Abstract
The prevalence of ESBL has been increasing worldwide. In this study, we investigated the molecular characteristics of ESBL among clinical isolates of Escherichia coli from a Japanese tertiary hospital. A total of 71 consecutive and nonduplicate clinical isolates of ESBL-positive E. coli collected at Tohoku University Hospital between January 2008 and March 2011 were studied. The antimicrobial susceptibility profile of these strains was determined. PCR and sequencing were performed to identify genes for β-lactamase (bla TEM, bla SHV, bla OXA-1-like, and bla CTX-M) and plasmid-mediated quinolone resistance determinants (PMQR). The isolates were also analyzed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Of the 71 strains, 68 were positive for CTX-M, 28 were positive for TEM, four were positive for OXA-1, and one was positive for SHV. Sequencing revealed that CTX-M-14 was the most prevalent (31/71), followed by CTX-M-27 (21/71) and then CTX-M-15 (9/71). Of the 28 TEM-positive strains, one was TEM-10 and the rest were TEM-1. One SHV-positive strain was SHV-12. The 21 CTX-M-27-producing isolates were divided into 14 unique PFGE types, while the 9 CTX-M-15 producers were divided into 8 types. Based on MLST, 9 CTX-M-14 procedures, 19 CTX-M-27 procedures, and 8 CTX-M-15 producers belonged to ST131. Thirty-five (94.6%) of the 37 ST131 E. coli strains showed resistance to levofloxacin, which was a higher rate than among non-ST131 strains (63.6%). Among ESBL-producing isolates, one, two, and six possessed qnrB, qnrS, qepA, and aac(6′)-Ib-cr, respectively. Of the 6 isolates with aac(6′)-Ib-cr, 4 carried the CTX-M-15 gene. Our data suggest that CTX-M-15-producing E. coli ST131 has emerged as a worldwide pandemic clone, while CTX-M-27 (a variant of CTX-M-14) is also spreading among E. coli ST131 in Japan.
Introduction
The ability of bacteria to produce extended-spectrum β-lactamases (ESBL) that hydrolyze penicillins, cephalosporins, and monobactams has resulted in intractable infections and serious consequences for infection control. Recently, the prevalence of ESBL procedures has been increasing and infections caused by these bacteria have become an emerging public health concern worldwide [1], [2].
ESBL can be classified into three main types, which are designated as TEM, SHV, and CTX-M. The CTX-M type of ESBL can be further classified into three groups, which are CTX-M-1, CTX-M-2, and CTX-M-9. In the 1990s, ESBL were generally found in Klebsiella pneumonia (TEM or SHV types) and most isolates were from nosocomial infections. Since 2000, however, the worldwide distribution of ESBL producers has shifted towards Escherichia coli with CTX-M type and isolates are obtained from both inpatients and outpatients [3]–[7]. In particular, CTX-M-15 (which belongs to the CTX-M-1 group) is widely distributed around the world [2]. In Japan, E. coli producing CTX-M type ESBL have also been emerging. In the early 2000s, the dominant CTX-M group underwent a shift from CTX-M-2 to CTX-M-9 [8].
There have also been reports about quinolone resistance among ESBL producers [7]. Quinolone resistance is usually caused by chromosomal mutations, but can also be related to plasmid-mediated quinolone resistance (PMQR) genes, including qnrA, qnrB, qnrC, qnrS, qepA, and aac(6′)-Ib-cr [9]. Several studies have indicated that the emergence of PMQR determinants in ESBL-producing Enterobacteriaceae poses a global threat [10], [11]. However, there have been few Japanese reports about the detection of PMQR and the prevalence of PMQR determinants among ESBL producers in Japan remains unclear [12].
In this study, we investigated the molecular characteristics and epidemiology of clinical isolates of ESBL-producing E. coli obtained at a Japanese tertiary hospital. This work was presented in part at the 52th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), San Francisco, 2010.
