Abstract
Sarcolipin (SLN) inhibits sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) pumps. To evaluate the physiological significance of SLN in skeletal muscle, we compared muscle contractility and SERCA activity between Sln-null and wild-type mice. SLN protein expression in wild-type mice was abundant in soleus and red gastrocnemius (RG), low in extensor digitorum longus (EDL), and absent from white gastrocnemius (WG). SERCA activity rates were increased in soleus and RG, but not in EDL or WG, from Sln-null muscles, compared with wild type. No differences were seen between wild-type and Sln-null EDL muscles in force-frequency curves or maximum rates of force development (+dF/dt). Maximum relaxation rates (−dF/dt) of EDL were higher in Sln-null than wild type across a range of submaximal stimulation frequencies, but not during a twitch or peak tetanic contraction. For soleus, no differences were seen between wild type and Sln-null in peak tetanic force or +dF/dt; however, force-frequency curves showed that peak force during a twitch and 10-Hz contraction was lower in Sln-null. Changes in the soleus force-frequency curve corresponded with faster rates of force relaxation at nearly all stimulation frequencies in Sln-null compared with wild type. Repeated tetanic stimulation of soleus caused increased (−dF/dt) in wild type, but not in Sln-null. No compensatory responses were detected in analysis of other Ca2+ regulatory proteins using Western blotting and immunohistochemistry or myosin heavy chain expression using immunofluorescence. These results show that 1) SLN regulates Ca2+-ATPase activity thereby regulating contractile kinetics in at least some skeletal muscles, 2) the functional significance of SLN is graded to the endogenous SLN expression level, and 3) SLN inhibitory effects on SERCA function are relieved in response to repeated contractions thus enhancing relaxation rates.
Keywords: knockout mouse, muscle contractility, isolated skeletal muscle, Ca2+ pump
Sarco (ENDO) plasmic reticulum (SER) Ca2+-ATPases (SERCAs) are ubiquitously expressed, integral membrane proteins that transport Ca2+ ions from the cytosol to the lumen of the SER. In the heart, a 52 amino acid transmembrane protein, phospholamban (PLN), interacts physically with SERCA2a, lowering the apparent Ca2+ affinity of the PLN-SERCA2a complex (19, 30). The inhibited complex is disrupted by phosphorylation of PLN or by elevation of cytosolic Ca2+, leading to the reversal of SERCA2a inhibition. Sarcolipin (SLN), a 31 amino acid protein, shares substantial identity with PLN in both primary sequence and gene structure (25, 35) and, like PLN, is an effective inhibitor of SERCA molecules (1–3, 16, 24). SLN was originally identified as a proteolipid that copurified with SERCA1a in rabbit fast-twitch skeletal muscle (20). Subsequently, SLN was found to be expressed highly in both human and rabbit fast-twitch skeletal muscle and to a lesser extent in slow-twitch and cardiac muscle, based on mRNA levels (25). SLN expression in the heart is largely restricted to the atrium (22), whereas PLN is highly expressed in the ventricle and not in the atrium (17). Recently, SLN-specific antibodies have been developed and used to determine the tissue-specific distribution of SLN in different muscles and species (4, 33). In mouse and rat, SLN is highly expressed in tongue, diaphragm, soleus, and atrium but not in ventricle, whereas in large mammals such as rabbit and dog, SLN is abundant in all skeletal muscles examined and in atrial but not ventricular tissues.
It became clear that PLN is a major regulator of cardiac contractility through functional analysis of Pln-null hearts (18) and hearts overexpressing either PLN or PLN mutants (37, 39). The physiological role of SLN in both cardiac and skeletal muscle has been uncovered more recently. Using intramuscular injection and electrotransfer of plasmid cDNA to overexpress SLN labeled with an NH2-terminal Flag epitope (NF-SLN) in rat soleus, we showed that SLN is an effective inhibitor of SERCA molecules in skeletal muscle (32). In that study, we found that NF-SLN reduced maximal Ca2+ transport activity in postnuclear homogenates by 31% and reduced maximal tetanic force and rates of contraction and relaxation. More recently, it has been shown that the slowing of sarcoplasmic reticulum (SR) Ca2+uptake and the speed of relaxation in both slow- and fast-twitch skeletal muscle from nebulin-null mice is associated with an upregulation of SLN protein by more than 20-fold (26). Overexpression of NF-SLN impaired Ca2+ cycling and contractility in mouse cardiac muscle, either in the presence or in the absence of endogenous PLN (2, 5, 16). The role of SLN in cardiac physiology also has been investigated in Sln-null mice (6). These studies showed that loss of SLN enhances SR Ca2+ transport and atrial contractility, establishing that SLN is a physiological regulator of SERCA2a in the atrium. The remaining question concerns the physiological role of SLN in the regulation of skeletal muscle. In the present study, we have approached this question through investigation of the physiological consequences of the loss of SLN in skeletal muscle from Sln-null mice.
We hypothesized that loss of SLN would increase the apparent affinity of SERCA for Ca2+ and increase the rate of relaxation in skeletal muscles that normally express SLN. To test these hypotheses, muscle contractility and SERCA activity were assessed in selected skeletal muscles: the white gastrocnemius and extensor digitorum longus (EDL), known to express very low levels of endogenous SLN (26), or the red gastrocnemius (RG) and soleus, known to express relatively high levels of SLN (4, 26). Soleus muscles were also subjected to a protocol of repetitive stimulation to determine whether SLN might contribute to positive lusitropy with repeated muscle contractions.
MATERIALS AND METHODS
Sln−/− mice
The generation of Sln-null mice has been described previously (6). Animals were housed 2– 4 per cage in an environmentally controlled room with a standard 12:12-h light-dark cycle and allowed access to food and water ad libitum. Experiments were performed on littermate Sln-null and wild-type males that were between the ages of 4 and 6 mo. All experimental protocols were approved by the Animal Care and Use Committees of the University of Toronto and the University of Waterloo and are consistent with the guidelines established by the Canadian Council on Animal Care.
Determination of SLN expression by RT-PCR
Total RNA was isolated from fresh muscle tissues using TRIzol reagent, essentially following the recommendations of the manufacturer (Invitrogen). RT-PCR analysis was performed using 50 ng of total RNA (14, 15), using primers specific for mouse Sln (forward: 5′-CTGAGGTCCTTGGTAGCCTG-3′ and reverse: 5′-AAGCTAAGGCTCACTGGCTG-3′). The PCR protocols were as follows: 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s (35 cycles) with a 72°C extension for 7 min.
