Conspectus
Living systems have evolved a variety of nanostructures to control the molecular interactions that mediate many functions including the recognition of targets by receptors, the binding of enzymes to substrates, and the regulation of enzymatic activity. Mimicking these structures outside of the cell requires methods that offer nanoscale control over the organization of individual network components. Advances in DNA nanotechnology have enabled the design and fabrication of sophisticated one-, two- and three-dimensional (1D, 2D and 3D) nanostructures that utilize spontaneous and sequence specific DNA hybridization. Compared to other self-assembling biopolymers, DNA nanostructures offer predictable and programmable interactions, and surface features to which other nanoparticles and bio-molecules can be precisely positioned.
The ability to control the spatial arrangement of the components while constructing highly-organized networks will lead to various applications of these systems. For example, DNA nanoarrays with surface displays of molecular probes can sense noncovalent hybridization interactions with DNA, RNA, and proteins and covalent chemical reactions. DNA nanostructures can also align external molecules into well-defined arrays, which may improve the resolution of many structural determination methods, such as X-ray diffraction, cryo-EM, NMR, and super-resolution fluorescence. Moreover, by constraining target entities to specific conformations, self-assembled DNA nanostructures can serve as molecular rulers to evaluate conformation-dependent activities.
This Account describes the most recent advances in the DNA nanostructure directed assembly of biomolecular networks and explores the possibility of applying this technology to other fields of study. Recently, several reports have demonstrated the DNA nanostructure directed assembly of spatially-interactive biomolecular networks. For example, researchers have constructed synthetic multi-enzyme cascades by organizing the position of the components using DNA nanoscaffolds in vitro, or by utilizing RNA matrices in vivo. These structures display enhanced efficiency compared to the corresponding unstructured enzyme mixtures. Such systems are designed to mimic cellular function, where substrate diffusion between enzymes is facilitated and reactions are catalyzed with high efficiency and specificity. In addition, researchers have assembled multiple choromophores into arrays using a DNA nanoscaffold that optimizes the relative distance between the dyes and their spatial organization. The resulting artificial light harvesting system exhibits efficient cascading energy transfers. Finally, DNA nanostructures have been used as assembly templates to construct nanodevices that execute rationally-designed behaviors, including cargo loading, transportation and route control.
Introduction
Biological systems use complex macromolecular nanostructure networks to mediate a range of cellular functions, such as biomolecular synthesis, signal transduction, and gene expression and regulation, all with high efficiency and specificity. Many of these macromolecular systems have evolved through the spontaneous self-assembly of components into highly organized spatial structures, where the position and orientation of molecules are precisely controlled to facilitate functionality. For example, the multi-enzyme cascades1 found in biochemical synthesis pathways and the light harvesting system in photosynthetic reaction centers2 both rely on very specific arrangements of components.
Over the past few decades, molecular self-assembly processes have been exploited to construct various nanostructures including vesicles, nanofibers and nanotubes from self-assembling lipids, peptides, nucleic acids and polysaccharides.3 However, it remains a challenge to accurately arrange multiple heterogeneous components into geometric patterns with nanometer precision, as in natural systems. Additional challenges include the development of novel assembly algorithms to increase structural complexity and improve the fidelity and yield of the assembly process.
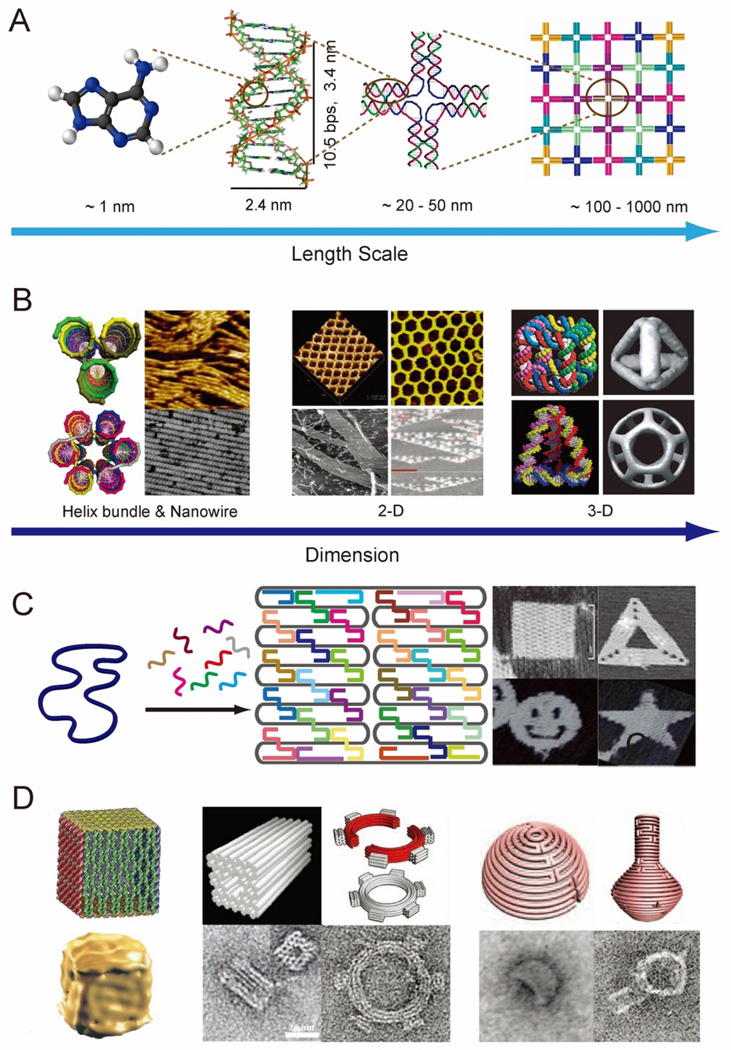
DNA is among the most promising biomolecules for the construction of complex, biomolecular networks.4 DNA is a self-assembling biopolymer that is directed by canonical Watson-Crick base pairing to form predictable, double helical secondary structures, which are stabilized by hydrogen-bonding, π-π stacking, and hydrophobic interactions. B-form DNA double helices have well-defined structural characteristics, including a helical repeat of ~ 3.4 nm, helical diameter of ~2.0 nm and ~ 34.3° twist angle between base-pairs in solution (Figure 1A).5 The use of double helical DNA molecules for nanoscale engineering pursuits began with Seeman’s construction of artificial branched DNA tiles, where four rationally designed oligomeric nucleic acid strands self-assembled into an immobile four-way junction.6 Double-crossover (DX) DNA tiles,7 with increased structural rigidity compared to four-way junction tiles, were developed later and were suitable for assembling more complex periodic nanostructures through sticky ends interactions.8 Tile-based DNA assembly has been demonstrated through the construction of a number of unique nanostructures, ranging from multi-helix bundles, nanotubes9–10 and 2D lattice arrays,11–12 to 3D geometric shapes including a cube,13 tetrahedron,14 and a buckyball (Figure 1B).15
Figure 1.
Introduction to structural DNA nanotechnology. (A) Self-assembly of nanostructures based on complementary DNA base pairing. (B) DNA helix bundles (left),9–10 2-D arrays (middle),11–12 and 3-D objects (right).13–15 (C) DNA origami for constructing 2-D nanostructures18 and (D) 3-D architectures - hollow box (left),19 multi-layer monolith and a square-toothed gear(middle),5, 22 and semi-sphere and a nanoflask (right).23 Figures are reproduced with permissions from ACS, AAAS and NPG.