Materials and Methods
Bacterial Strains
A total of 71 (2.9%) consecutive and non-duplicate clinical isolates of ESBL-producing E. coli were collected from among 2,488 E. coli isolates at Tohoku University Hospital during the period from January 2008 to March 2011. Each isolate was identified by using the VITEK 2 system (Sysmex bioMérieux Co., Ltd., Tokyo, Japan), and initial screening for ESBL was done with the VITEK 2 Advanced Expert System (Sysmex bioMérieux Co.) according to the manufacturer’s instructions. ESBL production was confirmed by the combined disk test according to CLSI guidelines [13], [14]. Among the 71 ESBL-producing strains of E. coli, 37 (52.1%) were isolated from urine, nine (12.7%) from sputum, five (7.0%) from blood, 3 (4.2%) from stools and abscesses, two (2.8%) from the pharynx and a wound, and 10 (14.1%) from other sites.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) of various antimicrobial agents was determined by the agar dilution method according to CLSI guidelines [13], [14]. The following antimicrobial agents were tested in this study: ampicillin, piperacillin, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, cefoxitin, imipenem, meropenem, aztreonam, levofloxacin, gentamicin, and amikacin. Quality control for the MIC analyses was performed with E. coli ATCC 35218 and E. coli ATCC 25922.
Detection of ESBL Genes and DNA Sequencing
PCR was performed with TaKaRa Ex Taq (Takara Bio Inc., Otsu, Japan) to identify ESBL genes, including bla TEM, bla SHV, bla OXA-1-like, and bla CTX-M [15], [16]. For CTX-M-positive strains, the CTX-M group was determined by PCR using CTX-M-1, CTX-M-2, and CTX-M-9 group-specific primers [16]. PCR products of the TEM, SHV, OXA-1-like, CTX-M-1 group, CTX-M-2 group, and CTX-M-9 group genes were sequenced on both strands with an ABI3730XL analyzer (Applied Biosystems, Foster City, CA, USA). BLAST version 2.2.24 (http://blast.ddbj.nig.ac.jp/) was used to process the sequencing data and identify genes. To differentiate bla CTX-M-15 from bla CTX-M-28 in the CTX-M-15-positive strains, the published primer pair was used for PCR and sequencing [17].
PMQR Gene Detection and DNA Sequencing
Detection of qnrA, qnrB, qnrC, qnrS, aac(6′)-Ib, and qepA in the isolates was performed by PCR, as described previously [15]. The aac(6′)-Ib amplicons were also sequenced to identify the -cr variant, as described above.
Pulsed-Field Gel Electrophoresis
For CTX-M-15- and CTX-M-27-positive strains, evaluation of chromosomal polymorphism was done by pulsed-field gel electrophoresis (PFGE) using the XbaI restriction enzyme (Takara Bio Inc.), as described previously [18]. Electrophoresis was performed on 1% PFGE agarose gel with a CHEF-DR III system (Bio-Rad Laboratories, Richmond, CA, USA), and electrophoretic patterns were analysed with GelCompar II software (Applied Maths, Kortrijik, Belgium). Isolates that showed >85% similarity were considered to reside within a single cluster [18].
Multilocus Sequence Typing
For CTX-M-14-, CTX-M-15-, and CTX-M-27-positive strains, multilocus sequence typing (MLST) was performed using seven housekeeping genes (adk, fumC, icd, purA, gyrB, recA, and mdh) according to the method of Jørgensen et al. [19]. DNA sequence variations were analyzed by using a MLST database for E. coli (http://mlst.ucc.ie/mlst/dbs/Ecoli).
Statistical Analysis
Statistical significance was evaluated by the chi-square test with Yates’ correction or Fisher’s exact test, and a p value of less than 0.05 was considered to be significant.
Ethics
This study focused on bacterial strains that were isolated for treatment. In addition, this study was completely anonymous and no identifiable information was obtained. According to the ethical guidelines for epidemiological studies released by the Ministry of Health, Labour, and Welfare in Japan [20], ethical approval and written or verbal informed consent are not required for this type of study.