Western blot analysis
Western blotting was performed to determine the expression of SLN, PLN, calsequestrin (CSQ), SERCA1a, and SERCA2a in skeletal muscle and heart tissues from both wild-type and Sln-null mice. Muscle tissues from Pln-null mice and wild-type littermates (18) were used as negative and positive controls for PLN protein, respectively. Tissue homogenates containing equal amount of protein (5 μg for SERCA1a, 10 μg for SERCA2a, CSQ, and SLN, and 40 μg for PLN) were separated using standard SDS-PAGE (10% for CSQ, SERCA1a, and SERCA2a) or tricine (13% for PLN and 16% for SLN). The proteins were transferred to nitrocellulose membranes and immunoprobed with specific primary antibodies—anti-rabbit CSQ1, anti-rabbit SERCA1a, anti-rabbit SERCA2a (Zymed); anti-mouse PLN (Abcam); and anti-rabbit SLN (4)—followed by horseradish peroxidase-conjugated secondary antibody. Signals were detected by Super Signal WestDura substrate (Pierce) and quantified by densitometry (ImageJ program; National Institutes of Health, Bethesda, MD).
Immunohistochemistry and immunofluorescent staining
Serial cross sections of the mounted soleus muscles (10 μm) from wild-type and Sln-null mice were cut in a cryostat maintained at −20°C. Immunohistochemical analyses of SERCA1a and SERCA2a expression were performed on individual frozen muscle sections air dried and fixed in a 100% cold acetone solution for 10 min, washed in PBS (10 mM, pH 7.2), and permeabilized using 0.5% Triton X-100. Following another wash, sections were blocked for 30 min in 1.5% horse serum at room temperature (RT). The SERCA1a [1:1,000; A52 (38)] and SERCA2a (1:250; Affinity Bioreagents) antibodies were applied to the individual sections for 1 h at RT. After the sections were washed in PBS, biotinylated horse anti-mouse immunoglobulin G (Vector Laboratories) was applied for 30 min at RT. Following another rinse in PBS, the sections were incubated for 30 min with a 1:500 dilution of a horseradish peroxidase-streptavidin conjugate (Vector Laboratories). SERCA1a and SERCA2a antibody binding were visualized using a horseradish peroxidase secondary detection system (NovaRED substrate kit, Vector Laboratories), which produces a brown-red precipitate.
To determine fiber type distribution and fiber type-specific SERCA1a and SERCA2a expression in soleus from wild-type and Sln-null mice, immunofluorescence analysis of myosin heavy chain (MHC) expression was performed as described previously (21). Briefly, cross sections were blocked with 10% goat serum followed by primary antibodies against MHCI (BA-F8), MHCIIa (SC-71), and MHCIIb (BF-F3) (Developmental Studies Hybridoma Bank). Sections were then washed in PBS and incubated with anti-mouse isotype-specific Alexa Fluor 350, Alexa Fluor 488, and Alexa Fluor 555 secondary antibodies (Molecular Probes). Additional experiments were performed on serial cross sections using an antibody against MHCIIx (6H1) (Developmental Studies Hybridoma Bank) to confirm the identity of type IIX fibers. Following another wash in PBS, sections were mounted with Prolong Gold antifade reagent (Molecular Probes). Slides were visualized with an Axio Observer Z1 structured-illumination fluorescent microscope equipped with standard Red/Green/Blue filters, an AxioCam HRm camera, and AxioVision software (Carl Zeiss). The immunofluorescent MHC-stained sections were matched with corresponding serial sections stained for SERCA1a and SERCA2a via a microscope (Nikon) linked to computer-based imaging analysis software (Image-Pro PLUS).
SERCA activity
Frozen tissues were homogenized in a solution containing (in mM) 250 sucrose, 5 HEPES, 10 NaN3, and 0.2 PMSF, using a Polytron homogenizer. Ca2+-dependent Ca2+-ATPase activity in homogenates was measured at 37°C, as described previously (12, 13). The data were analyzed by nonlinear regression with computer software (GraphPad Software), and the KCa values were calculated using an equation for a general cooperative model for substrate activation. For the smaller muscles (soleus and EDL), three muscle samples were pooled and the experiments were repeated a minimum of five times.
Electrical stimulation and muscle contractility measurements
Experiments were performed on adult 6-mo-old wild-type and Sln-null littermates (n = 6). The mice were euthanized by cervical dislocation, and the intact soleus and EDL muscles were removed and placed into oxygenated Tyrode solution (95% O2, 5% CO2) containing (in mM) 121 NaCl, 5 KCl, 0.5 MgCl2·6H2O, 0.4 NaH2PO4, 24 NaHCO3, 0.1 EDTA, 1.8 CaCl2, 5.5 glucose, pH 7.3, and maintained at 4 – 6°C. Electrically evoked muscle force was assessed in vitro at 25°C across a range of stimulation frequencies from 1 to 100 Hz, as described previously (27). Peak isometric force amplitude (mN) and the maximal rates of force development (+dF/dt) and relaxation (−dF/dt) were determined during a twitch and across a range of stimulation frequencies from 10 to 100 Hz. To determine whether SLN plays a role in mediating the lusitropic effects of skeletal muscle contractions, soleus muscles from wild-type and Sln-null mice were stimulated using a protocol consisting of 350-ms trains at 70 Hz once every 1 s for a total of 10 contractions, and comparisons were made between the 1st and 10th contractions for −dF/dt.
Statistics
Data are presented as means ± SE. In all cases, results shown are from a minimum of five to six separate experiments. Comparisons were made by Student’s t-test or ANOVA followed by appropriate post hoc test. P < 0.05 was considered significant.
RESULTS
The expression pattern of SLN in mouse cardiac and hindlimb muscles
Sln-null mice have already been partially characterized (6), but the effects of SLN ablation on skeletal muscle function have not been explored in this model. As expected, Sln mRNA was undetectable in cardiac and skeletal muscles from Sln-null mice but was readily detected in these muscles in wild-type mice using RT-PCR (not shown). Figure 1 provides semiquantitative Western blot analysis of SLN protein content in cardiac and selected hindlimb muscles in both wild-type and Sln-null mice. These analyses confirmed the absence of SLN in skeletal and cardiac muscles of Sln-null mice. For these analyses, cardiac muscle was analyzed separately as atrial and ventricular tissue to show that SLN is expressed in the atrium, but not in the ventricle in wild-type mice. Gastrocnemius muscle was fractionated into red (RG) and white (WG) portions to show that SLN is expressed in RG, but not in WG, in wild-type mice. Comparisons between skeletal muscles from wild-type mice show that SLN is expressed at relatively high levels in soleus and RG, at low levels in EDL, and not at all in WG.
