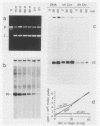
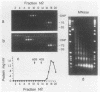
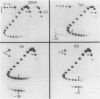
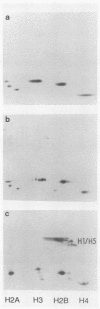
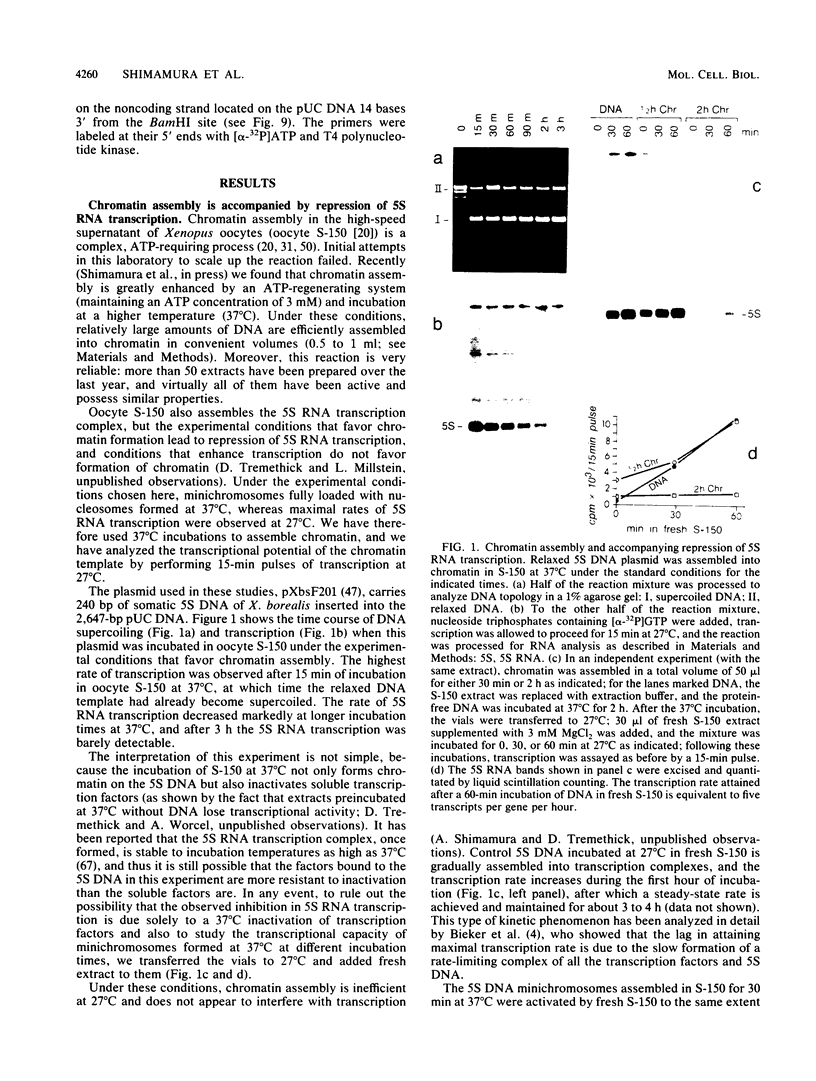
Abstract
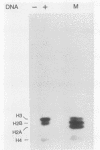
We describe an in vitro system, based on the Xenopus laevis oocyte supernatant of Glikin et al. (G. Glikin, I. Ruberti, and A. Worcel, Cell 37:33-41, 1984), that packages DNA into minichromosomes with regularly spaced nucleosomes containing histones H3, H4, H2A, and H2B but no histone H1. The same supernatant also assembles the 5S RNA transcription complex; however, under the conditions that favor chromatin assembly, transcription is inhibited and a phased nucleosome forms over the 5S RNA gene. The minichromosomes that are fully loaded with nucleosomes remain refractory to transcriptional activation by 5S RNA transcription factors. Our data suggest that this repression is caused by a nucleosome covering the 5S RNA gene and that histone H1 is not required for regular nucleosome spacing or for gene repression in this system.
