Abstract
Orofacial clefts are the most frequent craniofacial defects, which affect 1.5 in 1,000 newborns worldwide1,2. Orofacial clefting is caused by abnormal facial development3. In human and mouse, initial growth and patterning of the face relies on several small buds of tissue, the facial prominences4,5. The face is derived from six main prominences: paired frontal nasal processes (FNP), maxillary prominences (MxP) and mandibular prominences (MdP). These prominences consist of swellings of mesenchyme that are encased in an overlying epithelium. Studies in multiple species have shown that signaling crosstalk between facial ectoderm and mesenchyme is critical for shaping the face6. Yet, mechanistic details concerning the genes involved in these signaling relays are lacking. One way to gain a comprehensive understanding of gene expression, transcription factor binding, and chromatin marks associated with the developing facial ectoderm and mesenchyme is to isolate and characterize the separated tissue compartments.
Here we present a method for separating facial ectoderm and mesenchyme at embryonic day (E) 10.5, a critical developmental stage in mouse facial formation that precedes fusion of the prominences. Our method is adapted from the approach we have previously used for dissecting facial prominences7. In this earlier study we had employed inbred C57BL/6 mice as this strain has become a standard for genetics, genomics and facial morphology8. Here, though, due to the more limited quantities of tissue available, we have utilized the outbred CD-1 strain that is cheaper to purchase, more robust for husbandry, and tending to produce more embryos (12-18) per litter than any inbred mouse strain8. Following embryo isolation, neutral protease Dispase II was used to treat the whole embryo. Then, the facial prominences were dissected out, and the facial ectoderm was separated from the mesenchyme. This method keeps both the facial ectoderm and mesenchyme intact. The samples obtained using this methodology can be used for techniques including protein detection, chromatin immunoprecipitation (ChIP) assay, microarray studies, and RNA-seq.
Keywords: Developmental Biology, Issue 74, Biomedical Engineering, Bioengineering, Cellular Biology, Molecular Biology, Anatomy, Physiology, Surgery, Tissue Engineering, Embryo, Mammalian, Ectoderm, biology (general), Facial prominences, facial ectoderm, mesenchyme, Dispase II, orofacial clefts, facial development, mouse, animal model
Protocol
1. Prepare Dispase II
Prepare fresh Hepes-buffered Saline (HBS) (50 mM Hepes/KOH pH 7.4, 150 mM NaCl). Add 2 ml 0.5 M Hepes/KOH pH 7.4 and 1.2 ml 2.5 M NaCl into 16.8 ml Ultrapure H2O.
Make 10 mg/ml Dispase II. Weigh 0.2 g Dispase II (Roche, Cat# 4942078001) and dissolve in 20 ml HBS. Prepare 1.0 ml aliquots in individual Eppendorf tubes and store at -20 °C for long-term storage.
Dilute 10 mg/ml Dispase II 1:10 in PBS before use. Put the diluted Dispase II on ice.
2. Dissect CD-1 E10.5 Embryos from Pregnant Female
All animal experiments were performed in accordance with protocols approved by the University of Colorado Denver (UCD) Animal Care and Usage Committee. CD-1 (ICR) mice were obtained from Harlan Laboratories (Indianapolis, IN).
Clean the dissection area and tools with 70% ethanol.
Euthanize pregnant CD-1 female using approved protocols.
Spray the abdomen with 70% ethanol then open the abdomen. Use one set of forceps to pull the uterus and another to tear away the mesometrium from the uterus. Remove the uterus from the body. Repeat for uterine horn on other side of the abdomen.
Briefly rinse the uterus with ice cold PBS in a 10-cm Petri dish to wash out blood.
Transfer the uterus into a new Petri dish with ice cold PBS. Separate into one-embryo fragments by cutting between implantation sites.
Transfer one embryo into ice-cold PBS under a stereomicroscope. Use fine forceps to tear off the muscular wall of uterus starting at one of the cut sites to expose the embryo with the yolk sac. Peel off the yolk sac and amnion. Transfer dissected embryo to a 6-cm Petri dish with PBS on ice. Proceed until all embryos have been collected in this way.