Fig. 1.
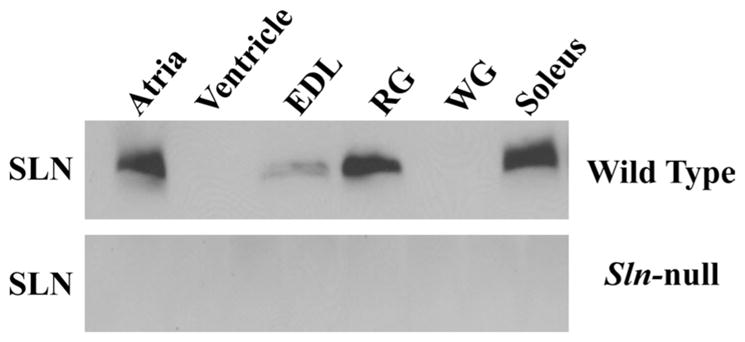
Quantification of sarcolipin (SLN) protein expression in cardiac and hindlimb muscles from wild-type and Sln-null mice. Immunoblot analyses of muscle homogenates from wild-type mice shows strong expression of SLN in atrium, red gastrocnemius (RG) and soleus muscles, low expression in extensor digitorum longus (EDL), and no expression in ventricle or white gastrocnemius (WG) muscles. As expected, muscle tissues from Sln-null mice do not contain any SLN protein.
Effect of Sln ablation on SERCA activity
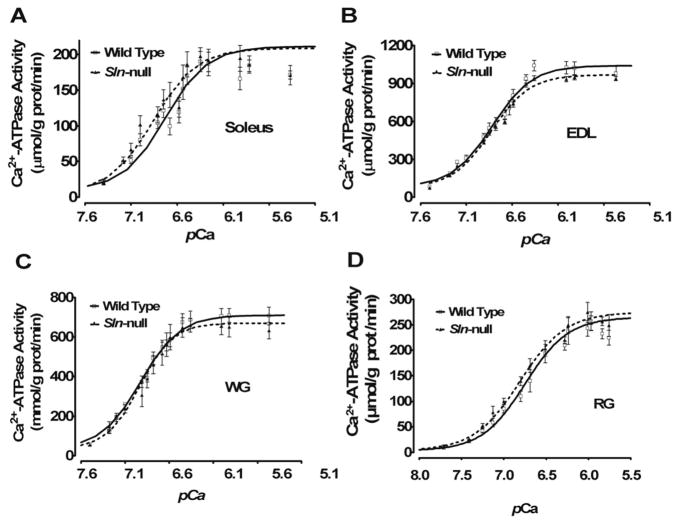
Ca2+-dependent Ca2+-ATPase activity was measured in muscle homogenates from different muscles to assess the effects of loss of SLN on SERCA function. For soleus and red gastrocnemius, with abundant SLN in wild type, loss of SLN increased the apparent affinity of SERCA for Ca2+ (Fig. 2, A and D, respectively), as indicated by a reduction (P < 0.05) in KCa and a corresponding leftward shift in SERCA activity-pCa curves (Table 1). For EDL and white gastrocnemius, with little to no SLN in wild type, no change in Ca2+ affinity was detected in Sln-null (Fig. 2, B and C, respectively).
Fig. 2.
Ca2+ dependence of sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) activity. SERCA Ca2+-dependent ATPase activity was assessed in muscle homogenates from wild-type and Sln-null mice in soleus (A), EDL (B), WG (C), and RG (D).
Table 1.
SERCA activity in mouse hindlimb muscles
| Muscle | Genotype | KCa, pCa | ΔKCa |
|---|---|---|---|
| Soleus | +/+ | 6.85 ± 0.01 | – |
| −/− | 6.95 ± 0.03* | 0.10 | |
| EDL | +/+ | 6.71 ± 0.02 | – |
| −/− | 6.73 ± 0.01 | 0.02 | |
| White gastrocnemius | +/+ | 6.80 ± 0.01 | – |
| −/− | 6.76 ± 0.03 | −0.04 | |
| Red gastrocnemius | +/+ | 6.72 ± 0.03 | – |
| −/− | 6.84 ± 0.03* | 0.12 |
Values are means ± SE. Homogenates were isolated from wild-type (+/+) and Sln-null (−/−) mouse hindlimb muscles and were analyzed for Ca2+-ATPase activity over Ca2+ concentrations ranging from pCa 8 to pCa 5 to obtain KCa. KCa is the negative logarithm of the Ca2+ concentration required to attain the half-maximal Ca2+-ATPase activity rate. SERCA, sarco(endo) plasmic reticulum Ca2+-ATPase; EDL, extensor digitorum longus.
P < 0.05, statistically significant (Sln-null vs. wild-type mice).
Effect of Sln ablation on muscle contractility
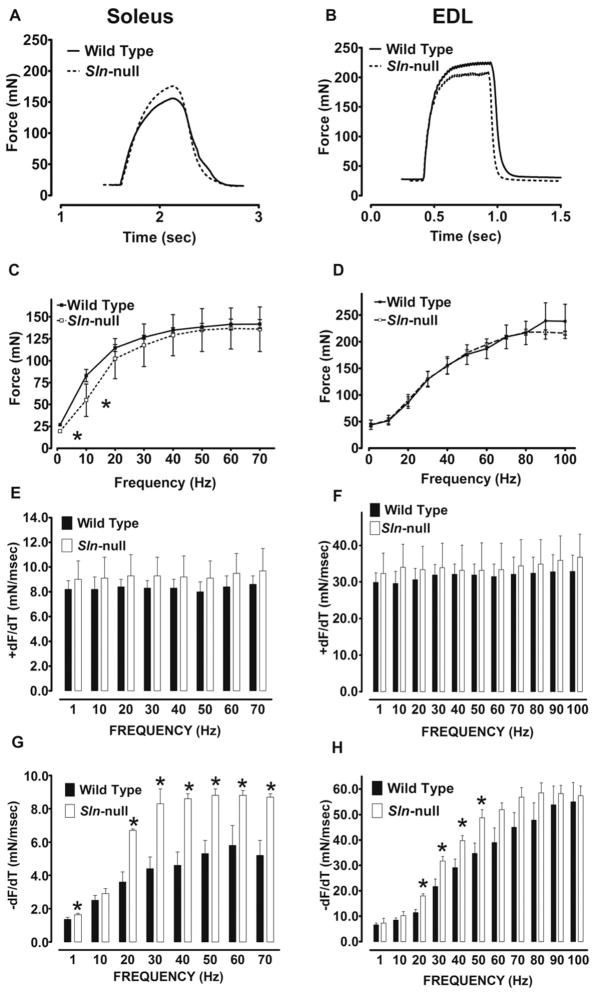
The effects of Sln ablation on skeletal muscle contractile function were assessed by measuring isometric contractile properties of isolated muscle preparations via electrical stimulation. We focused on the soleus and EDL, because these represented muscles with either low (EDL) or high (soleus) content of SLN expression. This allowed us to measure force for the entire muscle, thereby alleviating the necessity for isolating muscle bundles or surgically dissecting muscle strips.
