Abstract
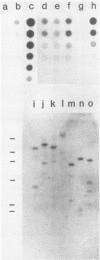
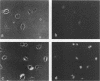
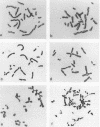
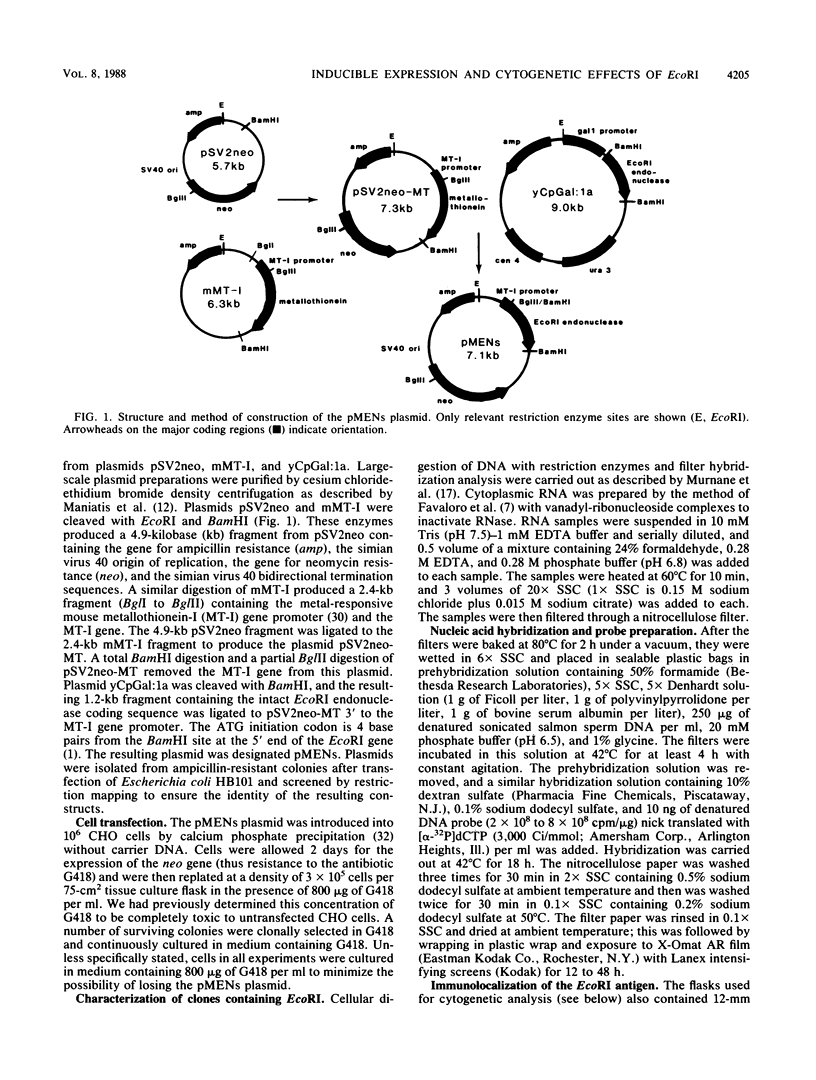
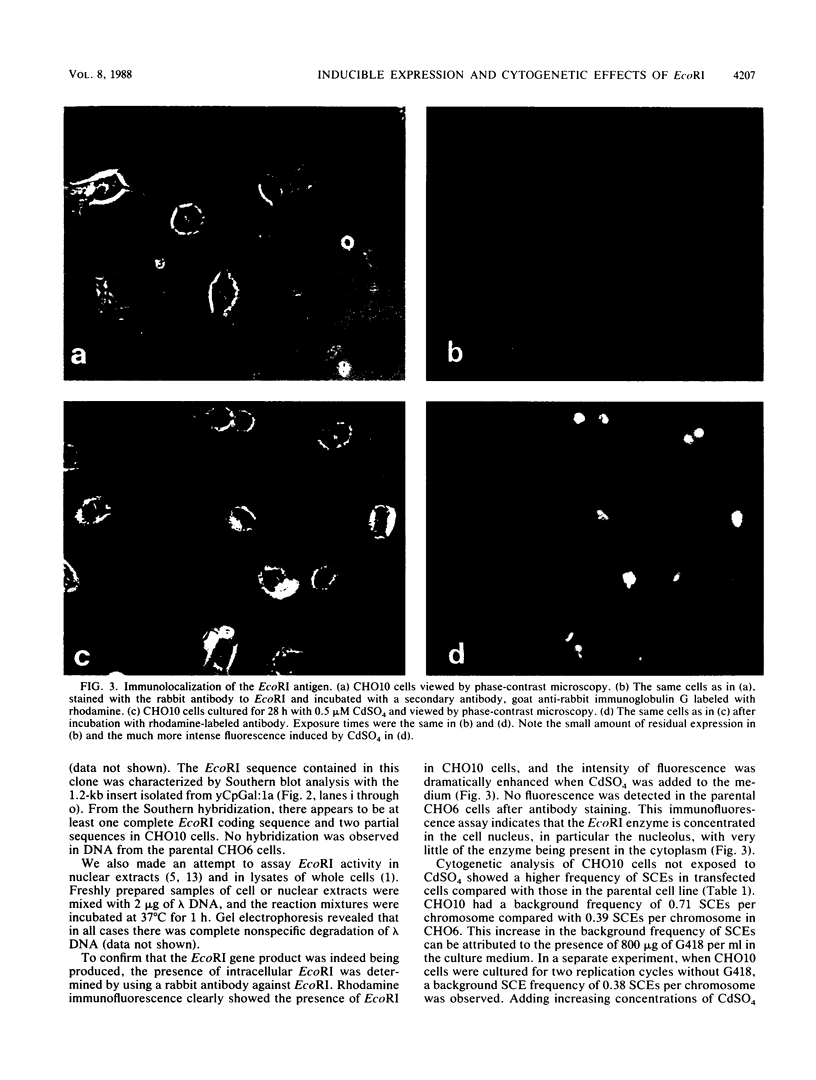
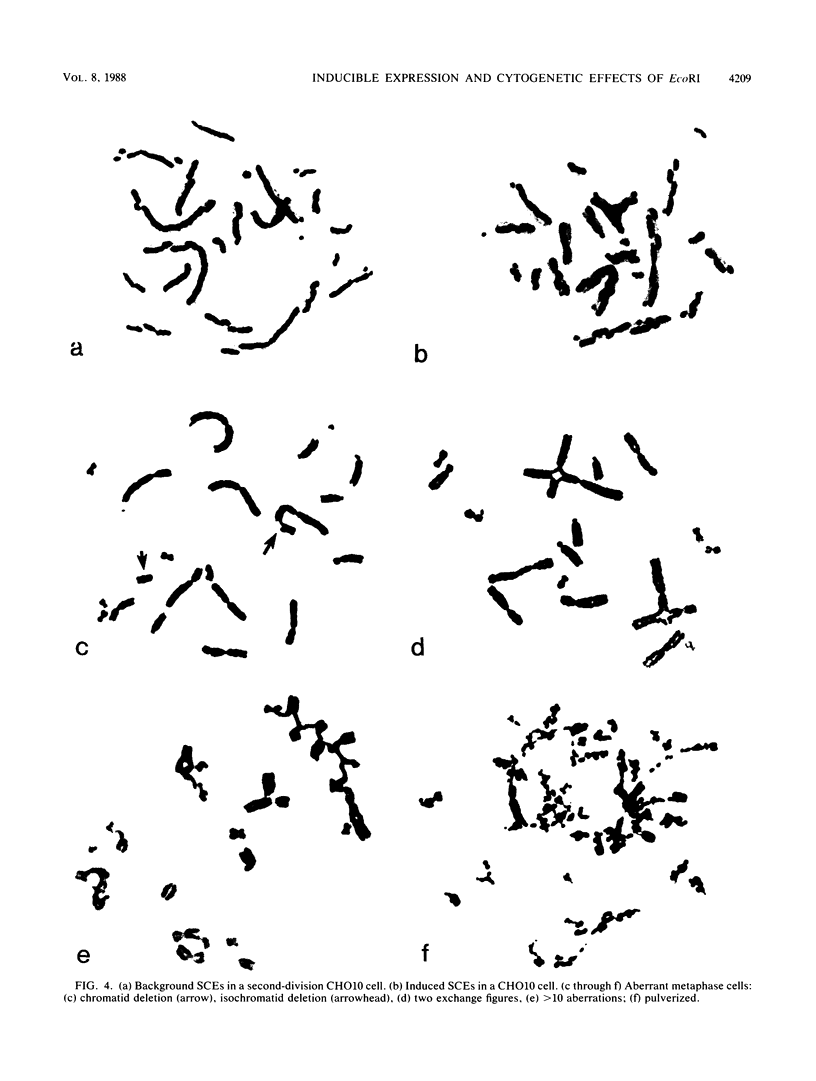
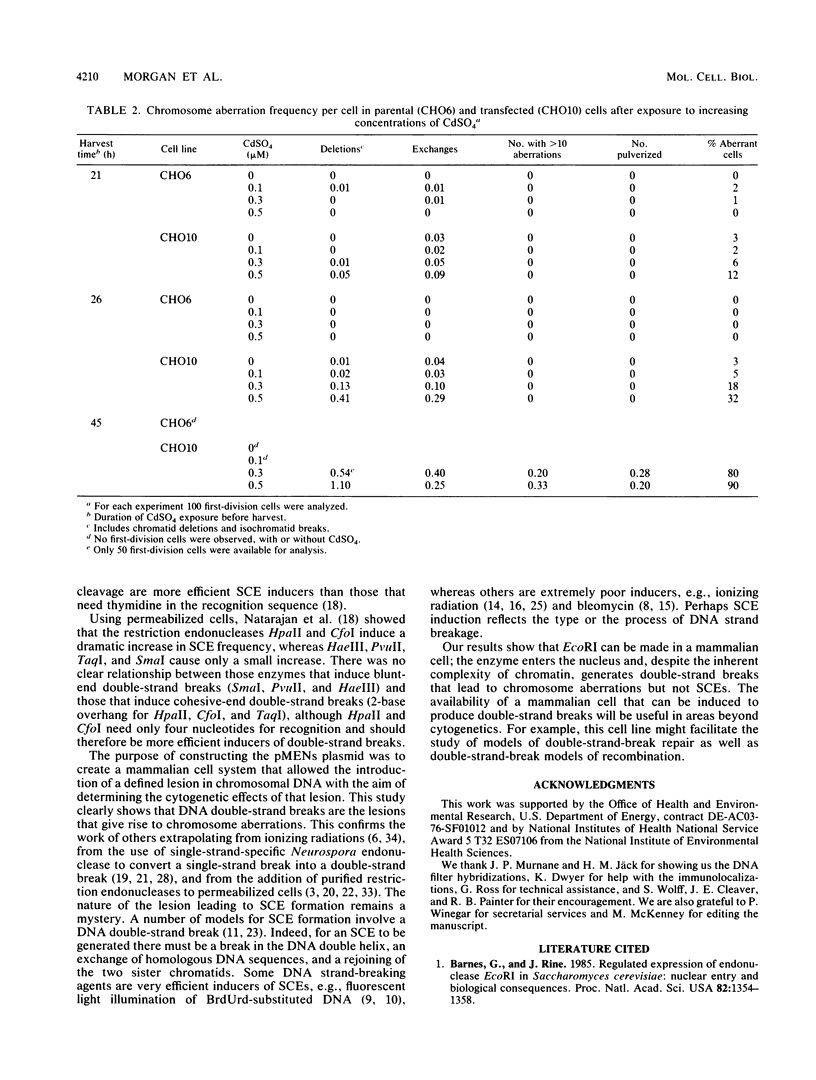
The cytogenetic endpoints sister chromatid exchange (SCE) and chromosome aberrations are widely used as indicators of DNA damage induced by mutagenic carcinogens. Chromosome aberrations appear to result directly from DNA double-strand breaks, but the lesion(s) giving rise to SCE formation remains unknown. Most compounds that induce SCEs induce a spectrum of lesions in DNA. To investigate the role of double-strand breakage in SCE formation, we constructed a plasmid that gives rise to one specific lesion, a staggered-end ("cohesive") DNA double-strand break. This plasmid, designated pMENs, contains a selectable marker, neo, which is a bacterial gene for neomycin resistance, and the coding sequence for the bacterial restriction endonuclease EcoRI attached to the mouse metallothionein gene promoter. EcoRI recognizes G decreases AATTC sequences in DNA and makes DNA double-strand breaks with four nucleotides overhanging as staggered ends. Cells transfected with pMENS were resistant to the antibiotic G418 and contained an integrated copy of the EcoRI gene, detectable by DNA filter hybridization. The addition of the heavy metal CdSO4 resulted in the intracellular production of EcoRI, as