An important milestone in structural DNA nanotechnology was the creation of aperiodic patterns using a scaffolding strategy. Early reports include the organization of DX tiles into 2D lattice barcode-patterns, directed by a long ligated DNA strand,16 and the assembly of a 3D octahedron, directed by a 1.7 kb DNA strand.17 In 2006, Paul Rothemund made a breakthrough in scaffold directed DNA nanostructure assembly; in the method he developed, referred to as DNA origami, a long single-stranded DNA scaffold (e.g. 7429-nt M13 phage genome DNA) is folded into arbitrary 2D shapes by following pre-determined folding paths that are specified by a collection of short oligonucleotide ‘staple’ strands (Figure 1C).18 Many 2D nanostructures including a square, rectangle, smiley face, triangle and star have been demonstrated using the DNA origami method. One of the most attractive properties of DNA origami structures is the addressability of the surface, the result of the unique sequence at each oligonucleotide staple position. Thus, various patterns can be displayed by selectively modifying staple strands at desired locations with single stranded probe extensions. The DNA-origami method has several advantages over “tile-based” assembly approaches: (1) scaffolded DNA can be folded into nearly any symmetric or asymmetric structure; (2) well-formed nanostructures are generated with high yield using unpurified oligonucleotides, as the scaffold imposes the correct stoichiometry between strands; (3) spatially-addressable assembly is achieved with a resolution of ~ 6 nm. The DNA-origami approach was further developed for the construction of 3D nanostructures. The Gothelf group assembled a hollow DNA box by joining six distinct (though connected by the scaffold) origami sheets through the action of staple strands bridging the edges (Figure 1 D left).19 The Shih group introduced a method to construct solid 3D shapes by packing scaffolded DNA double helices into pleated layers, constrained to a honeycomb or square lattice.5,20–21 Twisted and curved 3D objects were further developed through insertion or deletion of base pairs at selected positions within the helical layers (Figure 1D middle).22 The Yan group recently developed a strategy to construct DNA nanostructures with complex curvatures by nesting a collection of concentric DNA rings of decreasing circumference to generate the rounded contours of various 3D objects (Figure 1D right).23 In addition to these reports, several computational tools including caDNAno24 and CanDo25 have been developed to facilitate the design of DNA nanostructures, making structural DNA nanotechnology more accessible to researchers from other fields.
In addition to DNA, RNA nanotechnology has recently emerged as an attractive method to construct nanostructures with functional diversity.26 In order to form the variety of loops and structural motifs that are required for functionality, RNA nanostructures rely on the self-complementarity of single strands. One of the attractive features of RNA-based nanostructures is the potential for in vivo assembly, as single-stranded RNA molecules are readily transcribed in cells. DNA/RNA hybrid nanostructures are likely to have a synergistic potential that combines the predictability of DNA assembly with the functional diversity of RNA.27
DNA nanostructures are reliable directors in the organization of heterogeneous nanoscale-entities such as peptides,28 proteins29 and nanoparticles.30 Super-molecular networks of molecules that are scaffolded by DNA nanostructures exhibit well-controlled inter-component distances and relative numbers. This characteristic presents exciting opportunities for fundamental studies of distance-dependent molecular interactions, and for practical applications including biosensing, molecular biophysics, biocatalysis, drug delivery, and responsive nanodevices. Herein, we describe the progress that has been made in DNA directed assembly of biomolecular networks.
DNA-Directed Self-assembly
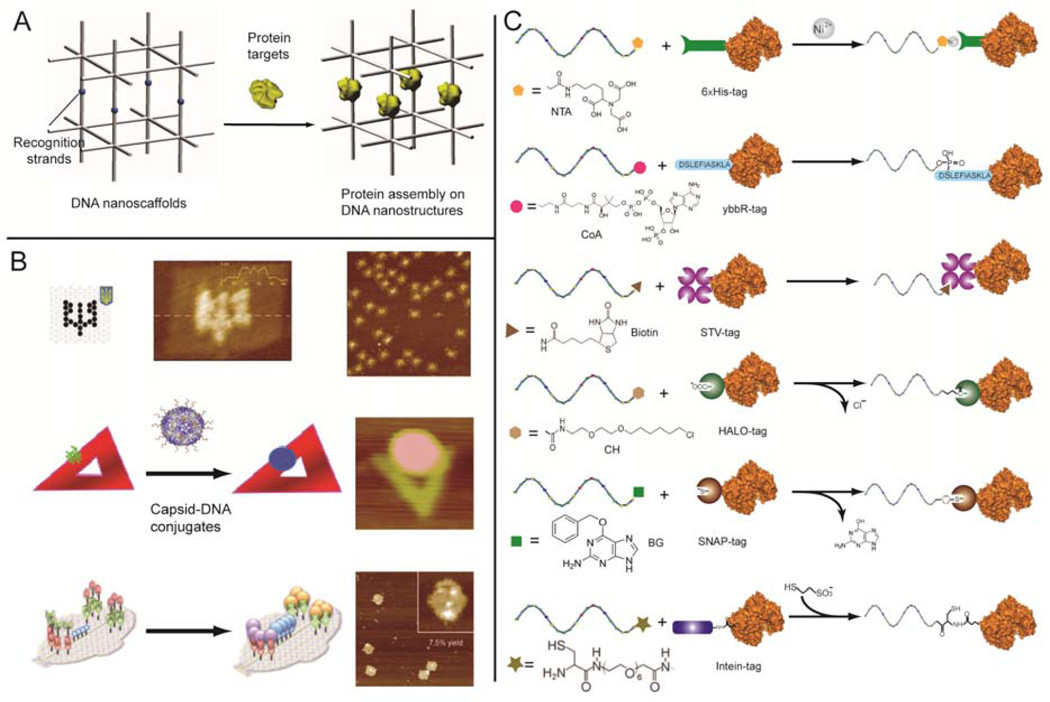
In Seeman’s original proposal, he suggested that a DNA nano-lattice could be used as a framework to organize proteins into 3D crystals, where the position and orientation of each protein could be controlled by elements of the DNA nanostructure (Figure 2A).6 Since that time, the sequence specificity of DNA hybridization has been exploited to assemble external bio-molecules at specific positions on addressable DNA nanostructures. Hybridization between the DNA functionalized bio-molecules and single stranded probe extensions of the DNA nanostructures generate networks of molecules with controlled inter-molecular distances and ratios. This approach was demonstrated by organizing smaller biomolecules, including aptamers31 and peptide,28 as well as larger macromolecules, including proteins29 and virus capsids32 on DNA nanostructures (Figure 2B).
Figure 2.
DNA-directed assembly. (A) Seeman’s proposal to organize macromolecules within a DNA nanoscaffold. (B) Patterning macromolecules on DNA origami: streptavidin (top),29 virus capsid (middle)32 and orthogonal protein decoration (bottom).36 Reproduce from ref (C) Site-specific protein-oligo conjugation using His-tag, ybbR-tag, STV-tag, HALO-tag, SNAP-tag and Intein-tag (from top to bottom). Figures are reproduced with permissions from IOP Publishing, ACS, and Wiley.
Critical to DNA-directed assembly efforts are the development of efficient oligonucleotide-biomolecule coupling methods. One of the attractive features of DNA scaffolds is that the constituent oligonucleotides can be modified with a variety of different functional groups for subsequent cross-linking reactions with other bio-molecules,33 amino and thiol modifications are among the most common. Despite their versatility, one of the drawbacks of conventional cross-linking methods is a lack of control over the conjugation site and stoichiometry of coupling. The presence of multiple lysine and cysteine residues on the surface of most proteins makes it difficult to generate a site-specific protein conjugation, which is required for certain applications.34 Genetic modification of proteins with reactive tags (His-tag and ybbR-tags for example), or the use of fusion domains (such as streptavidin, intein, SNAP and HALO), are alternative approaches to achieve site-specific protein-oligo conjugation with very high efficiency (Figure 2C).34–35 In addition to covalent coupling approaches, noncovalent binding between proteins and specific ligands can also be used for assembling protein nanoarrays.12,28,31 Orthogonal display of several proteins on DNA origami was demonstrated by employing three site-specific coupling strategies: SNAP-tag, HALO-tag, and biotin-streptavidin interactions.36 The use of strong domain interactions (Zinc-finger for example) is another approach for site-specific protein-oligo coupling and can be achieved by tethering one binding domain to a protein, and the other binding domain to an oligonucleotide.37–38 The versatility of DNA nanostructures has also been used to improve the binding affinity between molecules. The assembly of a multi-valent ligand complex was achieved by combining several individual ligands on a DNA nanostructure, where the distance between ligands was carefully controlled. The resulting binding affinity (Kd) between binding partners was in the low nanomolar range.39 It should be possible to achieve more precise control over the orientation of biomolecules by combining site-specific conjugation strategies with 3D DNA nanostructures that have specifically tailored cavities or cages to constrain the guest molecule through steric interactions.