Results
Antimicrobial Susceptibility Profile
The results of antimicrobial susceptibility testing are shown in Table 1. Meropenem was the most active agent (100% susceptibility; MIC range: ≤0.03–0.5 mg/L). Over 90% of the 71 strains showed intermediate resistance or resistance to ampicillin, piperacillin, cefotaxime, and aztreonam. In addition, 62.0% and 67.6% of the isolates showed intermediate resistance or resistance to ceftazidime and cefepime, respectively, while 32.4% displayed intermediate resistance or resistance to piperacillin-tazobactam.
Table 1. Susceptibility profile of ESBL-positive E. coli.
| Antimicrobial Agent | MIC (µg/ml) for all isolates (n = 71) | Percent non-susceptible a) | ||
| min | max | MIC90 | ||
| ampicillin | 256 | 256 | 256 | 100 |
| piperacillin | 64 | 256 | 256 | 100 |
| piperacillin-tazobactam | 2/4– | 256/4 | 256/4 | 32.4 |
| cefotaxime | 8 | 256 | 256 | 100 |
| ceftazidime | 1 | 256 | 64 | 93.0 |
| cefepime | 2 | 256 | 64 | 67.6 |
| cefoxitin | 4 | 256 | 64 | 62.0 |
| imipenem | 0.03 | 1 | 0.25 | 0 |
| meropenem | 0.03 | 0.5 | 0.03 | 0 |
| aztreonam | 2 | 256 | 128 | 90.1 |
| levofloxacin | 0.13 | 256 | 128 | 78.9 |
| gentamicin | 1 | 256 | 256 | 35.2 |
| amikacin | 4 | 256 | 16 | 9.9 |
Furthermore, 78.9% of all isolates demonstrated intermediate resistance or resistance to levofloxacin, while intermediate resistance or resistance to gentamicin and amikacin was noted in 35.2% and 9.9%, respectively.
ESBL Typing and Antimicrobial Susceptibility Profile
Among the 71 strains, 68 strains (95.8%) were positive for CTX-M, 28 strains (39.4%) were positive for TEM, four strains (5.6%) were positive for OXA-1-like, and one strain (1.4%) was positive for SHV. Among the 68 CTX-M-positive strains, 13 were from the CTX-M-1 group, one was from the CTX-M-2 group, and 54 were from the CTX-M-9 group.
DNA sequencing revealed that CTX-M-14 was most common (31 strains: 45.6%), followed by CTX-M-27 (21 strains: 30.9%) and then CTX-M-15 (9 strains: 13.2%). The only CTX-M-2 group-positive strain was CTX-M-2. Of the 28 TEM-positive strains, one was TEM-10 and the other 27 were TEM-1, and all 27 TEM-1-positive isolates possessed the CTX-M gene. One SHV-positive strain was SHV-12, and this isolate possessed CTX-M-14. All of the four OXA-1-like were OXA-1, and these isolates possessed CTX-M-15.
All 9 of the CTX-M-15-positive isolates showed intermediate resistance or resistance to ceftazidime, as did 18 (85.7%) of the 21 CTX-M-27-positive isolates. These rates of resistance were higher than those found in the other CTX-M-positive isolates (50%). The antimicrobial susceptibility profiles of CTX-M-15-positive strains are shown in Table 2.