measured by an anti-EcoRI antibody. Cytogenetic analysis after the addition of CdSO4 indicated a dramatic increase in the frequency of chromosome aberrations but very little effect on SCE frequency. Although there was some intercellular heterogeneity, these results confirm that DNA double-strand breaks do result in chromosome aberrations but that these breaks are not sufficient to give rise to SCE formation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes G., Rine J. Regulated expression of endonuclease EcoRI in Saccharomyces cerevisiae: nuclear entry and biological consequences. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1354–1358. doi: 10.1073/pnas.82.5.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7270–7278. [PubMed] [Google Scholar]
- Bryant P. E. Enzymatic restriction of mammalian cell DNA using Pvu II and Bam H1: evidence for the double-strand break origin of chromosomal aberrations. Int J Radiat Biol Relat Stud Phys Chem Med. 1984 Jul;46(1):57–65. doi: 10.1080/09553008414551061. [DOI] [PubMed] [Google Scholar]
- Crossen P. E., Morgan W. F. Analysis of human lymphocyte cell cycle time in culture measured by sister chromatid differential staining. Exp Cell Res. 1977 Feb;104(2):453–457. doi: 10.1016/0014-4827(77)90116-1. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gebhart E., Kappauf H. Bleomycin and sister-chromatid exchange in human lymphocyte chromosomes. Mutat Res. 1978 Sep;58(1):121–124. doi: 10.1016/0165-1218(78)90104-0. [DOI] [PubMed] [Google Scholar]
