Abstract
Promoters were isolated at random from the genome of Saccharomyces cerevisiae by using a plasmid that contains a divergently arrayed pair of promoterless reporter genes. A comprehensive library was constructed by inserting random (DNase I-generated) fragments into the intergenic region upstream from the reporter genes. Simple in vivo assays for either reporter gene product (alcohol dehydrogenase or beta-galactosidase) allowed the rapid identification of promoters from among these random fragments. Poly(dA-dT) homopolymer tracts were present in three of five randomly cloned promoters. With two exceptions, each RNA start site detected was 40 to 100 base pairs downstream from a TATA element. All of the randomly cloned promoters were capable of activating reporter gene transcription bidirectionally. Interestingly, one of the promoter fragments originated in a region of the S. cerevisiae rDNA spacer; regulated divergent transcription (presumably by RNA polymerase II) initiated in the same region.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Bernardi A., Gaillard C., Bernardi G. The specificity of five DNAases as studied by the analysis of 5'-terminal doublets. Eur J Biochem. 1975 Apr 1;52(3):451–457. doi: 10.1111/j.1432-1033.1975.tb04013.x. [DOI] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C., Fickett J. W., Goad W. B., Lewitter F. I., Rindone W. P., Swindell C. D., Tung C. S. The GenBank genetic sequence databank. Nucleic Acids Res. 1986 Jan 10;14(1):1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Brent R. Repression of transcription in yeast. Cell. 1985 Aug;42(1):3–4. doi: 10.1016/s0092-8674(85)80091-x. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Molecular cloning and expression in E. coli of a yeast gene coding for beta-galactosidase. Cell. 1978 Sep;15(1):123–130. doi: 10.1016/0092-8674(78)90088-0. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
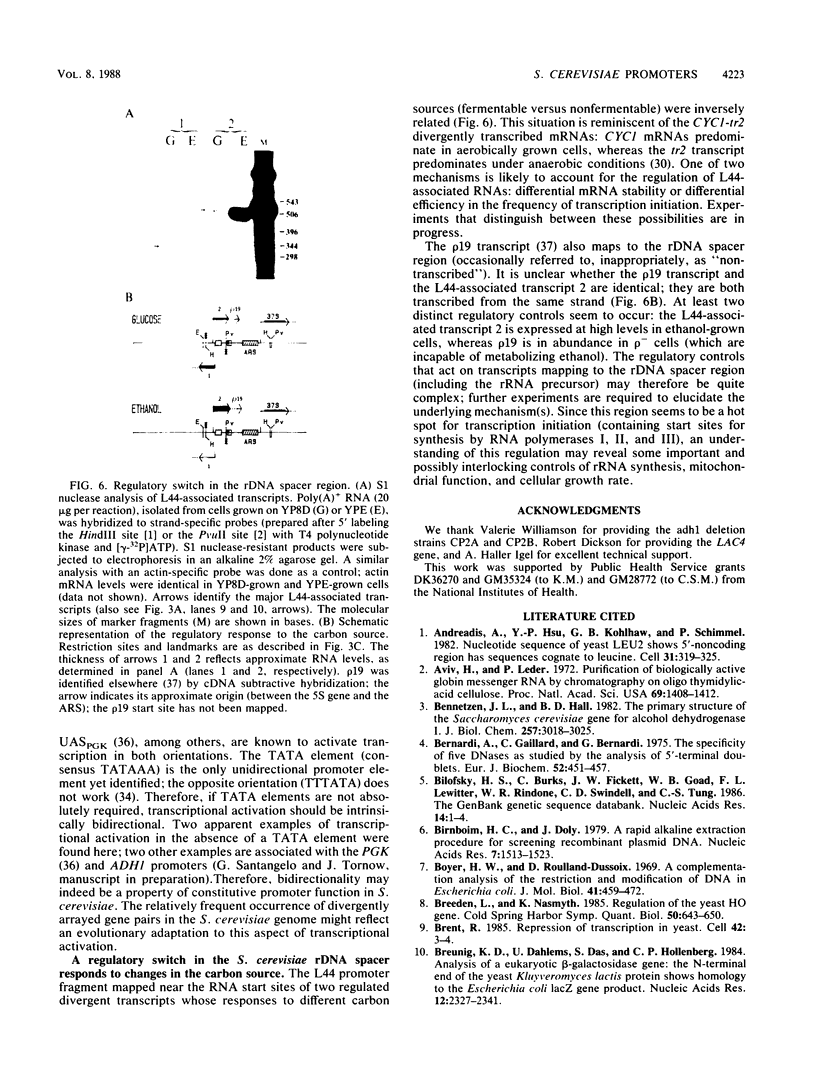
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fowler P. W., Ball A. J., Griffiths D. E. The control of alcohol dehydrogenase isozyme synthesis in Saccharomyces cerevisiae. Can J Biochem. 1972 Jan;50(1):35–43. doi: 10.1139/o72-007. [DOI] [PubMed] [Google Scholar]
- Gallis B. M., McDonnell J. P., Hopper J. E., Young E. T. Translation of poly(riboadenylic acid)-enriched messenger RNAs from the yeast, Saccharomyces cerevisiae, in heterologous cell-free systems. Biochemistry. 1975 Mar 11;14(5):1038–1046. doi: 10.1021/bi00676a024. [DOI] [PubMed] [Google Scholar]
- Garger S. J., Griffith O. M., Grill L. K. Rapid purification of plasmid DNA by a single centrifugation in a two-step cesium chloride-ethidium bromide gradient. Biochem Biophys Res Commun. 1983 Dec 28;117(3):835–842. doi: 10.1016/0006-291x(83)91672-8. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Weiss J. L., Walthall D. A., Zitomer R. S. Modulator sequences mediate oxygen regulation of CYC1 and a neighboring gene in yeast. Proc Natl Acad Sci U S A. 1983 Jan;80(1):151–155. doi: 10.1073/pnas.80.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. A bent helix in kinetoplast DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):279–283. doi: 10.1101/sqb.1983.047.01.033. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Saccharomyces cerevisiae CYC1 mRNA 5'-end positioning: analysis by in vitro mutagenesis, using synthetic duplexes with random mismatch base pairs. Mol Cell Biol. 1985 Dec;5(12):3545–3551. doi: 10.1128/mcb.5.12.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Ogden J. E., Stanway C., Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol. 1986 Dec;6(12):4335–4343. doi: 10.1128/mcb.6.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh V. S., Morgan M. M., Scott R., Clements L. S., Butow R. A. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987 Jan 30;235(4788):576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W., Murray A. W. Cloning regulated yeast genes from a pool of lacZ fusions. Methods Enzymol. 1983;101:253–269. doi: 10.1016/0076-6879(83)01019-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo G. M., Tornow J., Moldave K. Cloning of open reading frames and promoters from the Saccharomyces cerevisiae genome: construction of genomic libraries of random small fragments. Gene. 1986;46(2-3):181–186. doi: 10.1016/0378-1119(86)90402-6. [DOI] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Hoch F. L. ZINC, A COMPONENT OF YEAST ALCOHOL DEHYDROGENASE. Proc Natl Acad Sci U S A. 1955 Jun 15;41(6):327–338. doi: 10.1073/pnas.41.6.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Bennetzen J., Young E. T., Nasmyth K., Hall B. D. Isolation of the structural gene for alcohol dehydrogenase by genetic complementation in yeast. Nature. 1980 Jan 10;283(5743):214–216. doi: 10.1038/283214a0. [DOI] [PubMed] [Google Scholar]