Representative traces of tetanic contractions at 50 Hz are shown in Fig. 3, A (soleus) and B (EDL). Force-frequency relationships showed no difference between wild type and Sln-null for the EDL (Fig. 3D), whereas the curve was shifted to the right for soleus in Sln-null (Fig. 3C), owing to lower peak twitch force (P < 0.02) (Sln-null, 19.5 ± 01.6 vs. wild type, 26.7 ± 1.7) and 10 Hz force (P < 0.02) (Sln-null, 54.9 ± 18.6 vs. wild type, 83.0 ± 7.0). Maximum tetanic force was not different between wild type and Sln-null for either soleus or EDL.
Fig. 3.
Muscle contractility in soleus and EDL. Isolated skeletal muscles were subjected to force-frequency experiments, and representative tetanic contractions (single 500-ms train at 50 Hz) in soleus (A) and EDL (B) muscles from wild-type and Sln-null mice are shown. The maximum force generated (C and D) and the maximum rates of force development (+dF/dt) (E and F) and relaxation (−dF/dt) (G and H) were determined at each stimulation frequency in soleus (C, E, and G) and EDL (D, F, and H) muscles from wild-type and Sln-null mice. *Significant, P < 0.05 vs. wild type.
The maximal rates of force development (+dF/dt) and relaxation (−dF/dt) were assessed at each stimulation frequency. Loss of SLN had no effect on +dF/dt in either soleus (Fig. 3E) or EDL (Fig. 3F). However, loss of SLN improved (P < 0.05) −dF/dt in the EDL at several intermediate frequencies (Fig. 3H), whereas a much more marked improvement (P < 0.01) was seen in soleus across all stimulation frequencies except 10 Hz, but including the twitch (Fig. 3G).
Repeated tetanic stimulation (ten 350-ms trains at 70 Hz once every 1 s) of soleus caused an increase (P < 0.01) in −dF/dt in wild type (1st contraction, 5.4 ± 0.8 vs. 10th contraction, 7.9 ± 1.3), but not in Sln-null (1st contraction, 7.1 ± 1.8 vs. 10th contraction, 7.8 ± 1.2) (Fig. 4). Maximal relaxation rate measured during the last (10th) contraction was not different between wild type and Sln-null.
Fig. 4.
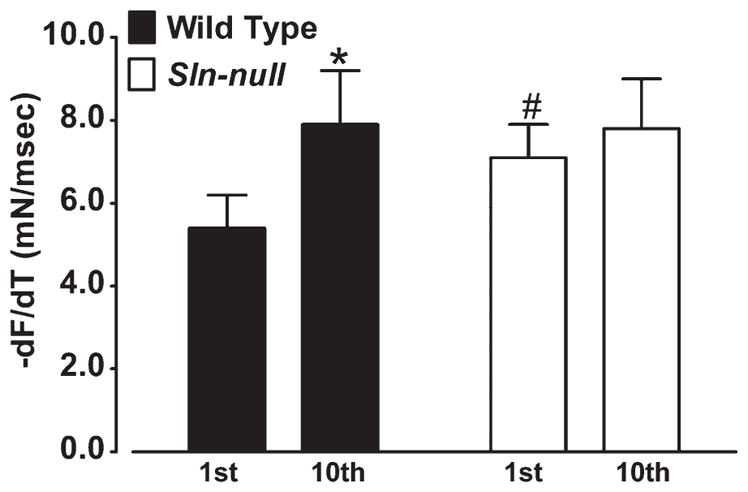
Effects of repeated contractions on maximum relaxation rate in soleus. Soleus muscles from wild-type and Sln-null mice were electrically stimulated repeatedly using a protocol consisting of 350-ms trains at 70 Hz once every 1 s for a total of 10 contractions, and comparisons were made between the 1st and 10th contractions for −dF/dt. *Significant, P < 0.05 vs. 1st contraction; #significant, P < 0.05 vs. wild type.
Effect of Sln ablation on myosin heavy chain and Ca2+ handling protein expression in soleus
Many factors can influence the rate of skeletal muscle relaxation including SERCA pump activity, the amount of calcium stored in the SR, and the properties of the contractile proteins. Therefore, it was important to determine whether adaptations involving other SR Ca2+-handling proteins or contractile proteins may have occurred to compensate for the loss of SLN.