Label-free detection of bimolecular interactions
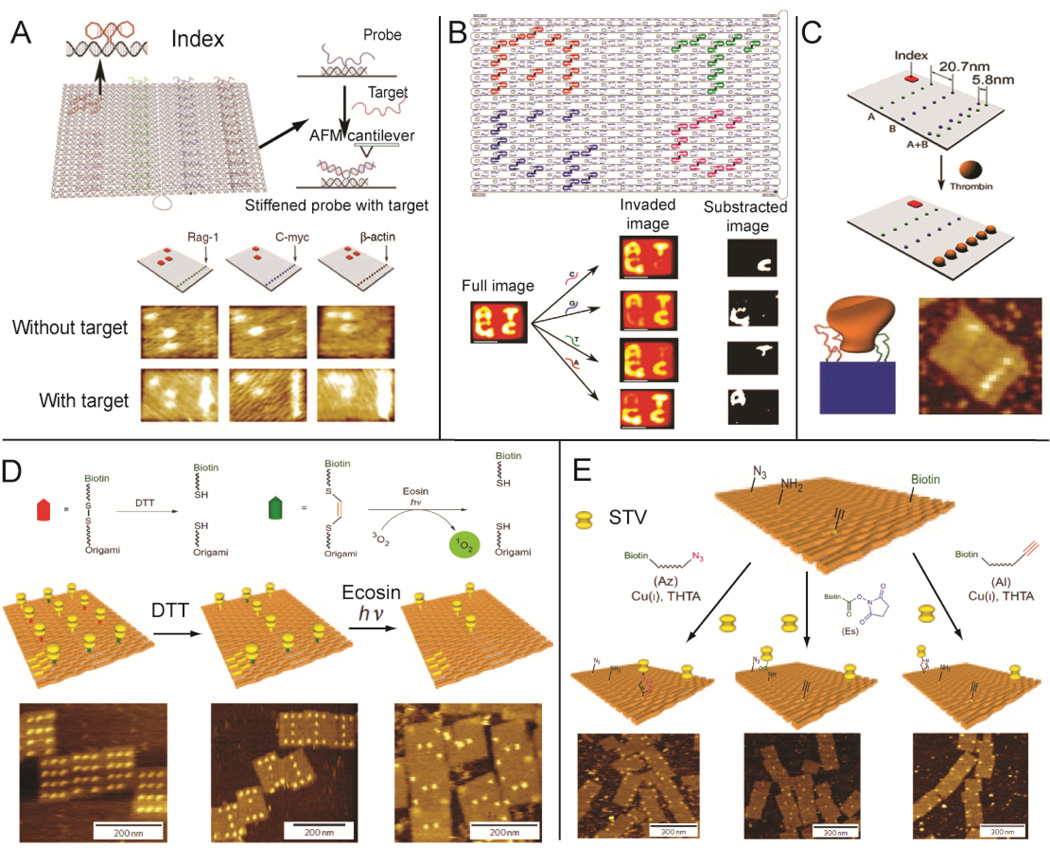
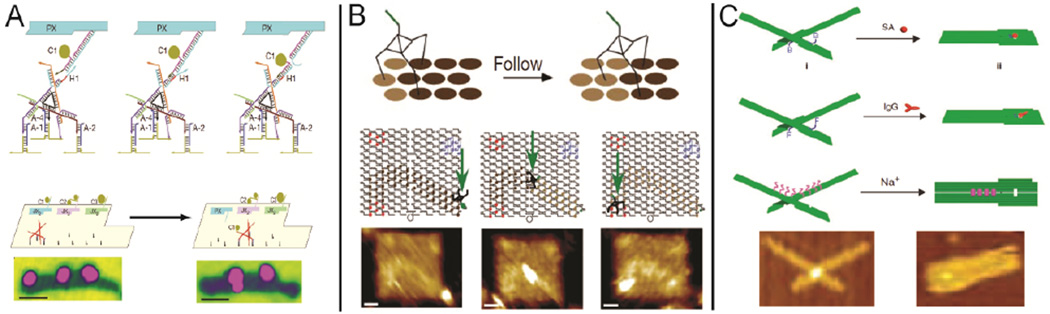
Label-free detection is becoming more and more attractive for biological assays, where bimolecular interactions (e.g. mass, dielectric and morphology) are characterized without the need for sample modification. DNA nanostructures have several features that make them promising agents of label-free detection. As water-soluble nanoscale ‘chips’, DNA nanostructures are capable of displaying multiple probes from their surface for the detection of various bimolecular interactions. For example, the interaction between probe extensions and target molecules can result in a measurable change in surface morphology (height), which can be distinguished by atomic force microscopy (AFM). This detection strategy was used to demonstrate label-free RNA hybridization on a DNA origami chip.40 In Figure 3A, multiple nucleic acid probes were designed to target specific RNA sequences and were precisely patterned on an underlying DNA origami scaffold. Before RNA hybridization, the single-stranded DNA probes were quite flexible and not clearly visible by AFM. Detection of specific RNA targets by the DNA probes resulted in the formation of double helical DNA-RNA V-shaped junctions with characteristic features that were obviously identified by AFM analysis. In addition, DNA origamis were decorated with “barcode loops” so that several nucleic acid detection tiles could be differentiated, allowing for simultaneous, multiple-target analysis. Beyond external molecule detection, DNA origamis have been used to demonstrate the detection of genomic, single nucleotide polymorphisms (SNPs) by direct AFM readout (Figure 3B).41 Letters corresponding to each of the four DNA nucleotides (A, T, C and G) were patterned on a DNA origami tile. Each letter was composed of a collection of single stranded DNA that was bound to probes that were extended from the DNA origami surface, creating an obviously topography that was easily detected by AFM. The single stranded DNA in each letter contained distinct binding sites that were complementary to the particular nucleotide they represented. In the presence of a DNA sequence containing the target nucleotide variation, the strands that represented the corresponding character were displaced, resulting in the disappearance of the underlying letter pattern.
Figure 3.
DNA nanostructures as a template for label-free detection of bimolecular interactions: (A) RNA hybridization assay;40 (B) SNP detection;41 (C) Spatially-dependent multivalent ligand-protein binding;42 (D) Chemical bond formation and (E) bond cleavage.46 Figures are reproduced with permissions from AAAS, ACS, NPG and Wiley.