- Ikushima T., Wolff S. Sister chromatid exchanges induced by light flashes to 5-bromodeoxyuridine- and 5-iododeoxyuridine substituted Chinese hamster chromosomes. Exp Cell Res. 1974 Jul;87(1):15–19. doi: 10.1016/0014-4827(74)90521-7. [DOI] [PubMed] [Google Scholar]
- Kato H. Mechanisms for sister chromatid exchanges and their relation to the production of chromosomal aberrations. Chromosoma. 1977 Feb 3;59(3):179–191. doi: 10.1007/BF00292776. [DOI] [PubMed] [Google Scholar]
- Morgan W. F., Crossen P. E. A comparison of induced sister chromatid exchange levels in Chinese hamster ovary cells and cultured human lymphocytes. Environ Mutagen. 1982;4(1):65–71. doi: 10.1002/em.2860040109. [DOI] [PubMed] [Google Scholar]
- Morgan W. F., Crossen P. E. X irradiation and sister chromatid exchange in cultured human lymphocytes. Environ Mutagen. 1980;2(2):149–155. doi: 10.1002/em.2860020207. [DOI] [PubMed] [Google Scholar]
- Morgan W. F., Djordjevic M. C., Jostes R. F., Pantelias G. E. Delayed repair of DNA single-strand breaks does not increase cytogenetic damage. Int J Radiat Biol Relat Stud Phys Chem Med. 1985 Nov;48(5):711–721. doi: 10.1080/09553008514551811. [DOI] [PubMed] [Google Scholar]
- Murnane J. P., Fuller L. F., Painter R. B. Establishment and characterization of a permanent pSV ori--transformed ataxia-telangiectasia cell line. Exp Cell Res. 1985 May;158(1):119–126. doi: 10.1016/0014-4827(85)90437-9. [DOI] [PubMed] [Google Scholar]