3. Dispase II Treatment
Wash all the embryos with PBS in a 6-cm Petri dish. Replace PBS with 10 ml diluted Dispase II in PBS. Cover the Petri dish and incubate at 37 °C for 25 min - a bacterial incubator or warm room should suffice - we have not used a CO2 tissue culture incubator for this step.
Take the Petri dish out and observe the embryo under a stereomicroscope. The ectoderm should have become loose, typified by a clear gap between this tissue layer and the underlying mesenchyme. If not, incubate for an additional 5 min at 37 °C. Then put the Petri dish on ice.
4. Dissect out the Intact Facial Prominences
Use a disposable glass pipette (VWR, Cat# 14672-380)to transfer one Dispase II treated embryo into ice-cold PBS in a 6-cm Petri dish under a stereomicroscope.
Use one pair of forceps to hold the embryo. Use another pair of fine forceps carefully to cut the boundaries of the facial prominences attached to head (Figure 1 shows the boundaries of the facial prominences). Start from one side of the MnP, then the MxP, and last the FNP. Do the same to the other side to dissect the facial prominences out intact (Figure 2A).
5. Separate Facial Ectoderm from Mesenchyme
Observe the facial prominences under a stereomicroscope with 3x working distance. Use a long glass Pasteur pipette to gently pipette the facial prominences up and down 5 times. After this, the ectoderm is easy to peel off.
Use one pair of forceps to hold the facial prominences. Use the other pair of forceps to peel off the ectoderm very slowly. Figure 2B shows the separated sheets of facial ectoderm.
Use a new long glass Pasteur pipette to transfer the facial ectoderm into a 1.5 ml Eppendorf tube with PBS on ice. Transfer the mesenchyme to another 1.5 ml Eppendorf tube with PBS on ice.
Use new ice cold PBS for each dissection. Collect samples.
Wash the samples with PBS. Centrifuge at 4 °C, 500 x g for 3 min.
Aspirate the PBS using long glass Pasteur pipettes. If the samples are to be used for ChIP assay, proceed to step 6.1. For RNA extraction, proceed to step 6.2. For Protein extraction, proceed to step 6.3.
6. Process Samples for Further Experimentation
- Samples for ChIP assay
- Add 1 ml PBS with 1% formaldehyde to crosslink. Homogenize the samples.
- Incubate at room temperature for 10 min with rotation.
- Add 110 μl of 1.25 M Glycine to each tube to quench unreacted formaldehyde.
- Mix and incubate at room temperature for 5 min.
- Spin at 500 x g at 4 °C for 3 min. Remove supernatant.
- Add 1 ml cold PBS to wash the tissues.
- Spin at 500 x g at 4 °C for 3 min.
- Remove PBS and repeat PBS wash once.
- Remove supernatant, Snap freeze the tissues in liquid nitrogen. Store the samples at -80 °C for subsequent pooling and processing.
- Samples for RNA extraction
- Place the facial ectoderm and mesenchyme in tubes containing RNAlater (Ambion).
- Incubate the samples in RNAlater Solution overnight at 4 °C to allow thorough penetration of the tissue.
- Store at -20 °C for subsequent pooling and processing.
Samples for protein extraction
Add lysis buffer to the sample for subsequent processing. Or snap freeze the tissues in liquid nitrogen. Store the samples at -80 °C for subsequent pooling and processing.
Notes
Time constraints. Plan on 4-6 hr from the start of dissection through to the beginning of section 6.
When proficient, it takes ~10-15 min per embryo to perform sections 4 and 5 (prominence dissection and ectoderm separation).
If the samples are for RNA isolation, every solution should be treated with Diethylpyrocarbonate first. Also clean the forceps and the working area with RNase Away.
Keep the samples on ice at all times except for the Dispase II treatment and dissections. Change to new ice cold PBS for each dissection to make sure the PBS used for dissection is cold.
Sharp forceps are critical to obtain precise dissection. Clean the forceps completely to prevent any contamination.
For collecting samples from different prominences, dissect each prominence separately after Dispase II treatment. Then separate the ectoderm and mesenchyme.
After treatment with Dispase II, the facial ectoderm is easy to peel off. Pay special attention to contamination from other ectoderm. Discard all potentially contaminated tissue.
Once you start, don't stop until the samples reach the appropriate point in Section 6. Finish separation of facial ectoderm and mesenchyme for all the embryos as quickly as you can.