Addressable DNA nanostructures, with the ability to organize a variety of ligands into specific spatial patterns, provide an opportunity to study the factors that govern protein-ligand binding. The distance dependent binding of a multivalent aptamer-protein complex was characterized using a DNA origami platform. Two aptamers were placed at several distances from a target protein to determine the spacing that resulted in the strongest binding (Figure 3C).42 By organizing bi-specific linkers, multi-functionalized DNA nanostructures can bring different cells into close proximity and induce specific cell-cell interactions for therapeutic applications.43 Distance-dependent ligand binding can also be used to probe the internal arrangement of protein domains. The ideal spatial arrangement of two tandem SH2 domains of Syk kinase was determined by organizing the domains with a double-stranded DNA nanoscaffold. The nanoscaffold displayed the two domain-binding ligands at various distances and flexibilities.44
DNA origami has been used to visualize chemical reactions on the single molecule level. In Figures 3D&E, origami structures act as addressable supports to monitor chemical formation and cleavage reactions with readout of chemical reactions achieved via biotin-streptavidin complexes. When biotin linkers were cleaved by disulfide bond-reduction or photo-generated singlet oxygen, the biotin-streptavidin conjugates were released from the origami surface. Using a similar approach, bond formation was also detected; incoming functional groups were linked to biotin and the incorporation of each group was visualized by the addition of streptavidin. Three functional groups commonly used for bioconjugation reactions were studied: alkyne, amine, and azide.45–46
Conformational biophysics
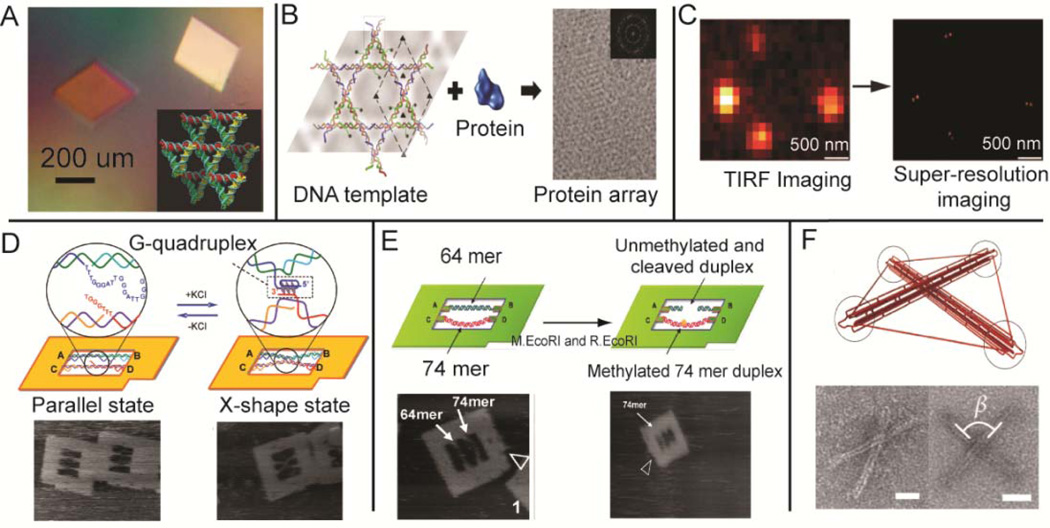
Nearly thirty years after the initial proposal of creating self-assembling 3D crystals using DNA junction structures,6 Seeman and co-workers demonstrated a 4 Å-resolution crystal structure of a self-assembled DNA tensegrity triangle (Figure 4A).47 In addition to X-ray diffraction experiments, DNA nanostructures have also been used to align and localize macromolecules in particular nano-environments for other structural determination methods. Self-assembled DNA nano-affinity templates were used to facilitate data collection in single-particle electron cryomicroscopy (cryo-EM) by creating dense and non-overlapping arrays of protein molecules (Figure 4B).48 Detergent-resistant, DNA-nanotube liquid crystals were employed to introduce weak alignment of membrane proteins for their structural determination by NMR.49 This method was recently used for NMR structural determination of mitochondrial uncoupling protein 2 (UCP2).50 Reconstruction of super-resolution fluorescence images were reported, where DNA origami served as a molecular ruler to locally organize several fluorophores for imaging calibration (Figure 4C).51
Figure 4.
DNA nanostructures as biophysical study tools: (A) X-ray diffraction,47 (B) cyro-EM48 and (C) super-resolution imaging.51 Conformational studies of (D) G-quadruplex formation,52 (E) DNA methylation53 and (F) constrained intermolecular forces.55 Figures are reproduced with permissions from NPG, ACS and Wiley.
Beyond structural determination methods, DNA nanostructures can be used to study conformation dependent biological activities by constraining macromolecules to specific environments with controlled arrangements and molecular forces. Real-time observations of a G-quadruplex, a structure that is associated with the telomeric region of chromosomes, were facilitated by stretching two corresponding G-strands across the inner cavity of a DNA origami frame structure (Figure 4D).52 The formation and disruption of the G-quadruplex structure was visulized by fast-scaning AFM through the addition or removal of K+ ion. In Figure 4E, conformation-dependent DNA methylation was also studied by using a DNA origami frame structure to control the tension of two double helical substrates ( 64-nts and the 74-nts ).53 AFM images of enzyme-substrate binding and cleavage revealed that enzyme-catalyzed methylation occurs more frequently for the structurally relaxed 74-nt substrate. Other DNA nanostructures have also been designed to measure the biophysical properties of molecules. DNA-based nanomechanical scissors were used to measure the force experienced when MutS binds to unpaired and bulged bases. In this system, the MutS binding force was determined by analyzing the interruption of sticky-end hybridization.54 Toward exploiting intermolecular forces such as tensional integrity to perform work, pre-stressed 3D tensegrity DNA nanostructures in which rigid bundles of double helices resist compressive forces were assembled. The forces generated by the prestressing mechanism may be be used to bend DNA bundles or actuate enzymatic cleavage at specific sites (Figure 4F).55
Organization of multi-enzyme reaction pathways
The metabolism of living systems involves complex synthetic pathways with numerous multistep reactions that possess extraordinary yields and specificities. Many of the enzyme systems carrying out these reaction pathways are highly organized complexes with precisely controlled enzyme positions and orientations, facilitating efficient diffusion of substrates between the enzymes.1 Artificial synthesis of these multi-enzyme systems is generally achieved by genetic fusion,56 chemical crosslinking and co-immobilization,57 however, precise control over spatial organization of components is lacking for these methods.
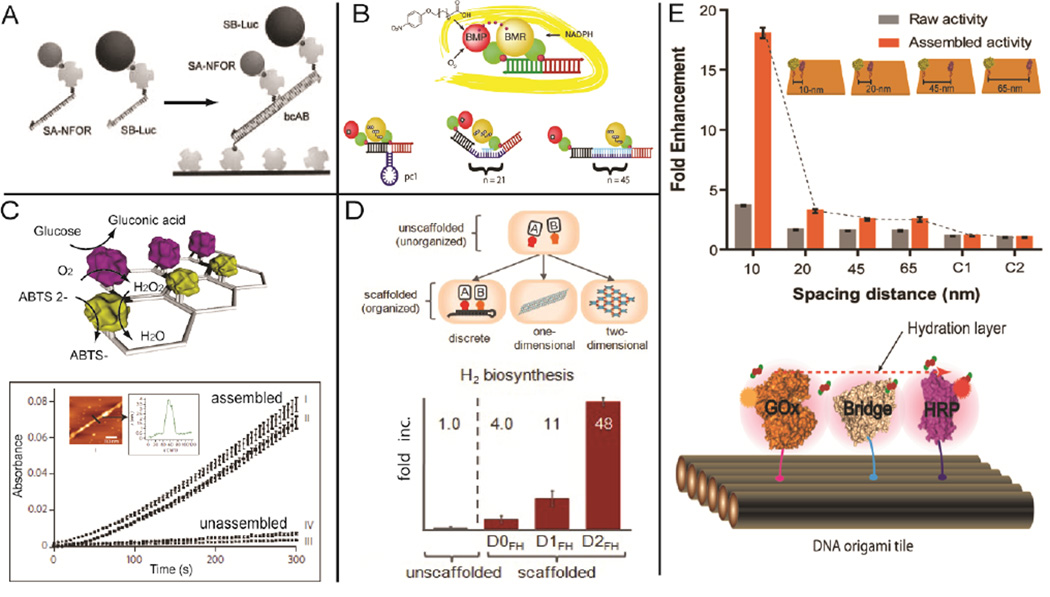
Utilizing DNA nanostructures as assembly scaffolds, it has become feasible to organize multiple enzymes with controlled spacing in linear as well as 2D or 3D geometric patterns, which enables the study of cascade activity.58 One of the first demonstrations was the assembly of a bio-enzymatic NAD(P)H:FMN oxidoreductase and luciferase cascade on a double-stranded DNA scaffold with an observed ~ 3-fold increase in activity compared to the corresponding unassembled enzyme pair (Figure 5A).59 This strategy was later applied to probing the distance-dependent activity of multi-domain complexes of Cytochrome P450 BM3 by varying the length of spacing scaffolds between the BMR reductase domain and the BMP porphyrin domain (Figure 5B).60 2D DNA nanostructures provide an even greater opportunity to organize multi-enzyme systems into more complicated geometric patterns. There was a report of the self-assembly of a glucose oxidase (GOx) and horseradish peroxidase (HRP) enzyme cascade on 2D hexagonal DNA strips, with the distance between the two enzymes controlled by the underlying nanostructure (Figure 5C).61 A greater than 10-fold activity enhancement was observed compared to the corresponding unstructured enzymes. In addition to in vitro assembly, multi-enzyme pathways can also be organized by introducing nucleic acids nanostructures as assembly scaffolds in vivo, an approach facilitated by recent advances in RNA nanotechnology.26 This idea was demonstrated by the assembly of an intracellular reaction pathway ([FeFe]-hydrogenase and ferredoxin) for enhancing bacteria hydrogen production.62 In Figure 5D, discrete, 1D and 2D RNA scaffolds were assembled in vivo through the incorporation of aptamers for capturing the target enzyme cascade. Remarkably, a 48-fold enhancement of hydrogen production was observed for the RNA-templated [FeFe]-hydrogenase and ferredoxin network. This study suggests that a metabolic engineering approach can be used to introduce structural nucleic acids nanostructures inside cells for the organization of multi-enzyme reaction pathways. Recently, a GOx/HRP cascade was organized on DNA origami tiles with precisely controlled spatial positions, which was applied to investigating the distance-dependent inter-enzyme substrate diffusion (Figure 5E).63 The study revealed that substrate transfer between enzymes might occur at the connected hydration shells for closely paced enzymes, and demonstrated this idea by constructing a protein bridge to facilitate the intermediate transfer across protein surfaces.