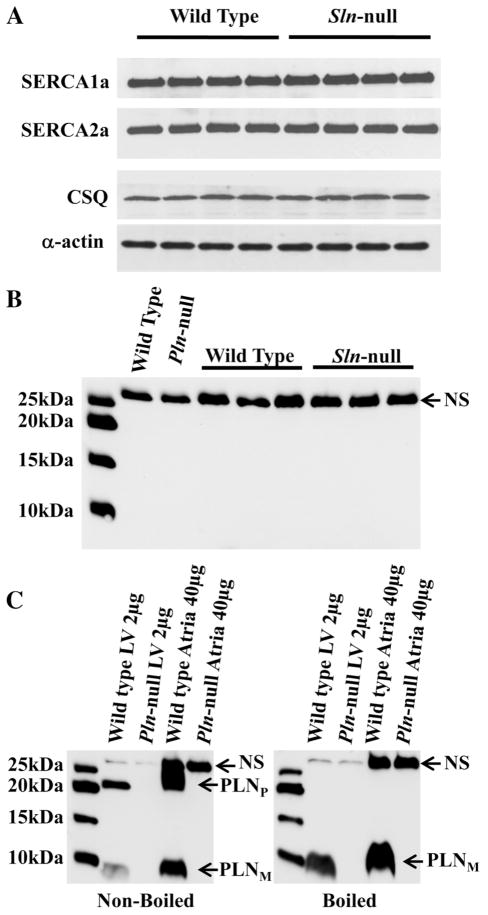
Western blotting analyses showed that the loss of SLN induced no compensatory changes in the expression of SERCA1a, SERCA2a, or CSQ in soleus (Fig. 5A) or in any of the other muscles examined (data not shown). As shown in Fig. 5B, the PLN antibody reacted with a nonspecific protein (slightly larger than 25 kDa) that is present in Pln-null soleus, but we could not detect any PLN protein in soleus from wild-type or Sln-null mice. There was also no PLN in any of the other muscles examined from wild-type or Sln-null mice (data not shown). Figure 5C shows, as expected, that PLN in both left ventricle and atria is present in pentameric and monomeric form in nonboiled samples but just monomeric form in boiled samples from wild-type mice but not Pln-null mice. It appears that the protein in soleus that reacts nonspecifically with the PLN antibody is also in left ventricle and atria and is unaffected by boiling. These results indicate clearly that there is no detectable PLN protein in mouse skeletal muscle, which means that SLN and PLN are unlikely to be coexpressed in the same muscle fibers.
Fig. 5.
Western blot analysis of SERCA1a, SERCA2a, calsequestrin (CSQ) and phospholamban (PLN). Total homogenate preparations were subjected to semiquantitative Western blotting as described in MATERIALS AND METHODS. A: the levels of SERCA1a, SERCA2a, and CSQ in soleus were not detectably different between wild-type and Sln-null mice. α-Actin was used as a loading control. The loading control sample from only one of the membranes is shown. B: soleus samples from one wild-type and Pln-null littermates and three wild-type and Sln-null littermates were incubated with anti-PLN antibody. A nonspecific (NS) protein band was detected, but PLN was not expressed in these muscles. Left lane: Precision Plus Protein Western C Standards (Bio-Rad). C: nonboiled (left) and boiled (right) left ventricle (LV) and atria samples from one wild-type and Pln-null littermates were incubated with the anti-PLN antibody. Bands indicating a nonspecific protein and the monomer (PLNM) and pentamer (PLNP) forms of PLN are shown. Left lane: Precision Plus Protein Western C Standards.
Myosin heavy chain expression of mouse soleus from wild-type and Sln-null mice was determined by immunofluorescence analysis, and representative sections are shown in Fig. 6, A and B. Fiber type analysis confirmed that there were no differences between wild-type and Sln-null mice in the relative percentage of type I (wild type, 45 ± 2.5 vs. Sln-null, 44 ± 3.3), type IIA (wild type, 46 ± 1.7 vs. Sln-null, 48 ± 2.2), type IIA/X (wild type, 2.6 ± 0.7 vs. Sln-null, 2.4 ± 0.7), or type IIX (wild type, 6.2 ± 1.2 vs. Sln-null, 5.4 ± 1.6) fibers. In EDL, the relative percentages of type IIA (wild type, 10 ± 1.5 vs. Sln-null, 10 ± 1.5), type IIA/X (wild type, 2.0 ± 0.3 vs. Sln-null, 2.5 ± 0.8), type IIX (wild type, 21 ± 2.9 vs. Sln-null, 19 ± 3.6), type IIX/B (wild type, 3.8 ± 0.8 vs. Sln-null, 3.3 ± 0.5), and type IIB (wild type, 65 ± 5.0 vs. Sln-null, 67 ± 5.0) fibers were also not different between wild-type and Sln-null mice. Immunohistochemical staining of serial cross sections was used to determine the fiber type-specific expression pattern of SERCA1a (Fig. 6, C and D) and SERCA2a (Fig. 6, E and F) in soleus from wild-type (Fig. 6, C and E) and Sln-null mice (Fig. 6, D and F). As can be seen in Fig. 6, generally, SERCA1a is confined to type II fibers and SERCA2a is confined to type I fibers in soleus from both wild-type and Sln-null mice. However, there were a few type IIA fibers in wild-type and Sln-null soleus that expressed both SERCA1a and SERCA2a (denoted by an asterisk in Fig. 6). The fiber type-specific distribution of SERCA isoform expression followed a similar pattern in EDL (data not shown).
Fig. 6.
Myosin heavy chain and SERCA isoform staining of soleus cross sections from wild-type and Sln-null mice. A and B: representative cross sections (×200) of so-leus from wild-type (A) and Sln-null (B) mice stained with myosin heavy chain antibodies to identify type I (blue), type IIA (green), and type IIX (unstained) fibers. C–F: serial cross sections showing SERCA1a (C and D) and SERCA2a (E and F) protein expression in soleus from wild-type (C and E) and Sln-null (D and F) mice. No differences in fiber type distribution or the fiber type-specific pattern of SERCA isoform expression were observed between wild-type and Sln-null mice. Asterisk (*) represents examples of type IIA fibers that stain heavily for SERCA1a but also stain lightly for SERCA2a.
DISCUSSION
It is well established that protein-protein interactions between SLN and either SERCA1a or SERCA2a lowers the apparent affinity of the SERCAs for Ca2+ (1, 24). It has been demonstrated that the rate of relaxation of force for both cardiac myocytes (2, 5, 7, 16) and slow-twitch skeletal muscle (32) that overexpress SLN is slower, thus demonstrating that Ca2+ removal by the SR limits the rate of muscle relaxation in vivo. Although these studies show that increases in SLN expression can result in altered or impaired function, it was important to determine the effects of loss of SLN on muscle function, given the likelihood that endogenous SLN protein levels are optimal for physiological function. Recently, Babu et al. (6) generated a Sln-null mouse line and demonstrated that SLN acts as a major regulator of SERCA2a, mediating the β-adrenergic responses in atria, but not in ventricles.