Table 2. β-lactamases, PMQR, and antimicrobial susceptibility profiles of CTX-M-15-positive isolates.
| No. of strains | β-lacrtamases | PMQR | ST | PIPC | PIPC/TAZ | CAZ | CTX | CFPM | AZT | MEPM | LVFX | GM | AMK |
| 9 | CTX-M-15, TEM-1 | – | ST131 | ?256 | 128 | 32 | ?256 | 32 | 64 | ?0.03 | 2 | 128 | 8 |
| 24 | CTX-M-15, OXA-1 | aac(6′)-Ib-cr | ST131 | ?256 | 16 | 64 | ?256 | 32 | 64 | ?0.03 | 32 | 64 | 32 |
| 40 | CTX-M-15, OXA-1 | aac(6′)-Ib-cr | ST131 | ?256 | 8 | 32 | ?256 | 16 | 64 | ?0.03 | 32 | 128 | 16 |
| 45 | CTX-M-15, TEM-1, OXA-1 | aac(6′)-Ib-cr | ST131 | ?256 | 128 | 8 | 64 | 16 | 64 | ?0.03 | 64 | 4 | 32 |
| 55 | CTX-M-15, OXA-1 | aac(6′)-Ib-cr | ST131 | ?256 | 128 | 128 | ?256 | 128 | 128 | ?0.03 | 32 | 4 | 32 |
| 56 | CTX-M-15 | – | ST131 | ?256 | ?256 | ?256 | ?256 | ?256 | ?256 | ?0.03 | 32 | 4 | 32 |
| 57 | CTX-M-15 | – | ST131 | ?256 | ?256 | ?256 | ?256 | ?256 | ?256 | ?0.03 | 64 | ?256 | 16 |
| 62 | CTX-M-15 | – | ST131 | ?256 | ?256 | ?256 | ?256 | ?256 | ?256 | ?0.03 | 64 | 2 | 16 |
| 72 | CTX-M-15, TEM-1 | – | ST58 | ?256 | 64 | 64 | ?256 | 128 | 128 | ?0.03 | 0.13 | 1 | 4 |
PMQR Gene Typing
One isolate was positive for qnrB, one for qnrS, and two for qepA in this study. None of the isolates were positive for qnrA or qepA. Among a total of 71 isolates, 7 carried aac(6′)-Ib genes, and sequencing revealed aac(6′)-Ib-cr in six of these 7 strains.
Analysis of PMQR-Positive Strains
Among the 10 PMQR-positive isolates, six possessed resistance genes from the CTX-M-1 group. Of the six isolates with aac(6′)-Ib-cr genes, four (66.7%) carried the CTX-M-15 gene. The SHV-12-positive strains possessed qnrB genes. All of the PMQR-positive strains were resistant to levofloxacin (MIC: 32 - ≥ 256 mg/L).
PFGE
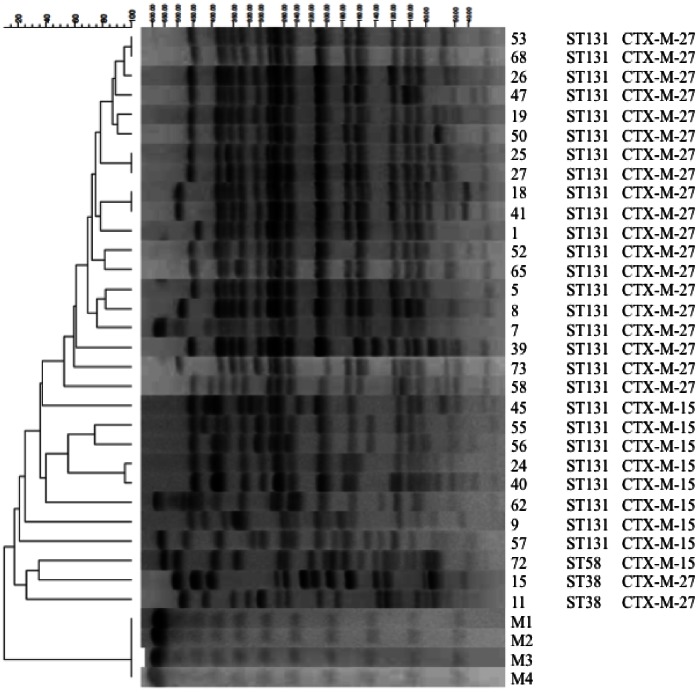
Figures 1 shows a dendrogram and PFGE of XbaI-digested genomic DNA from 9 CTX-M-15-producing and 21 CTX-M-27-producing strains of E. coli. The 9 CTX-M-15-producing isolates were divided into 8 unique PFGE types, and the commonest type was found in 2 patients. The 21 CTX-M-27-producing isolates could be divided into 14 unique PFGE types, with the most common type being found in 6 patients.