Figure 5.
DNA/RNA nanostructures for engineering multi-enzyme systems. A linear, double-stranded DNA scaffold for (A) assembling an enzyme cascade: NAD(P)H:FMN (NFOR) oxidoreductase and luciferase (Luc);59 and (B) evaluating the distance-dependent activity of Cytochrome P450 BM3 by varying the spacing between the BMR reductase domain and the BMP porphyrin domain.60 (C) 2D DNA strip for organizing GOx/HRP cascades.61 (D) In vivo assembly of RNA nanostructures to organize the [FeFe]-hydrogenase and ferredoxin enzyme pathway for improved hydrogen production.62 (E) Organization of a GOx/HRP cascade on DNA origami tiles with controlled spatial positions (top), and a protein bridge for facilitating surface-limited intermediate diffusion between enzymes (bottom).63 Figures are reproduced with permissions from Wiley, ACS, NPG and AAAS.
Light-harvesting networks
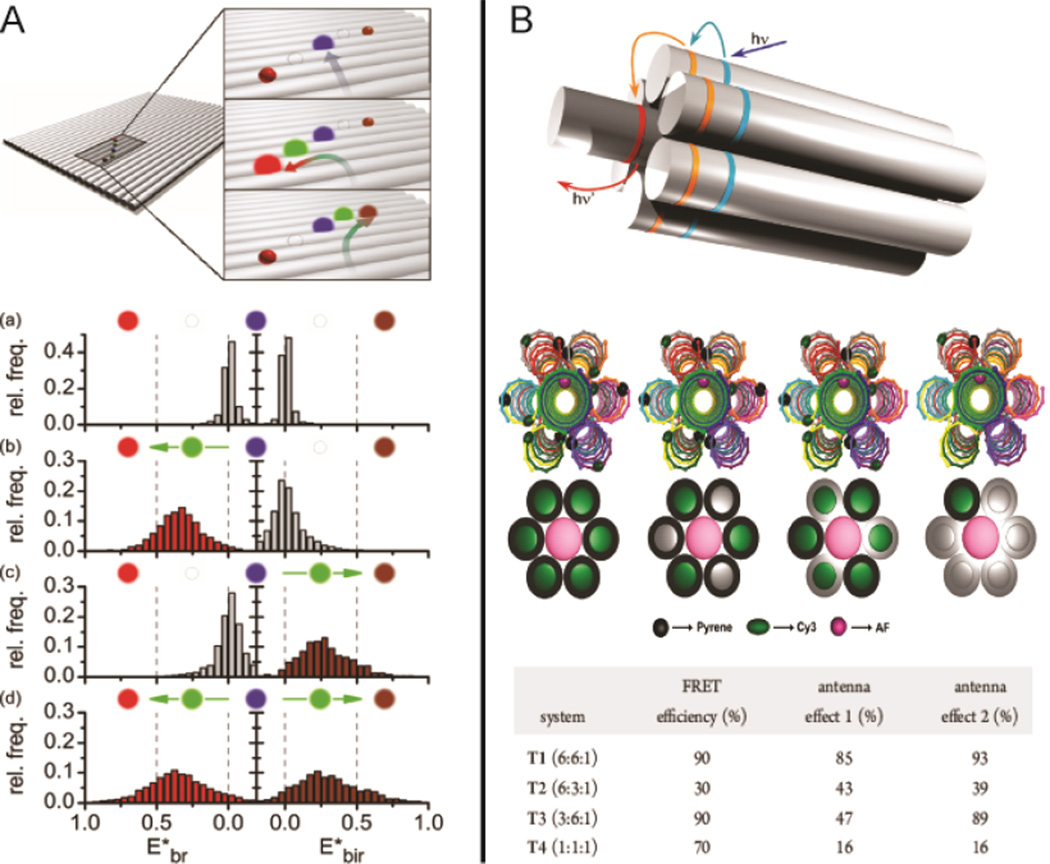
In natural photosynthesis, light is harvested by antenna systems that consist of networks of spatially-organized chromophores to facilitate unidirectional energy transfer to a redox center.64 In artificial systems, DNA nanostructures can be used to arrange multiple pairs of fluorescence donors and acceptors into precise geometric patterns to achieve efficient energy-transfer. In Figure 6A, a DNA origami tile was used to organize several distinct fluorophores into closely packed linear arrangements to achieve multi-color energy transfer, observable at the single-molecule level.65 Energy transfer was directed along a path from a blue- to red- dye or from a blue- to IR-dye by placing a “jumper dye” between the primary donor and the final acceptor. As shown in Figure 6B, an artificial light-harvesting antenna was constructed by assembling multiple donor-acceptor pairs on a seven-helix DNA bundle.66 Steady-state and time-resolved fluorescence spectroscopy was used to measure the efficiency of energy transfer for networks with various ratios of donor to acceptor dyes.
Figure 6.
Energy-transfer within DNA nanostructures. (A) Four-color FRET65 and (B) artificial light-harvesting network.66 Figures are reproduced with permissions from ACS.