In this study, we examined the physiological function of SLN in skeletal muscle using the Sln-null mouse line (6). We found faster muscle relaxation rates in soleus in Sln-null mice compared with wild-type mice. This resulted from an increased affinity of SERCA for Ca2+, as demonstrated by a leftward shift in the Ca2+ activity-pCa curves measured in crude homogenates (Fig. 2). This also explains the lower twitch and 10-Hz force amplitude observed in the soleus from Sln-null mice (31). By contrast, loss of SLN resulted in only minor increases in the maximum rate of muscle relaxation in EDL, and no changes were detectable in SERCA Ca2+ affinity, as measured in crude homogenates prepared from EDL. There were also no differences in the force-frequency responses of EDL between wild-type and Sln-null mice. In keeping with these observations, SLN is expressed at much higher relative levels in soleus than in EDL. Nevertheless, the loss of SLN in EDL did enhance muscle relaxation, suggesting that the low level of SLN present in EDL has functional significance. In RG (Fig. 2D), but not WG (Fig. 2C), the SERCA activity-pCa curve was shifted to the left in Sln-null mice compared with wild type, an effect that must simply reflect differences in endogenous SLN content between these muscles. It is important to note that although there were no differences between wild-type and Sln-null mice in fiber type distribution, SERCA isoforms, PLN or CSQ expression in any muscle examined, compensatory adaptations are possible in knockout mice and we cannot rule out the possibility that other compensatory adaptations may have occurred in Sln-null mice which could have influenced our results.
SLN antibodies generated in two laboratories (4, 33) have been used to examine the level of SLN expression in mouse heart, diaphragm, quadriceps, soleus, and EDL. Studies using both antibodies and mRNA (22) have shown that SLN is expressed abundantly in atria but is low in ventricle, and these findings are confirmed in our present study. With respect to skeletal muscles, the two antibodies gave slightly different results: SLN was found to be highly expressed in diaphragm and soleus and weakly detectable in EDL using an antibody directed against the 100% conserved COOH terminus of SLN (4, 26), but it was undetectable in either soleus or EDL using an antibody directed against the variable NH2 terminus of SLN (33). We propose that the antibody directed against the highly conserved COOH terminus is the more sensitive and, therefore, the more reliable antibody. In this study we repeated measurements of the relative levels of expression of SLN in various mouse skeletal muscles using the antibody directed against the COOH terminus of SLN (4). We found that SLN was expressed abundantly in soleus and RG, to a lesser extent in EDL, and was absent from WG (Fig. 1). Compared with soleus, SLN levels were 3.8-fold lower in EDL.
It is well accepted that SERCA1a is expressed in fast-twitch skeletal muscle fibers, whereas SERCA2a is expressed in slow-twitch fibers in rats (23, 36). Here, we show that the different SERCA isoforms are, generally, also confined to entirely different muscle fibers in mouse skeletal muscle with SERCA1a expressed in fast-twitch (type II) fibers and SERCA2a expressed in slow-twitch (type I) fibers. Interestingly, a small percentage of type IIA fibers also express SERCA2a but at much lower levels than type I fibers.
On the basis of the copurification of substantial levels of SLN with SERCA1a from rabbit fast-twitch skeletal muscle (20) and its overall expression profile, we proposed that SLN may act as the counterpart of PLN in fast-twitch skeletal muscle by regulating SERCA1a function (24). However, it is now clear that SLN also acts as a physiological regulator of SERCA2a in the atrium (2, 6, 16, 22), and coinduction of Sln and Atp2a2 in skeletal muscles of α-tocopherol-deficient mice suggests that expression of SLN and SERCA2a mRNAs can be coregulated (34). Biochemical support for the view that SLN and PLN can play a similar role in the regulation of either SERCA1a or SERCA2a comes from our extensive analysis in a model system (1, 24), which showed that SLN and PLN can each regulate either SERCA1a or SERCA2a. We have also shown that, when expressed together, SLN and PLN form a superinhibitory dimeric complex (1). Thus both biochemical and physiological studies imply that SLN and PLN are alternative regulators of SERCA2a. The results of our current study now suggest that SLN can also regulate SERCA2a in some skeletal muscles. Our current observations are that WG does not express SLN or SERCA2a, EDL expresses very low levels of SLN and SERCA2a, whereas RG and soleus express higher levels of SLN and SERCA2a. However, given that whole muscles were examined in our study, it is not possible to say whether SLN associates with SERCA1a, SERCA2a, or both, since both SERCA isoforms are expressed in muscles that express SLN. We attempted to perform immunohistochemistry with the SLN antibody in this study, but the results were unclear because of nonspecific binding. This is a limitation of our current study. In any event, our results imply that SLN is a homologue of PLN that regulates both SERCA2a and SERCA1a in a variety of muscle tissues.
It would seem to be critical that the expression of SLN and the expression of PLN occur in different fibers, since their coexpression results in the formation of a superinhibitory PLN-SLN dimer that would be expected to disrupt what we would currently regard as physiological regulation of SERCA2a (1). This potential problem appears to be resolved at least for mouse skeletal muscle because we were not able to detect any PLN protein in the mouse muscles we examined in this study. Nevertheless, superinhibition of SERCA2a by PLN-SLN complexes could occur in the atria and, perhaps with different techniques, such as high-resolution immunohistochemistry, future studies may show that this does, in fact, occur physiologically. A further question to be investigated is the fact that Sln appears to be much more responsive to upregulation than the Pln gene (10, 26, 29). Further investigation of this phenomenon could uncover another layer of regulation by SLN.
Here, we assessed the lusitropic effects of repeated tetanic stimulation of soleus from wild-type and Sln-null mice to determine whether SLN plays a role in the dynamic regulation of SERCA function in skeletal muscle. We found that the relaxation rate was increased in soleus from wild-type mice in response to repeated tetanic stimulation. The soleus relaxation rates in wild-type mice were enhanced by repeated stimulation to levels matching the rates observed in Sln-null mice during a single contraction. These lusitropic effects would appear to be mediated by SLN, given that soleus muscles from Sln-null mice fail to show these effects of repeated stimulation on muscle relaxation rate and that no compensatory adaptations in the expression of PLN or SERCAs were observed in soleus from Sln-null mice. These results support the view that SLN inhibitory effects on SERCA function are relieved in response to repeated contractions.
We have shown that phosphorylation of SLN at Thr5 by serine/threonine kinase 16 can relieve SLN inhibition of SERCA2a function and improve Ca2+ dynamics in cardiomyocytes (16). Recently, phosphorylation of Thr5 by Ca2+/calmodulin-dependent protein kinase II (CaMKII) was shown to be a key mechanism involved in the regulation of SLN function in cardiac myocytes (8). Although the precise signaling path-way(s) involved in the lusitropic response to repeated contractions in soleus are unknown, given that CaMKII is enriched in the SR membrane in skeletal muscle (11), that it becomes activated during repeated muscle contractions (28), and that Thr5 is a target of CaMKII signaling, we propose that CaMKII is of primary importance (9).