- Natarajan A. T., Mullenders L. H., Meijers M., Mukherjee U. Induction of sister-chromatid exchanges by restriction endonucleases. Mutat Res. 1985 Sep;144(1):33–39. doi: 10.1016/0165-7992(85)90121-6. [DOI] [PubMed] [Google Scholar]
- Natarajan A. T., Obe G. Molecular mechanisms involved in the production of chromosomal aberrations. I. Utilization of Neurospora endonuclease for the study of aberration production in G2 stage of the cell cycle. Mutat Res. 1978 Oct;52(1):137–149. doi: 10.1016/0027-5107(78)90102-1. [DOI] [PubMed] [Google Scholar]
- Natarajan A. T., Obe G. Molecular mechanisms involved in the production of chromosomal aberrations. III. Restriction endonucleases. Chromosoma. 1984;90(2):120–127. doi: 10.1007/BF00292448. [DOI] [PubMed] [Google Scholar]
- Natarajan A. T., Obe G., van Zeeland A. A., Palitti F., Meijers M., Verdegaal-Immerzeel E. A. Molecular mechanisms involved in the production of chromosomal aberrations. II. Utilization of Neurospora endonuclease for the study of aberration production by X-rays in G1 and G2 stages of the cell cycle. Mutat Res. 1980 Feb;69(2):293–305. doi: 10.1016/0027-5107(80)90094-9. [DOI] [PubMed] [Google Scholar]
- Obe G., Palitti F., Tanzarella C., Degrassi F., De Salvia R. Chromosomal aberrations induced by restriction endonucleases. Mutat Res. 1985 Jun-Jul;150(1-2):359–368. doi: 10.1016/0027-5107(85)90133-2. [DOI] [PubMed] [Google Scholar]