Responsive nanodevice
In 1966, the science fiction movie “Fantastic Voyage” described a shrunken, micrometer-sized submarine that could be injected into the human circulatory system to search for and destroy a threatening blood clot in the brain. The enormous potential of DNA nanotechnology is bringing us closer to this dream. Autonomous DNA walkers are early demonstrations of functional nanorobots, where the motion of the legs are coordinated and driven by either strand displacement67 or deoxyribozyme(DNAzyme)-substrate binding and cleavage.68 Recent advances in DNA origami make it possible to construct integrated nanosystems that combine walkers, cargo, tracks, and drive mechanisms to achieve complex motions on 2D or 3D surfaces. There was a report of an integrated system that executed cargo loading, transportation and destination control functions.69 In Figure 7A, the hands of the DNA walker bound to specific nanoparticle cargo when the cassette was switched from an “OFF” to “ON” state. Fuel strands were employed to initiate the walker’s stepwise movement, with a 120 degree rotation for each step. The cargo-transportation system was programmed to reach eight different destinations by controlling the states of the three loading cassettes, and the movement along the tracks. In parallel, a spider-like molecular walker was developed with the ability to travel along a 2D oligonucleotide substrate track assembled on a DNA origami tile.70 The walker was composed of an inert streptavidin protein body, with three catalytic DNAzyme legs and a single capture leg for loading the molecular spider on the surface of the origami (Figure 7B). For movement along a predetermined path, the molecular walker was first loaded at the START position via hybridization of the capture leg to a partially complementary probe extended form the DNA origami surface. The walker was subsequently released by the addition of a 27-nt single-stranded DNA trigger that was fully complementary to the START probe, displacing the capture leg and allowing the walker to move to the substrate track. The catalytic action of the DNAzyme legs, binding to and cleaving the underlying deoxyribonucleic acid substrate track, drove the spider toward uncleaved substrate until it reached a STOP site, where further movement was inhibited by strong binding between a noncleavable probe and the DNAzyme legs.
Figure 7.
Responsive DNA nanodevices (A) a cargo transportation system consisting of an assembly template, cargo loading apparatus and DNA walker,69 (B) walker movement along a 2-D deoxyribonucleotide substrate surface70 and (C) forceps for sensing various noncovalent interactions.74 Figures are reproduced with permissions from NPG.
In addition to walkers, other responsive DNA nanodevices such as tweezers,71 I-motif switches,72 and hybridization-chain-reaction systems73 have been developed. These devices are capable of sensing the presence of specific DNA, changes in pH, and mRNA expression. Recently, origami-based forceps with the ability to switch between ‘open’ and ‘closed’, positions were reported. The action of the forceps was triggered by noncovalent interactions including metal ion-nucleotide, biotin-streptavidin and antigen-antibody binding interactions (Figure 7C).74
Future Perspective
Self-assembled DNA nanostructures can now be used to organize a variety of heterogeneous elements into precise patterns on rationally-designed 2D and 3D nanoarchitectures. Future challenges include identifying how to harness this power to construct functional, spatially-interactive biomolecule complexes. Here, we identify several potential applications of DNA nanotechnology in constructing artificial bio-nanosystems.
Bottom-up engineering of multi-component complexes
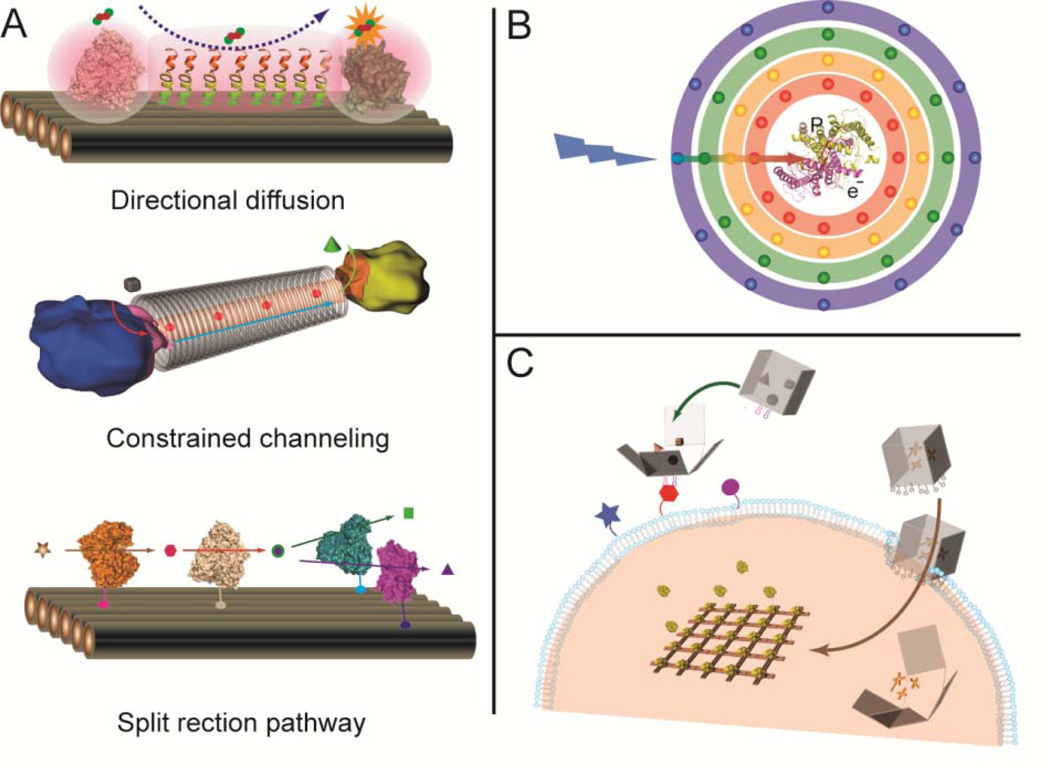
Translating biochemical reaction pathways to non-cellular environments is of great scientific interest. Exerting control over these pathways beyond nature’s repertoire would enable enzyme-catalyzed production of novel molecules and energy conversion optimized for ambient and extreme environments. Engineering functional multi-enzyme complexes requires a method to reliably organize the individual protein components with control over the relative position, orientation, and quantity of the participating molecules. The combination of self-assembled DNA nanostructures and common bio-conjugation strategies makes it possible to rationally design and organize multi-protein pathways, as well as modulate the local environment and influence the corresponding chemical reactions (Figure 8A). For example, the direct transfer of a substrate from one enzyme to a proximal enzyme (substrate channeling), is one of the primary ways that natural systems facilitate highly efficient enzyme activity.75 Similar channeling effects can be replicated in a DNA nanostructure system by optimizing the relative position and orientation of the catalytic components. Directed diffusion over longer distances can be achieved by modifying the environment between two enzymes with specific properties (polarity or hydrophobicity) that encourage substrate diffusion. It is also possible to constrain the diffusion between two enzymes by constructing a DNA cavities or nanotubes. Further, enzyme pathway feedback mechanisms may be realized by constructing branched reaction pathways, where the catalytic activities are regulated by activation or deactivation of a specific pathway.
Figure 8.
(A) Engineering enzyme pathways to achieve directional substrate diffusion (top), constrained substrate tunneling (middle) and split enzyme pathways as feedback mechanisms (bottom). (B) Schematic illustration of an artificial photosynthesis system that couples light harvesting and charge separation components within a multi-layer DNA nanostructure. (C) DNA nanocontainer for target-specific drug delivery and in vivo regulation of cellular activities.
Artificial macromolecular photosynthetic complex
Natural photosynthetic systems harvest light energy and convert it into chemically useful forms. Artificial photosynthetic complexes that execute light harvesting and charge separation have been constructed by incorporating chromophores and electron donors or acceptors into supramolecular structures.76 However, these systems exhibit little spatial control and often involve complex synthetic chemistry. DNA nanostructures can serve as scaffolds for the assembly of biohybrid systems, where efficient light harvesting apparatus can be coupled with charge separation complexes with nanometer-scale precision (Figure 8B). In particular, an artificial light-harvesting complex must be capable of wide-spectrum absorbance and contain an efficient energy transfer pathway, both of which can be satisfied by DNA nanostructures directed assembly. DNA nanostructures can be used to arrange multiple chromophores into 2D or 3D patterns with optimized stoichiometric ratios and inter-component distances. Units of charge-separation can be held in close proximity to the light-harvesting components for efficient conversion of light energy.