In summary, we have found increased SERCA activity rates and increased contractile kinetics in Sln-null skeletal muscles such as soleus that normally express relatively high levels of SLN. Furthermore, our study provides evidence that SLN plays a major role in mediating the lusitropic effects of repeated contractions in skeletal muscle. These results demonstrate that SLN is a key modulator of SERCA function and skeletal muscle relaxation in vivo.
Acknowledgments
We thank Wenping Li for excellent technical assistance.
GRANTS
This work was supported by research grants from the Heart and Stroke Foundation (HSF) Ontario to D. H. MacLennan (T-5042) and A. O. Gramolini (T-6281) and by the Canadian Institutes of Health Research to D. H. Mac-Lennan (MOP 12545) and A. R. Tupling (MOP 86618 and MOP 47296). We also acknowledge fellowship support from TACTICS to M. G. Trivieri. P. H. Backx is a Career Investigator of HSF Ontario and A. O. Gramolini is a New Investigator of HSF Canada. A. O. Gramolini holds a Canada Research Chair.
Footnotes
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
References
- 1.Asahi M, Kurzydlowski K, Tada M, MacLennan DH. Sarcolipin inhibits polymerization of phospholamban to induce superinhibition of sarco(endo)plasmic reticulum Ca2+-ATPases (SERCAs) J Biol Chem. 2002;277:26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]
- 2.Asahi M, Otsu K, Nakayama H, Hikoso S, Takeda T, Gramolini AO, Trivieri MG, Oudit GY, Morita T, Kusakari Y, Hirano S, Hongo K, Hirotani S, Yamaguchi O, Peterson A, Backx PH, Kurihara S, Hori M, MacLennan DH. Cardiac-specific overexpression of sarcolipin inhibits sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs cardiac function in mice. Proc Natl Acad Sci USA. 2004;101:9199–9204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahi M, Sugita Y, Kurzydlowski K, de Leon S, Tada M, Toyoshima C, MacLennan DH. Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc Natl Acad Sci USA. 2003;100:5040–5045. doi: 10.1073/pnas.0330962100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babu GJ, Bhupathy P, Carnes CA, Billman GE, Periasamy M. Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J Mol Cell Cardiol. 2007;42:215–222. doi: 10.1016/j.yjmcc.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babu GJ, Bhupathy P, Petrashevskaya NN, Wang H, Raman S, Wheeler D, Jagatheesan G, Wieczorek D, Schwartz A, Janssen PM, Ziolo MT, Periasamy M. Targeted overexpression of sarcolipin in the mouse heart decreases sarcoplasmic reticulum calcium transport and cardiac contractility. J Biol Chem. 2006;281:3972–3979. doi: 10.1074/jbc.M508998200. [DOI] [PubMed] [Google Scholar]
- 6.Babu GJ, Bhupathy P, Timofeyev V, Petrashevskaya NN, Reiser PJ, Chiamvimonvat N, Periasamy M. Ablation of sarcolipin enhances sarcoplasmic reticulum calcium transport and atrial contractility. Proc Natl Acad Sci USA. 2007;104:17867–17872. doi: 10.1073/pnas.0707722104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babu GJ, Zheng Z, Natarajan P, Wheeler D, Janssen PM, Periasamy M. Overexpression of sarcolipin decreases myocyte contractility and calcium transient. Cardiovasc Res. 2005;65:177–186. doi: 10.1016/j.cardiores.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Bhupathy P, Babu GJ, Ito M, Periasamy M. Threonine-5 at the N-terminus can modulate sarcolipin function in cardiac myocytes. J Mol Cell Cardiol. 2009;47:723–729. doi: 10.1016/j.yjmcc.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhupathy P, Babu GJ, Periasamy M. Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca2+ ATPase. J Mol Cell Cardiol. 2007;42:903–911. doi: 10.1016/j.yjmcc.2007.03.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campanaro S, Romualdi C, Fanin M, Celegato B, Pacchioni B, Trevisan S, Laveder P, De Pitta C, Pegoraro E, Hayashi YK, Valle G, Angelini C, Lanfranchi G. Gene expression profiling in dysferlinopathies using a dedicated muscle microarray. Hum Mol Genet. 2002;11:3283–3298. doi: 10.1093/hmg/11.26.3283. [DOI] [PubMed] [Google Scholar]
- 11.Damiani E, Sacchetto R, Margreth A. Variation of phospholamban in slow-twitch muscle sarcoplasmic reticulum between mammalian species and a link to the substrate specificity of endogenous Ca(2+)-calmodulin-dependent protein kinase. Biochim Biophys Acta. 2000;1464:231–241. doi: 10.1016/s0005-2736(00)00153-x. [DOI] [PubMed] [Google Scholar]
- 12.Duhamel TA, Green HJ, Stewart RD, Foley KP, Smith IC, Ouyang J. Muscle metabolic, SR Ca2+-cycling responses to prolonged cycling, with and without glucose supplementation. J Appl Physiol. 2007;103:1986–1998. doi: 10.1152/japplphysiol.01440.2006. [DOI] [PubMed] [Google Scholar]
- 13.Fu M, Tupling AR. Protective effects of Hsp70 on the structure and function of SERCA2a expressed in HEK-293 cells during heat stress. Am J Physiol Heart Circ Physiol. 2009;296:H1175–H1183. doi: 10.1152/ajpheart.01276.2008. [DOI] [PubMed] [Google Scholar]
- 14.Gramolini AO, Belanger G, Thompson JM, Chakkalakal JV, Jasmin BJ. Increased expression of utrophin in a slow vs. a fast muscle involves posttranscriptional events. Am J Physiol Cell Physiol. 2001;281:C1300–C1309. doi: 10.1152/ajpcell.2001.281.4.C1300. [DOI] [PubMed] [Google Scholar]
- 15.Gramolini AO, Jasmin BJ. Expression of the utrophin gene during myogenic differentiation. Nucleic Acids Res. 1999;27:3603–3609. doi: 10.1093/nar/27.17.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gramolini AO, Trivieri MG, Oudit GY, Kislinger T, Li W, Patel MM, Emili A, Kranias EG, Backx PH, MacLennan DH. Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proc Natl Acad Sci USA. 2006;103:2446–2451. doi: 10.1073/pnas.0510883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koss KL, Ponniah S, Jones WK, Grupp IL, Kranias EG. Differential phospholamban gene expression in murine cardiac compartments. Molecular and physiological analyses. Circ Res. 1995;77:342–353. doi: 10.1161/01.res.77.2.342. [DOI] [PubMed] [Google Scholar]
