that the smallest staple binding domain was 13 nts (instead of 8 nts). The annealing program was also adjusted to reflect the thermodynamic requirements of the longer staples (Details can be found in SI). Note that number of unique sequences for a 13 nt domain is 413 (~67.1 million), which is far greater than the length of the λDNA. A triangular design also ensures the final origami structure is relatively rigid, improving the cooperative assembly. The AFM images shown in Figure 3B reveal that the assembled structure has periodic cavities between neighboring helices, which are expected to be 16.8 nm (52 bps) in length. The pattern observed in the AFM images matches the designed folding path of the scaffold (Fig. 3A). Due to the extra scaffold crossovers between helices, the cavity size along these lines is expected to be smaller. The zoom out images shown in Figures 3C and S7 further confirms the successful assembly of the designed structure. The molecular weight and dimension of the 1/4 λDNA based triangle is 3.4 and 2.8 times larger, respectively, than the triangle (128 nm edges) designed by Rothemund in 2006 that utilized an M13mp18 scaffold.1 Considering the variety of natural dsDNA sources, this result underscores the potential of fabricating larger and more complex structures using this strategy.
Figure 3.
Assembling larger, more rigid triangle structures from a dsDNA scaffold. A. The schematic diagram illustrates the designed folding path using 1/4 of the full length λDNA genome. B. The corresponding AFM image reveals assembled products that exhibit the expected surface pattern. C. Zoom out images of the large triangle origami structures. Scale bars are 200 nm.
Conclusion
We demonstrated that dsDNA scaffolds can be used to fabricate integrated DNA origami structures that incorporate both of the constituent ssDNA molecules. We placed special emphasis on developing general design strategies to prevent the unwanted recombination of the long dsDNA scaffold and evaluating how various experimental conditions influence the assembly process. We applied two basic principles toward the design of scaffold folding paths – folding path asymmetry and periodic convergence of the two ssDNA scaffold strands. We assert that asymmetry in the folding path minimizes the unwanted complementarity between staples, and that incorporating an offset between the folding paths of each ssDNA scaffold strand reduces the chances that complementary portions of the strands are brought into close proximity with one another, both of which decrease the likelihood of dsDNA scaffold recovery. Meanwhile, in order to promote the assembly of a single, fully integrated structure rather than two individual ones, we permitted the folding paths of the two ssDNA scaffold strands to periodically converge. Our results reveal that this basic strategy can be used to reliably assemble integrated DNA nanostructures from dsDNA templates. However, our study also calls attention to the limitations of dsDNA as a source of scaffolds for DNA origami structure assembly. Despite the systematic manipulation of various experimental conditions, including annealing program, concentration of components, etc., we found that large DNA origami structures with flexible connection points could not be reliably assembled using this method. Consequently, there is a significant bottleneck in using very long dsDNA templates to scale up DNA origami assembly. In order for this technique to be successful, various energetic and structural barriers must still be addressed.
Methods
λDNA fragments purification
Fractions λ5 and λ6 from lambda DNA-Hind III digestion were purified from 1.0% agarose gel (1×TBE, 0.5 μg/mL ethidium bromide) at 180 V for 40 min. The bands were cut and recovered by using a Gel Extraction Kit purchased from Fermentas Molecular Biology (http://www.fermentas.com).
Agarose Gel electrophoresis
The folding products from λDNA fragments were subject to electrophoresis on 0.8% agarose gel (1×TAE-Mg2+, 0.5 μg/mL ethidium bromide) at 75-80 V for two to three hours in ice bath and visualized under UV light.
AFM imaging
In liquid scan
the sample (2 μL) was deposited onto a freshly cleaved mica surface (Ted Pella, Inc.) and left to adsorb for 2 min. 50 μL buffer (1 × TAE-Mg2+, plus 2 μL 100mM NiCl2) was added onto the mica, and the sample was scanned in the Scanasyst in Fluid mode using scanasyst in fluid+ tips (Veeco,Inc.), on a Veeco 8 AFM with the assistant of its fluid cell.
In air scan
the sample (2 μL) was deposited onto a freshly cleaved mica surface (Ted Pella, Inc.) and left to adsorb for 2 min. 50 μL buffer was added onto the mica and was blown away immediately by condensed air. Then a second wash with 50 μL pure H2O was applied in the same way. The dry sample was then scanned in Scanasyst in Air mode using scanasyst in air tips (Veeco,Inc.), on a Veeco 8 AFM.