Figure 1. MLST and PFGE of XbaI-digested genomic DNA from CTX-M-15- and CTX-M-27-producing E. coli strains.
The 9 CTX-M-15-producing isolates were divided into 8 unique PFGE types. The 21 CTX-M-27-producing isolates were divided into 14 unique PFGE types, with the most common type being found in 6 patients. M1∼4: Lambda Ladder.
MLST
Based on the results of MLST analysis, 8 (88.9%) of the 9 CTX-M-15-producing E. coli strains belonged to ST131 and one strain (11.1%) belonged to ST58. Among the 21 CTX-M-27-producing strains, 19 (90.5%) belonged to ST131 and two (9.5%) belonged to ST38. The CTX-M-14-producing strains were divided into 8 unique MLST types, and 10 (32.2%) of these strains were ST131 (Table 3). Of the 37 ST131 isolates, 35 (94.6%) showed resistance to levofloxacin, and this rate of resistance was significantly higher than that among non-ST131 strains (58.3%).
Table 3. MLST types of CTX-M-14-, CTX-M-15-, and CTX-M-27-positive strains.
| CTX type | n | Sequence type | ||||||||
| 10 | 38 | 58 | 68 | 92 | 95 | 131 | 357 | 648 | ||
| CTX-M-14 | 31 | 1 | 6 | 5 | 1 | 2 | 10 | 2 | 4 | |
| CTX-M-15 | 9 | 1 | 8 | |||||||
| CTX-M-27 | 21 | 2 | 19 | |||||||
Discussion
Currently, the CTX-M type is predominant among ESBL producers around the world. Many epidemiological studies of CTX-M type ESBL have been performed in different countries [1], [2], and CTX-M-15-producing E. coli ST131 have spread worldwide [2]. In the present study, a total of nine CTX-M-15-positive strains (eight belonging to ST131) were isolated. There has been one previous report about detection of CTX-M-15- producing E. coli ST131 in Japanese patients [21]. The present study demonstrated that these strains can be frequently isolated in a Japanese tertiary hospital.
In the present study, four of the six aac(6′)-Ib-cr positive strains possessed CTX-M-15, and we demonstrated a significant association between the CTX-M-15 and aac(6′)-Ib-cr genes. It has been reported that isolates with the aac(6′)-Ib-cr gene often possess a CTX-M-15-producing plasmid [5], [22]. This suggests that spread of the aac(6′)-Ib-cr gene might occur concurrently with the CTX-M-15 gene. The aac(6′)-Ib-cr gene also confers resistance to aminoglycosides, so CTX-M-15 producers can develop multidrug resistance. In addition, most E. coli ST131 isolates identified around the world show resistance to quinolones confirmed by chromosomal mutations. In our study, 35 (94.6%) of the 37 E. coli ST131 isolates showed resistance to levofloxacin, and 7 of the 8 CTX-M-15-producing E. coli ST131 strains were resistant to levofloxacin. Quinolones are frequently used to treat infections among outpatients in many countries, including Japan, but there is concern that these drugs will not be effective against CTX-M-15 producers and will apply selection pressure to these isolates.
The 9 CTX-M-15-producing isolates could be divided into 8 unique PFGE types, while the 21 CTX-M-27-producing isolates were divided into 14 unique PFGE types with the most common type being found in 6 patients. These results suggested that certain clones were spreading in our hospital and that nosocomial infection was occurring, so improved infection control and surveillance is required.