- 18.Luo W, Grupp IL, Harrer J, Ponniah S, Grupp G, Duffy JJ, Doetschman T, Kranias EG. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 19.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 20.MacLennan DH, Yip CC, Iles GH, Seeman P. Isolation of sarcoplasmic reticulum proteins. Cold Spring Harb Symp Quant Biol. 1972;37:469–478. [Google Scholar]
- 21.McMillan EM, Quadrilatero J. Differential apoptosis-related protein expression, mitochondrial properties, proteolytic enzyme activity, and DNA fragmentation between skeletal muscles. Am J Physiol Regul Integr Comp Physiol. 2011;300:R531–R543. doi: 10.1152/ajpregu.00488.2010. [DOI] [PubMed] [Google Scholar]
- 22.Minamisawa S, Wang Y, Chen J, Ishikawa Y, Chien KR, Matsuoka R. Atrial chamber-specific expression of sarcolipin is regulated during development and hypertrophic remodeling. J Biol Chem. 2003;278:9570–9575. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- 23.Murphy RM, Larkins NT, Mollica JP, Beard NA, Lamb GD. Calsequestrin content and SERCA determine normal and maximal Ca2+ storage levels in sarcoplasmic reticulum of fast- and slow-twitch fibres of rat. J Physiol. 2009;587:443–460. doi: 10.1113/jphysiol.2008.163162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odermatt A, Becker S, Khanna VK, Kurzydlowski K, Leisner E, Pette D, MacLennan DH. Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem. 1998;273:12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 25.Odermatt A, Taschner PE, Scherer SW, Beatty B, Khanna VK, Cornblath DR, Chaudhry V, Yee WC, Schrank B, Karpati G, Breuning MH, Knoers N, MacLennan DH. Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics. 1997;45:541–553. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 26.Ottenheijm CAC, Fong C, Vangheluwe P, Wuytack F, Babu GJ, Periasamy M, Witt CC, Labeit S, Granzier H. Sarcoplasmic reticulum calcium uptake and speed of relaxation are depressed in nebulin-free skeletal muscle. FASEB J. 2008;22:2912–2919. doi: 10.1096/fj.07-104372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y, Zvaritch E, Tupling AR, Rice WJ, de Leon S, Rudnicki M, McKerlie C, Banwell BL, MacLennan DH. Targeted disruption of the ATP2A1 gene encoding the sarco(endo)plasmic reticulum Ca2+ ATPase isoform 1 (SERCA1) impairs diaphragm function and is lethal in neonatal mice. J Biol Chem. 2003;278:13367–13375. doi: 10.1074/jbc.M213228200. [DOI] [PubMed] [Google Scholar]
- 28.Rose AJ, Kiens B, Richter EA. Ca2+-calmodulin-dependent protein kinase expression and signalling in skeletal muscle during exercise. J Physiol. 2006;574:889–903. doi: 10.1113/jphysiol.2006.111757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schertzer JD, van der Poel C, Shavlakadze T, Grounds MD, Lynch GS. Muscle-specific overexpression of IGF-I improves E-C coupling in skeletal muscle fibers from dystrophic mdx mice. Am J Physiol Cell Physiol. 2008;294:C161–C168. doi: 10.1152/ajpcell.00399.2007. [DOI] [PubMed] [Google Scholar]
- 30.Simmerman HK, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 31.Sun YB, Lou F, Edman KA. The relationship between the intracellular Ca2+ transient and the isometric twitch force in frog muscle fibres. Exp Physiol. 1996;81:711–724. doi: 10.1113/expphysiol.1996.sp003971. [DOI] [PubMed] [Google Scholar]
- 32.Tupling AR, Asahi M, MacLennan DH. Sarcolipin overexpression in rat slow twitch muscle inhibits sarcoplasmic reticulum Ca2+ uptake and impairs contractile function. J Biol Chem. 2002;277:44740–44746. doi: 10.1074/jbc.M206171200. [DOI] [PubMed] [Google Scholar]
- 33.Vangheluwe P, Schuermans M, Zador E, Waelkens E, Raeymaekers L, Wuytack F. Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem J. 2005;389:151–159. doi: 10.1042/BJ20050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasu VT, Ott S, Hobson B, Rashidi V, Oommen S, Cross CE, Gohil K. Sarcolipin and ubiquitin carboxy-terminal hydrolase 1 mRNAs are over-expressed in skeletal muscles of α-tocopherol deficient mice. Free Radic Res. 2009;43:106–116. doi: 10.1080/10715760802616676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wawrzynow A, Theibert JL, Murphy C, Jona I, Martonosi A, Collins JH. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch Biochem Biophys. 1992;298:620–623. doi: 10.1016/0003-9861(92)90457-8. [DOI] [PubMed] [Google Scholar]
- 36.Wu KD, Lytton J. Molecular cloning and quantification of sarcoplasmic reticulum Ca(2+)-ATPase isoforms in rat muscles. Am J Physiol Cell Physiol. 1993;264:C333–C341. doi: 10.1152/ajpcell.1993.264.2.C333. [DOI] [PubMed] [Google Scholar]
- 37.Zhai J, Schmidt AG, Hoit BD, Kimura Y, MacLennan DH, Kranias EG. Cardiac-specific overexpression of a superinhibitory pentameric phospholamban mutant enhances inhibition of cardiac function in vivo. J Biol Chem. 2000;275:10538–10544. doi: 10.1074/jbc.275.14.10538. [DOI] [PubMed] [Google Scholar]
- 38.Zubrzycka-Gaarn E, Phillips L, MacLennan DH. Monoclonal antibodies to the Ca2+ + Mg2+-dependent ATPase of skeletal muscle sarcoplasmic reticulum–cross-reactivity with ATPase isozymes and other Ca2+-binding proteins. Prog Clin Biol Res. 1984;168:19–23. [PubMed] [Google Scholar]
- 39.Zvaritch E, Backx PH, Jirik F, Kimura Y, de Leon S, Schmidt AG, Hoit BD, Lester JW, Kranias EG, MacLennan DH. The transgenic expression of highly inhibitory monomeric forms of phospholamban in mouse heart impairs cardiac contractility. J Biol Chem. 2000;275:14985–14991. doi: 10.1074/jbc.275.20.14985. [DOI] [PubMed] [Google Scholar]