In vivo delivery and regulation
Nanotechnology has been applied to target-specific drug delivery, in vivo regulation, visualization and sensing. Structural DNA nanotechnology may be used to construct more effective drug-delivery vehicles through the implementation of complex control mechanisms to sense specific targets, respond to environmental conditions, release molecular payloads and trigger additional responses to regulate biological functions that impede disease progression. DNA-based nanocontainers, such as DNA boxes with switchable lids that open and close,19 and nanocages with the ability to encapsulate or release nanoparticles,77 have demonstrated potential as drug delivery vehicles. An autonomous DNA nanorobot controlled by an aptamer-encoded logic gate was recently reported to transport molecular payloads to cells, sense cell surface inputs for triggered activation, and transform its structure for payload delivery.78 However, additional research is needed to improve these DNA nanodevices. First, new structural switching mechanisms (rather than strand displacement) should be implemented to control drug release in specific biological conditions. One possibility is to use structural-switching aptamers79 to introduce a locking mechanisms to DNA nanocontainers,78 which are triggered by aptamer-target binding. Second, the resistance of DNA nanostructures to the components of serum and cell lysate must be increased so that they may withstand in vivo delivery conditions. A recent study has shown that certain DNA origami structures maintain their structural integrity after incubation with cell lysate for 12 hours, a significant increase in stability compared to natural single- and double-stranded DNA.80 Finally, it is a challenge to transfer DNA nanostructures across biological membranes, since most cell membranes will only permit free passage of small molecules. Some recent studies have shown that DNA nanostructures modified with CPG81–82 or aptamers83 can be taken by cells. The display of certain ligands (amphiphilic molecules for example) from the surface of a DNA nanostructure may facilitate tissue penetration and cellular uptake of DNA nanodevices. Combining DNA/RNA nanotechnology with molecular biology may result in the development of novel ways to regulate cellular response. It may be feasible to construct artificial intracellular or extracellular nanomatrices that are designed to influence gene expression or modulate biological pathways (Figure 8C).
Concluding Remarks
Self-assembled DNA nanostructures are excellent scaffolds to direct the assembly of highly organized, spatially-interactive biomolecule networks with enhanced functionality. Combining the promise of structural DNA nanotechnology with biology, chemistry, computer science, physics and materials science, will likely result in the emergence of new and exciting discoveries beyond the limited scope discussed here.
Acknowledgement
The authors acknowledge the funding supports from the Army Research Office, National Science Foundation, Office of Naval Research, National Institute of Health, Department of Energy and Sloan Foundation. The authors would like to thank Jeanette Nangreave and Carole Flores for the help in editing the manuscript.
Biographies
Jinglin Fu received his Ph.D degree in Chemistry (2010) under the supervision of Dr. Neal Woodbury from Arizona State University. Currently he is carrying out postdoctoral studies as a member of Prof. Hao Yan’s group with research focus on the enzymology of multi-enzyme systems on self-assembled nanostructures.
Minghui Liu received her bachelor’s degree (2008) from Wuhan University, China. Currently she is a Ph.D student in Hao Yan’s Lab at Arizona State University, with research focus on DNA-nanostructures directed protein assembly.
Yan Liu received her Ph.D degree in Chemistry (2000) with Prof. Kenneth Eisenthal from Columbia University. She is currently an Assistant Professor in the department of Chemistry and Biochemistry at Arizona State University, with research focus on DNA nanotechnology and quantum material synthesis.
Hao Yan received his Ph.D degree in Chemistry (2001) with Prof. Nadrian Seeman from New York University. He is currently a Professor in Chemistry and Biochemistry at Arizona State University. His research interests are aimed at the construction of DNA based molecular devices that can function as molecular assemblers to control chemical synthesis and macromoleculear interactions.
Reference
- 1.Savage DF, Afonso B, Chen AH, Silver PA. Spatially Ordered Dynamics of the Bacterial Carbon Fixation Machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 2.Cogdell RJ, Gall A, Köhler J. The architecture and function of the light-harvesting apparatus of purple bacteria: from single molecules to in vivo membranes. Q. Rev. Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 3.Stupp SI. Self-Assembly and Biomaterials. Nano Lett. 2010;10:4783–4786. doi: 10.1021/nl103567y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C, Liu Y, Yan H. Designer DNA Nanoarchitectures. Biochemistry. 2009;48:1663–1674. doi: 10.1021/bi802324w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seeman NC. Nucleic acid junctions, lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 7.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 8.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 9.Park SH, Barish R, Li H, Reif JH, Finkelstein G, Yan H, LaBean TH. Three-Helix Bundle DNA Tiles Self-Assemble into 2D Lattice or 1D Templates for Silver Nanowires. Nano Lett. 2005;5:693–696. doi: 10.1021/nl050108i. [DOI] [PubMed] [Google Scholar]
- 10.Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, Seeman NC. Six-Helix Bundles Designed from DNA. Nano Lett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao C, Sun W, Seeman NC. Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy. J. Am. Chem. Soc. 1999;121:5437–5443. [Google Scholar]
- 12.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-Templated Self-Assembly of Protein Arrays and Highly Conductive Nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 14.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 15.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 16.Yan H, LaBean TH, Feng L, Reif JH. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8103–8108. doi: 10.1073/pnas.1032954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 18.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 19.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 20.Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H. J. Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke Y, Voigt NV, Gothelf KV, Shih WM. J. Am. Chem. Soc. 2011;134:1770–1774. doi: 10.1021/ja209719k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietz H, Douglas SM, Shih WM. Folding DNA into Twisted and Curved Nanoscale Shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA Origami with Complex Curvatures in Three-Dimensional Space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 24.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro CE, et al. A primer to scaffolded DNA origami. Nat. Methods. 2011;8:221–229. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 26.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko SH, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Synergistic self-assembly of RNA and DNA molecules. Nat. Chem. 2010;2:1050–1055. doi: 10.1038/nchem.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams BAR, Lund K, Liu Y, Yan H, Chaput JC. Self-Assembled Peptide Nanoarrays: An Approach to Studying Protein–Protein Interactions. Angew. Chem., Int. Ed. 2007;46:3051–3054. doi: 10.1002/anie.200603919. [DOI] [PubMed] [Google Scholar]
- 29.Kuzyk A, Kimura M, Numajiri K, Koshi N, Ohnishi TO, kada F, Komiyama M. DNA origami as a nanoscale template for protein assembly. Nanotechnology. 2009;20:235305. doi: 10.1088/0957-4484/20/23/235305. [DOI] [PubMed] [Google Scholar]