The enzyme activities of Dispase II might be different between batches and brands. It is necessary to adjust the treatment time for different brands and batches of Dispase II.
Representative Results
After Dispase II treatment, the ectoderm of the embryo tends to be loose. The facial prominences should be intact after dissection (Figure 2A). The isolated facial ectoderm is clear and free of mesenchyme tissue (Figure 2B). To determine the efficacy of the protocol and to detect cross contamination we assayed for forebrain expressed Zic39, ectodermal specific cdh15 and mesenchymal specific sox1010 using reverse transcriptase PCR. Our studies confirmed that both facial ectoderm and mesenchyme expressed the expected genes and were free of cross contamination (Figure 3).
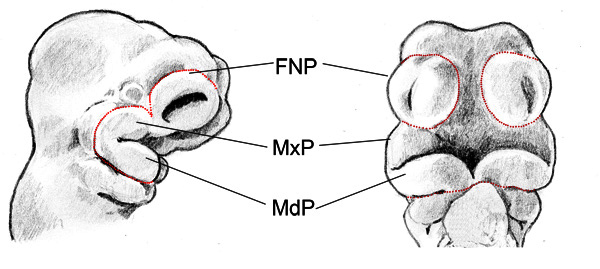 Figure 1. Schematic diagram showing mouse E10.5 facial prominences. Lateral view (left) of the head and rostral view (right) of the E10.5 embryo head. Red dotted lines indicate the boundary of the facial prominences. Frontonasal prominence (FNP), Maxillary prominence (MxP), Mandibular prominence (MdP).
Figure 1. Schematic diagram showing mouse E10.5 facial prominences. Lateral view (left) of the head and rostral view (right) of the E10.5 embryo head. Red dotted lines indicate the boundary of the facial prominences. Frontonasal prominence (FNP), Maxillary prominence (MxP), Mandibular prominence (MdP).
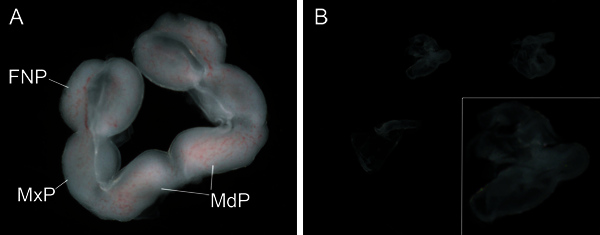 Figure 2. Appearance of dissected whole E10.5 facial prominences and facial ectoderm.(A) Whole facial prominences. (B) Facial ectoderm. Panels A and B are same magnification. Insert in B shows higher magnification of one of the facial ectoderm in panel B. Frontonasal prominence (FNP), Maxillary prominence (MxP), Mandibular prominence (MdP).
Figure 2. Appearance of dissected whole E10.5 facial prominences and facial ectoderm.(A) Whole facial prominences. (B) Facial ectoderm. Panels A and B are same magnification. Insert in B shows higher magnification of one of the facial ectoderm in panel B. Frontonasal prominence (FNP), Maxillary prominence (MxP), Mandibular prominence (MdP).
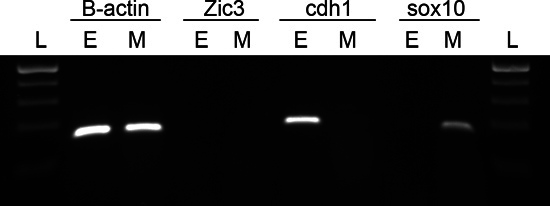 Figure 3. Reverse transcriptase PCR confirmed that both facial ectoderm and mesenchyme are free of cross contamination. Total RNA was extracted from facial ectoderm (E) and mesenchyme (M). The RNA was reverse transcribed into its complementary DNA (cDNA), followed by PCR with specific primers to detect the expression of beta-actin (B-actin), Zic3, cdh1 and sox10. The primer sequences in Table 1 were obtained from primer bank (http://pga.mgh.harvard.edu/primerbank/). The PCR products were separated on a 2% agarose gel. DNA Ladder (L).