- PUCK T. T., CIECIURA S. J., ROBINSON A. Genetics of somatic mammalian cells. III. Long-term cultivation of euploid cells from human and animal subjects. J Exp Med. 1958 Dec 1;108(6):945–956. doi: 10.1084/jem.108.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter R. B. A replication model for sister-chromatid exchange. Mutat Res. 1980 May;70(3):337–341. doi: 10.1016/0027-5107(80)90023-8. [DOI] [PubMed] [Google Scholar]
- Perry P., Evans H. J. Cytological detection of mutagen-carcinogen exposure by sister chromatid exchange. Nature. 1975 Nov 13;258(5531):121–125. doi: 10.1038/258121a0. [DOI] [PubMed] [Google Scholar]
- Perry P., Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974 Sep 13;251(5471):156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- Sen P., Hittelman W. N. Induction of chromosome damage by Neurospora endonuclease in repair-inhibited quiescent normal human fibroblasts. Mutat Res. 1984 Dec;129(3):359–364. doi: 10.1016/0027-5107(84)90090-3. [DOI] [PubMed] [Google Scholar]
- Singer B., Kuśmierek J. T. Chemical mutagenesis. Annu Rev Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- Stuart G. W., Searle P. F., Chen H. Y., Brinster R. L., Palmiter R. D. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7318–7322. doi: 10.1073/pnas.81.23.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Winegar R. A., Preston R. J. The induction of chromosome aberrations by restriction endonucleases that produce blunt-end or cohesive-end double-strand breaks. Mutat Res. 1988 Jan;197(1):141–149. doi: 10.1016/0027-5107(88)90150-9. [DOI] [PubMed] [Google Scholar]