- 30.Tan SJ, Campolongo MJ, Luo D, Cheng W. Building plasmonic nanostructures with DNA. Nat. Nanotechnol. 2011;6:268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 31.Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H. Spatially Addressable Multiprotein Nanoarrays Templated by Aptamer-Tagged DNA Nanoarchitectures. J. Am. Chem. Soc. 2007;129:10304–10305. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- 32.Stephanopoulos N, Liu M, Tong GJ, Li Z, Liu Y, Yan H, Francis MB. Immobilization and One-Dimensional Arrangement of Virus Capsids with Nanoscale Precision Using DNA Origami. Nano Lett. 2010;10:2714–2720. doi: 10.1021/nl1018468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodchild J. Conjugates of oligonucleotides and modified oligonucleotides: a review of their synthesis and properties. Bioconjugate Chem. 1990;1:165–187. doi: 10.1021/bc00003a001. [DOI] [PubMed] [Google Scholar]
- 34.Niemeyer CM. Semisynthetic DNA–Protein Conjugates for Biosensing and Nanofabrication. Angew. Chem., Int. Ed. 2010;49:1200–1216. doi: 10.1002/anie.200904930. [DOI] [PubMed] [Google Scholar]
- 35.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 36.Saccà B, et al. Orthogonal Protein Decoration of DNA Origami. Angew. Chem., Int. Ed. 2010;49:9378–9383. doi: 10.1002/anie.201005931. [DOI] [PubMed] [Google Scholar]
- 37.Schweller RM, Constantinou PE, Frankel NW, Narayan P, Diehl MR. Design of DNA-Conjugated Polypeptide-Based Capture Probes for the Anchoring of Proteins to DNA Matrices. Bioconjugate Chem. 2008;19:2304–2307. doi: 10.1021/bc8003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakata E, Liew FF, Uwatoko C, Kiyonaka S, Mori Y, Katsuda Y, Endo M, Sugiyama H, Morii T. Angew. Chem., Int. Ed. 2012;51:2421–2424. doi: 10.1002/anie.201108199. [DOI] [PubMed] [Google Scholar]
- 39.Williams BAR, Diehnelt CW, Belcher P, Greving M, Woodbury NW, Johnston SA, Chaput JC. Creating Protein Affinity Reagents by Combining Peptide Ligands on Synthetic DNA Scaffolds. J. Am. Chem. Soc. 2009;131:17233–17241. doi: 10.1021/ja9051735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-Assembled Water-Soluble Nucleic Acid Probe Tiles for Label-Free RNA Hybridization Assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian HKK, Chakraborty B, Sha R, Seeman NC. The Label-Free Unambiguous Detection and Symbolic Display of Single Nucleotide Polymorphisms on DNA Origami. Nano Lett. 2011;11:910–913. doi: 10.1021/nl104555t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Yan H, Liu Y, Chang Y. Targeted Cell–Cell Interactions by DNA Nanoscaffold-Templated Multivalent Bispecific Aptamers. Small. 2011;7:1673–1682. doi: 10.1002/smll.201002292. [DOI] [PubMed] [Google Scholar]
- 44.Eberhard H, Diezmann F, Seitz O. DNA as a Molecular Ruler: Interrogation of a Tandem SH2 Domain with Self-Assembled, Bivalent DNA–Peptide Complexes. Angew. Chem., Int. Ed. 2011;50:4146–4150. doi: 10.1002/anie.201007593. [DOI] [PubMed] [Google Scholar]
- 45.Helmig S, et al. Single Molecule Atomic Force Microscopy Studies of Photosensitized Singlet Oxygen Behavior on a DNA Origami Template. ACS Nano. 2010;4:7475–7480. doi: 10.1021/nn102701f. [DOI] [PubMed] [Google Scholar]
- 46.Voigt NV, et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 47.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selmi DN, Adamson RJ, Attrill H, Goddard AD, Gilbert RC, Watts A, Turberfield AJ. DNA-Templated Protein Arrays for Single-Molecule Imaging. Nano Lett. 2011;11:657–660. doi: 10.1021/nl1037769. [DOI] [PubMed] [Google Scholar]
- 49.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476:109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinhauer C, Jungmann R, Sobey TL, Simmel FC, Tinnefeld P. DNA Origami as a Nanoscopic Ruler for Super-Resolution Microscopy. Angew. Chem., Int. Ed. 2009;48:8870–8873. doi: 10.1002/anie.200903308. [DOI] [PubMed] [Google Scholar]
- 52.Sannohe Y, Endo M, Katsuda Y, Hidaka K, Sugiyama H. Visualization of Dynamic Conformational Switching of the G-Quadruplex in a DNA Nanostructure. J. Am. Chem. Soc. 2010;132:16311–16313. doi: 10.1021/ja1058907. [DOI] [PubMed] [Google Scholar]
- 53.Endo M, Katsuda Y, Hidaka K, Sugiyama H. Regulation of DNA Methylation Using Different Tensions of Double Strands Constructed in a Defined DNA Nanostructure. J. Am. Chem. Soc. 2010;132:1592–1597. doi: 10.1021/ja907649w. [DOI] [PubMed] [Google Scholar]
- 54.Gu H, Yang W, Seeman NC. DNA Scissors Device Used to Measure MutS Binding to DNA Mis-pairs. J. Am. Chem. Soc. 2010;132:4352–4357. doi: 10.1021/ja910188p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liedl T, Hogberg B, Tytell J, Ingber DE, Shih WM. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 57.Sheldon RA. Enzyme Immobilization: The Quest for Optimum Performance. Adv. Synth. Catal. 2007;349:1289–1307. [Google Scholar]
- 58.Teller C, Willner I. Organizing protein-DNA hybrids as nanostructures with programmed functionalities. Trends Biotechnol. 2010;28:619–628. doi: 10.1016/j.tibtech.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Niemeyer CM, Koehler J, Wuerdemann C. DNA-Directed Assembly of Bienzymic Complexes from In Vivo Biotinylated NAD(P)H:FMN Oxidoreductase and Luciferase. ChemBioChem. 2002;3:242–245. doi: 10.1002/1439-7633(20020301)3:2/3<242::AID-CBIC242>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 60.Erkelenz M, Kuo CH, Niemeyer CM. DNA-Mediated Assembly of Cytochrome P450 BM3 Subdomains. J. Am. Chem. Soc. 2011;133:16111–16118. doi: 10.1021/ja204993s. [DOI] [PubMed] [Google Scholar]
- 61.Wilner OI, Weizmann Y, Gill R, Lioubashevski O, Freeman R, Willner I. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 62.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 63.Fu J, Liu M, Liu Y, Woodbury NW, Yan H. J. Am. Chem. Soc. 2012;134:5516–5519. doi: 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gust D, Moore TA, Moore AL. Mimicking Photosynthetic Solar Energy Transduction. J. Acc. Chem. Res. 2000;34:40–48. doi: 10.1021/ar9801301. [DOI] [PubMed] [Google Scholar]
- 65.Stein IH, Steinhauer C, Tinnefeld P. Single-Molecule Four-Color FRET Visualizes Energy-Transfer Paths on DNA Origami. J. Am. Chem. Soc. 2011;133:4193–4195. doi: 10.1021/ja1105464. [DOI] [PubMed] [Google Scholar]
- 66.Dutta PK, Varghese R, Nangreave J, Lin S, Yan H, Liu Y. DNA-Directed Artificial Light-Harvesting Antenna. J. Am. Chem. Soc. 2011;133:11985–11993. doi: 10.1021/ja1115138. [DOI] [PubMed] [Google Scholar]
- 67.Omabegho T, Sha R, Seeman NC. A Bipedal DNA Brownian Motor with Coordinated Legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He Y, Liu DR. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat. Nanotechnol. 2010;5:778–782. doi: 10.1038/nnano.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu H, Chao J, Xiao SJ, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chhabra R, Sharma J, Liu Y, Yan H. Addressable Molecular Tweezers for DNA-Templated Coupling Reactions. Nano Lett. 2006;6:978–983. doi: 10.1021/nl060212f. [DOI] [PubMed] [Google Scholar]
- 72.Modi S, Swetha MG, Goswami D, Gupta GD, Mayor S, Krishnan Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 73.Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuzuya A, Sakai Y, Yamazaki T, Xu Y, Komiyama M. Nanomechanical DNA origami 'single-molecule beacons' directly imaged by atomic force microscopy. Nat. Commun. 2011;2:449. doi: 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miles EW, Rhee S, Davies DR. The Molecular Basis of Substrate Channeling. J. Biol. Chem. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 76.Wasielewski MR. Self-Assembly Strategies for Integrating Light Harvesting and Charge Separation in Artificial Photosynthetic Systems. Acc. Chem. Res. 2009;42:1910–1921. doi: 10.1021/ar9001735. [DOI] [PubMed] [Google Scholar]
- 77.Zhao Z, Jacovetty EL, Liu Y, Yan H. Encapsulation of Gold Nanoparticles in a DNA Origami Cage. Angew. Chem., Int. Ed. 2011;50:2041–2044. doi: 10.1002/anie.201006818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 79.Oh SS, Plakos K, Lou X, Xiao Y, Soh HT. In vitro selection of structure-switching, self-reporting aptamers. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14053–14058. doi: 10.1073/pnas.1009172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mei Q, et al. Stability of DNA Origami Nanoarrays in Cell Lysate. Nano Lett. 2011;11:1477–1482. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schüller VJ, Heidegger S, Sandholzer N, Nickels PC, Suhartha NA, Endres S, Bourquin C, Liedl T. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 82.Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 83.Chang M, Yang CS, Huang DM. ACS Nano. 2011;5:6156–6163. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]