Annealing Program
For small structures
90°C (15 min); 25°C (2 min); 45°C to 4°C (20 min/°C)
For big triangles
90°C (15 min); 25°C (2 min); 60°C (1 min) - 59°C (1 min) - 58°C (2 min) - 57°C (3 min) - 56°C (5 min) - 55°C (10 min) - 54°C (15 min) - 53°C (20 min) - 52°C (30 min) - 51°C (40 min) - 50°C (50 min); 49°C to 4°C (20 min/°C)
Supplementary Material
Acknowledgement
This research was partly supported by grants from the National Science Foundation, Office of Naval Research, Army Research Office, Department of Energy, and National Institutes of Health to H.Y. and Y. L. H.Y. was also supported by the Presidential Strategic Initiative Fund from Arizona State University.
Footnotes
Supporting Information Available: Supporting material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rothemund PW. Folding DNA to Create Nanoscale Shapes and Patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.Andersen ES, Dong M, Nielsen MM, Jahn K, Lind-Thomsen A, Mamdouh W, Gothelf KV, Besenbacher F, Kjems J. DNA Origami Design of Dolphin-Shaped Structures with Flexible Tails. ACS Nano. 2008;2:1213–1218. doi: 10.1021/nn800215j. [DOI] [PubMed] [Google Scholar]
- 3.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Self-assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietz H, Douglas SM, Shih WM. Folding DNA into Twisted and Curved Nanoscale Shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H. Multilayer DNA Origami Packed on a Square Lattice. J. Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 7.Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Scaffolded DNA Origami of a DNA Tetrahedron Molecular Container. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 8.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA Origami with Complex Curvatures in Three-Dimensional Space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 9.Han D, Pal S, Liu Y, Yan H. Folding and Cutting DNA into Reconfigurable Topological Nanostructures. Nat. Nanotechnol. 2010;5:712–717. doi: 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pound E, Ashton JR, Becerril HCA, Woolley AT. Polymerase Chain Reaction Based Scaffold Preparation for the Production of Thin, Branched DNA Origami Nanostructures of Arbitrary Sizes. Nano Lett. 2009;9:4302–4305. doi: 10.1021/nl902535q. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Chao J, Pan D, Liu H, Huang Q, Fan C. Folding Super-Sized DNA Origami with Scaffold Strands from Long-Range PCR. Chem. Commun. 2012;48:6405–6407. doi: 10.1039/c2cc32204h. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Z, Liu Y, Yan H. Organizing DNA Origami Tiles into Larger Structures Using Preformed Scaffold Frames. Nano Lett. 2011;11:2997–3002. doi: 10.1021/nl201603a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Liu M, Wang L, Nangreave J, Yan H, Liu Y. Molecular Behavior of DNA Origami in Higher-Order Self-Assembly. J. Am. Chem. Soc. 2010;132:13545–13552. doi: 10.1021/ja106292x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Zhong H, Wang R, Seeman NC. Crystalline Two-Dimensional DNA-Origami Arrays. Angew. Chem. Int. Edit. 2011;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo S, Rothemund PW. Programmable Molecular Recognition Based on the Geometry of DNA Nanostructures. Nat. Chem. 2011;3:620–626. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 16.Högberg BR, Liedl T, Shih WM. Folding DNA Origami from a Double-Stranded Source of Scaffold. J. Am. Chem. Soc. 2009;131:9154–9155. doi: 10.1021/ja902569x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro CE, Kilchherr F, Kim DN, Shiao EL, Wauer T, Wortmann P, Bathe M, Dietz H. A Primer to Scaffolded DNA Origami. Nat. Methods. 2011;8:221–229. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 18.Wartell RM, Benight AS. Thermal Denaturation of DNA Molecules: A Comparison of Theory with Experiment. Phys. Rep. 1985;126:67–107. [Google Scholar]
- 19.Howley PM, Israel MF, Law M-F, Martin MA. A Rapid Method for Detecting and Mapping Homology between Heterologous DNAs. J. Biol. Chem. 254(11):4876–4883. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.