We found 21 isolates that produced CTX-M-27 in this study. CTX-M-27 is a variant of CTX-M-14, which only differs by the substitution D240G [23]. Based on MLST analysis, CTX-M-14-producing strains were divided into 8 unique MLST types, and 10 (32.2%) of these strains were ST131. Most of the CTX-M-27-producing strains (19 strains: 90.5%) also belonged to ST131 (Table 3). Furthermore, the 14 unique clones of CTX-M-27-positive strains identified by PFGE analysis, 12 belonged to ST131. We previously reported that CTX-M-27 producers were frequently isolated in the clinical setting in Japan [21]. There have been no reports about a cluster of CTX-M-27-producing pathogens in other countries, so CTX-M-27-producing E. coli may have arisen in Japan due to a point mutation of the CTX-M-14 gene in E. coli ST131.
Several case reports of unusually severe or fatal extraintestinal infections due to E. coli ST131 [24]-[27] have suggested that the rapid and extensive emergence of such strains may be partly due to high virulence compared with other E. coli types. In addition, it was previously demonstrated that CTX-M-27 confers stronger resistance to CAZ than CTX-M-14 [23]. Thus, we have to be concerned that CTX-M-27-producing E. coli ST131 could become predominant over CTX-M-14 in Japan because of its high virulence and selection pressure due to use of CAZ.
A limitation of this study is that we did not investigate the CTX-M-8 and CTX-M-25 groups among CTX-M-type ESBL groups. However, these two groups have not yet been identified in Japan. The dominant group of ESBL-producing E. coli was CTX-M-2 until 2000, while CTX-M-9 has been the dominant group since 2000. This study showed increasing emergence of CTX-M-15 from the CTX-M-1 group, which was rarely reported previously in Japan. The dominant type of ESBL may change again in the future, so that investigation of the CTX-M-8 and CTX-M-25 groups, as well as rarely identified PER, VEB, and IBC type ESBL, may be necessary.
In conclusion, this study revealed that CTX-M-15-producing E. coli ST131 (a worldwide pandemic clone) has emerged in Japan. Our findings also suggest that CTX-M-27 (a variant of CTX-M-14) is spreading among clinical isolates of E. coli ST131 in the Japanese tertiary hospital setting.
Funding Statement
The authors have no support or funding to report.
References
- 1. Cantón R, Coque TM (2006) The CTX-M β-lactamase pandemic. Current Opinion in Microbiology 9: 466–75. [DOI] [PubMed] [Google Scholar]
- 2. Pitout JDD, Laupland KB (2008) Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8: 159–166. [DOI] [PubMed] [Google Scholar]
- 3. Bonnet R (2004) Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 48: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heffernana HM, Woodhousea RE, Popea CE, Blackmore TK (2009) Prevalence and types of extended-spectrum β-lactamases among urinary Escherichia coli and Klebsiella spp. in New Zealand. Int J Antimicrob Agents 34: 544–549. [DOI] [PubMed] [Google Scholar]
- 5. Peirano G, Pitout JDD (2010) Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35: 316–321. [DOI] [PubMed] [Google Scholar]
- 6. Pitout JD, Church DL, Gregson DB, Chow BL, McCracken M, Mulvey MR, et al. (2007) Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob Agents Chemother 51: 1281–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pitout JDD, Hanson ND, Church DL, Laupland KB (2004) Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum β-lactamases: importance of community isolates with bla CTX-M genes. Clin Infect Dis 38: 1736–1741. [DOI] [PubMed] [Google Scholar]
- 8. Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, et al. (2009) Change in the prevalence of extended-spectrum β-actamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 63: 72–79. [DOI] [PubMed] [Google Scholar]
- 9. Martinez-Martinez L, Pascual A, Jacoby GA (1998) Quinolone resistance from a transferable plasmid. Lancet 351: 797–799. [DOI] [PubMed] [Google Scholar]
- 10. Lautenbach E, Strom BL, Bilker WB, Patel JB, Edelstein PH, et al. (2001) Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae . Clin Infect Dis 33: 1288–1294. [DOI] [PubMed] [Google Scholar]
- 11. Robicsek A, Jacoby GA, Hooper DC (2006) The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6: 629–640. [DOI] [PubMed] [Google Scholar]
- 12. Ode T, Saito R, Kumita W, Sato K, Okugawa S, et al. (20099 Analysis of plasmid-mediated multidrug resistance in Escherichia coli and Klebsiella oxytoca isolates from clinical specimens in Japan. Int J Antimicrob Agents 34: 347–350. [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. (2009) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Eighth Edition. M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA, USA.