Figure 3. Reverse transcriptase PCR confirmed that both facial ectoderm and mesenchyme are free of cross contamination. Total RNA was extracted from facial ectoderm (E) and mesenchyme (M). The RNA was reverse transcribed into its complementary DNA (cDNA), followed by PCR with specific primers to detect the expression of beta-actin (B-actin), Zic3, cdh1 and sox10. The primer sequences in Table 1 were obtained from primer bank (http://pga.mgh.harvard.edu/primerbank/). The PCR products were separated on a 2% agarose gel. DNA Ladder (L).
| Gene | Primer name | sequences | Product size |
| Beta-actin | mActb-F | 5'-GGCTGTATTCCCCTCCATCG-3' | 154 bp |
| mActb-R | 5'-CCAGTTGGTAACAATGCCATGT-3' | ||
| zic3 | mZic3-F | 5'-TCCCTTCGGGGACTCAACC-3' | 138 bp |
| mZic3-R | 5'-GCATTGGCATAACCTGAACCC-3' | ||
| cdh1 | mcdh1-F | 5'-CAGGTCTCCTCATGGCTTTGC-3' | 175 bp |
| mcdh1-R | 5'-CTTCCGAAAAGAAGGCTGTCC-3' | ||
| sox10 | msox10-F | 5'-ACACCTTGGGACACGGTTTTC-3' | 165 bp |
| msox10-R | 5'-TAGGTCTTGTTCCTCGGCCAT-3' |
Table 1. Primer sequences.
Discussion
This protocol provides a straightforward method to separate embryonic mouse facial ectoderm and mesenchyme based on an initial Dispase II treatment step. In previous studies, we have performed dissection of the facial prominences prior to Dispase II treatment, but we have consistently found that the mesenchyme becomes "sticky" and more difficult to manipulate. Our new protocol avoids this problem. Combination Dispase II treatment with gentle physical force by pipetting up and down makes separation of ectoderm and mesenchyme easier. Further, this protocol also works well for the separation of other embryonic ectoderm and mesenchyme samples, such as limb bud ectoderm and mesenchyme with shorter Dispase II treatment. Similarly, it can be used at other stages of development - for example, for separating E11.5 facial ectoderm and mesenchyme with a longer Dispase II treatment step.
The sample size obtained from an E10.5 mouse facial prominence is quite small and therefore we tend to store batches for eventually processing and analysis. In this respect it is necessary to pool ectodermal samples from 100-150 embryos to obtain 10 mg of tissue. A larger amount of mesenchyme is derived from each embryo requiring less pooling. The yield of sonicated genomic DNA for ChIP assay is about 80 μg from 10 mg facial ectoderm. The total RNA yield is about 20 μg per 10 mg facial ectoderm. From a comparative Bradford protein assay, we estimate that we can obtain up to 1 mg protein per 10 mg facial ectoderm. Once the RNA, chromatin, or protein samples are in hand, they can be utilized for numerous assays including protein detection, ChIP assay, microarray analysis, and RNA-seq. Therefore, this protocol enables a thorough analysis of differential gene expression, epigenetic modification, and transcription factor binding responsible for the molecular crosstalk between facial ectoderm and mesenchyme during embryonic development.
Disclosures
The authors have no competing interests or other conflicts associated with the contents of this article.
Acknowledgments
The authors would like to thank Irene Choi for illustrating embryo heads in Fig 1 and the other members of the laboratory for help and discussion. This work is supported by NIH grant DE012728 (T.W.)
References
- Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat. Rev. Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Chapter 2 - The Etiopathogenesis of cleft lip and cleft palate: usefulness and caveats of mouse models. Current Topics in Developmental Biology. 2008;84:37–138. doi: 10.1016/S0070-2153(08)00602-9. [DOI] [PubMed] [Google Scholar]
- Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Human Molecular Genetics. 1999;8:1–7. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: Morphogenetic and molecular mechanisms. Dev. Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BS, Yang H, Melvin VS, Taketo MM, Williams T. Ectodermal WNT/β-catenin signaling shapes the mouse face. Developmental Biology. 2011;349:261–269. doi: 10.1016/j.ydbio.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, et al. Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS ONE. 2009;4:e8066. doi: 10.1371/journal.pone.0008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MF, Fisher EM. The origins and uses of mouse outbred stocks. Nat. Genet. 2005;37(11):1181–1186. doi: 10.1038/ng1665. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, et al. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Developmental Biology. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Britsch S, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15(1):66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]