- 14.Clinical and Laboratory Standards Institute. (2009) Performance standards for antimicrobial susceptibility testing; Nineteenth informational supplement. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA, USA.
- 15. Kanamori H, Yano H, Hirakata Y, Endo S, Arai K, et al. (2011) High prevalence of extended-spectrum β-lactamases and qnr determinants in Citrobacter species from Japan: Dissemination of CTX-M-2. J Antimicrob Chemother 66: 2255–2262. [DOI] [PubMed] [Google Scholar]
- 16. Karisik E, Ellington MJ, Pike R, Warren RE, Livermore DM, et al. (2006) Molecular characterization of plasmids encoding CTX-M-15 β-lactamases from Escherichia coli strains in the United Kingdom. J Antimicrob Chemother 58: 665–668. [DOI] [PubMed] [Google Scholar]
- 17. Muzaheed, Doi Y, Adams-Haduch JM, Endimiani A, Sidjabat HE, et al. (2008) High prevalence of CTX-M-15-producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. J Antimicrob Chemother 61: 1393–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, et al. (2010) Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-β-lactamase-producing Escherichia coli among companion animals. J Antimicrob Chemother 65: 651–660. [DOI] [PubMed] [Google Scholar]
- 19. Jørgensen RL, Nielsen JB, Friis-Møller A, Fjeldsøe-Nielsen H, Schønning K (2010) Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J Antimicrob Chemother 65: 460–464. [DOI] [PubMed] [Google Scholar]
- 20.Ministry of Health, Labour, and Welfare. (2011) Ethical guidelines of epidemiology study [in Japanese].
- 21. Kuroda H, Yano H, Hirakata Y, Arai K, Endo S, et al. (2012) Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Japan: Emergence of CTX-M-15-producing E. coli ST131. Diagn Microbiol Infect Dis 74: 201–203. [DOI] [PubMed] [Google Scholar]
- 22. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A (2009) Plasmid-mediated quinolone resistance: a multifaceted threat. Clinical Microbiology Reviews 22: 664–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonnet R, Recule C, Baraduc R, Chanal C, Sirot D, et al. (2003) Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother 52: 29–35. [DOI] [PubMed] [Google Scholar]
- 24. Ender PT, Gajanana D, Johnston B, Clabots C, Tamarkin FJ, et al. (2009) Transmission of extended-spectrum β-lactamase-producing Escherichia coli (sequence type ST131) between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J Clin Microbiol 47: 3780–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson JR, Anderson JT, Clabots C, Johnston B, Cooperstock M (2010) Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr Infect Dis J 29: 474–475. [DOI] [PubMed] [Google Scholar]
- 26. Owens RC Jr, Johnson JR, Stogsdill P, Yarmus L, Lolans K, et al. (2011) Community transmission in the United States of a CTX-M-15-producing sequence type ST131 Escherichia coli strain resulting in death. J Clin Microbiol 49: 3406–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vigil KJ, Johnson JR, Johnston BD, Kontoyiannis DP, Mulanovich VE, et al. (2010) Escherichia coli pyomyositis: an emerging infectious disease among patients with hematologic malignancies. Clin Infect Dis 50: 374–380. [DOI] [PubMed] [Google Scholar]