Full text
PDF



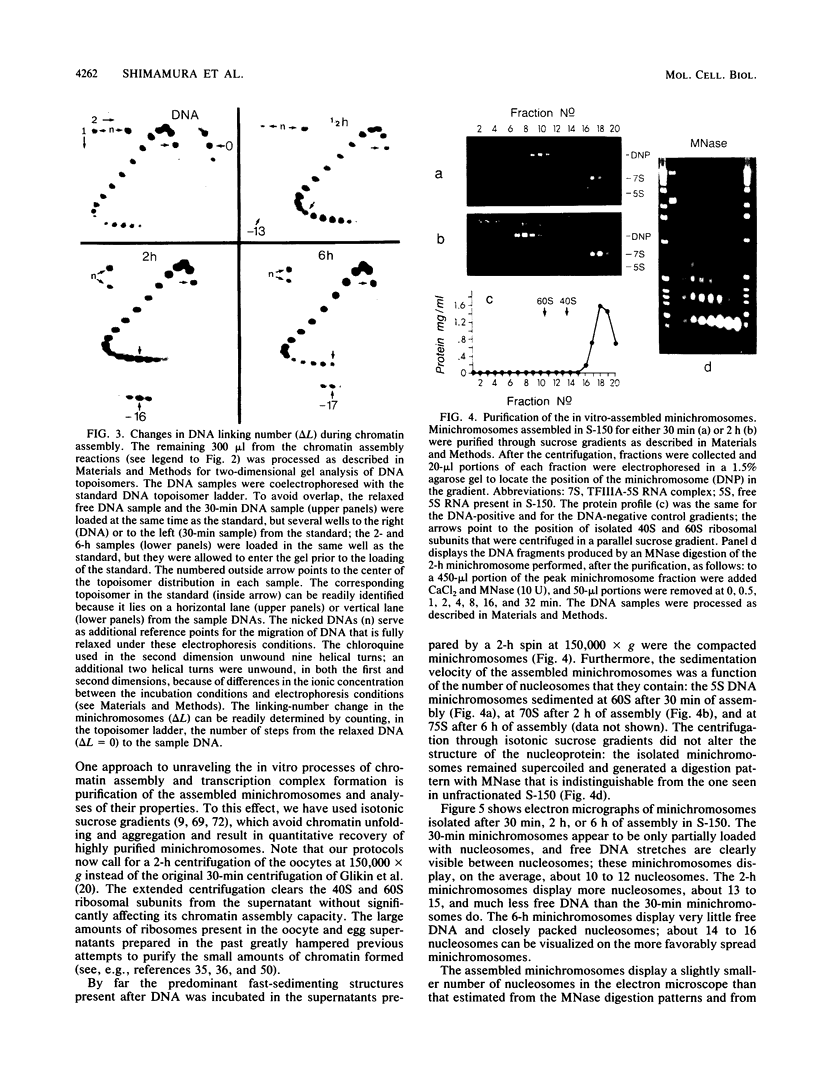
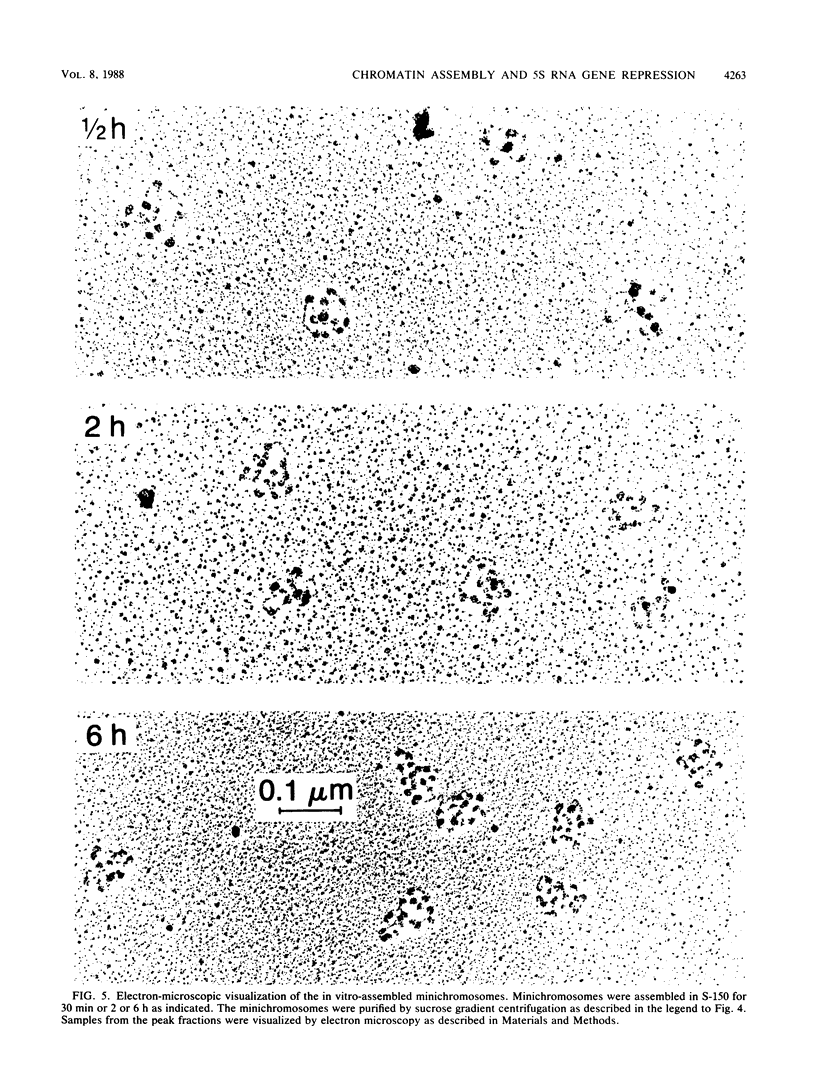


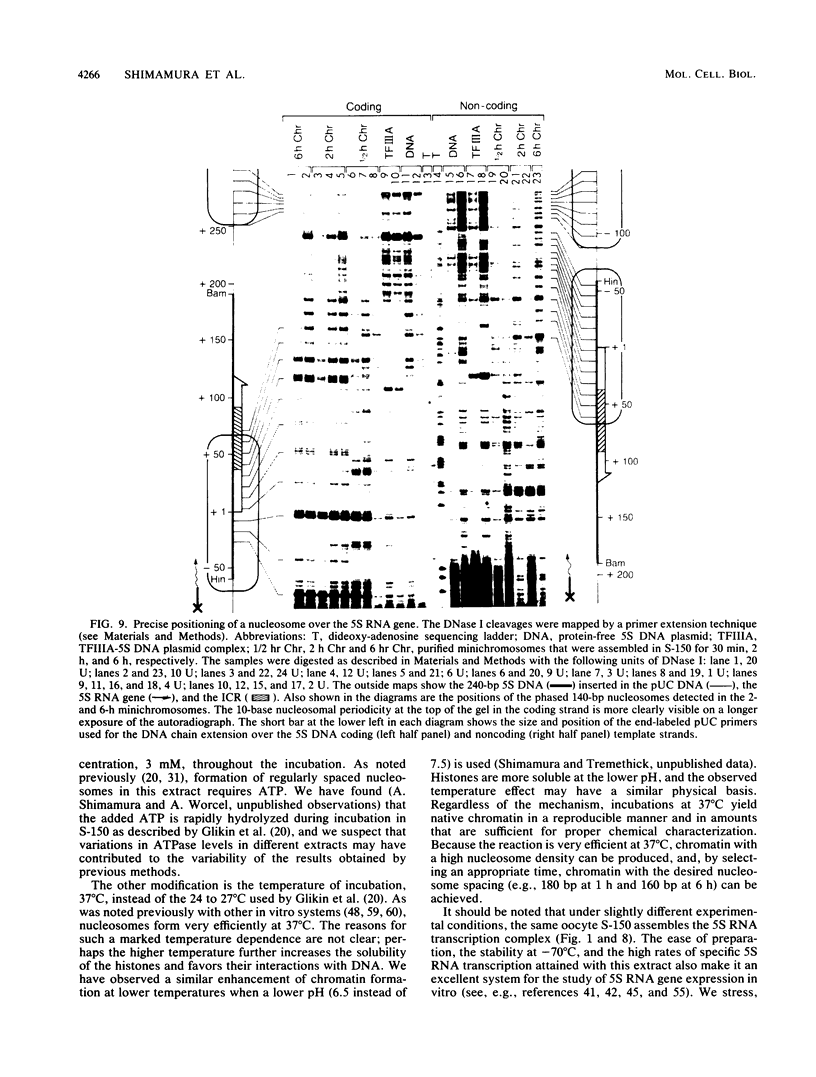



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamson E. D., Woodland H. R. Histone synthesis in early amphibian development: histone and DNA syntheses are not co-ordinated. J Mol Biol. 1974 Sep 15;88(2):263–285. doi: 10.1016/0022-2836(74)90481-1. [DOI] [PubMed] [Google Scholar]
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Boulikas T. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 1988 Jan;7(1):57–67. doi: 10.1002/j.1460-2075.1988.tb02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Certa U., Colavito-Shepanski M., Grunstein M. Yeast may not contain histone H1: the only known 'histone H1-like' protein in Saccharomyces cerevisiae is a mitochondrial protein. Nucleic Acids Res. 1984 Nov 12;12(21):7975–7985. doi: 10.1093/nar/12.21.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth S. M., Black S. J., Laskey R. A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987 Dec 24;51(6):1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Honda B. M., Laskey R. A., Thomas J. O. Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell. 1980 Sep;21(2):373–383. doi: 10.1016/0092-8674(80)90474-2. [DOI] [PubMed] [Google Scholar]
- Ebralidse K. K., Grachev S. A., Mirzabekov A. D. A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nature. 1988 Jan 28;331(6154):365–367. doi: 10.1038/331365a0. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Razvi F., Ruberti I., Mohr I., Worcel A. Chromatin-specific hypersensitive sites are assembled on a Xenopus histone gene injected into Xenopus oocytes. J Mol Biol. 1985 Feb 5;181(3):333–349. doi: 10.1016/0022-2836(85)90223-2. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Razvi F., Worcel A. Assembly of transcriptionally active chromatin in Xenopus oocytes requires specific DNA binding factors. Cell. 1984 Sep;38(2):511–521. doi: 10.1016/0092-8674(84)90506-3. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Wasserman W., Worcel A. Properties of the chromatin assembled on DNA injected into Xenopus oocytes and eggs. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):549–556. doi: 10.1101/sqb.1983.047.01.064. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Worcel A. Analysis of the chromatin assembled in germinal vesicles of Xenopus oocytes. J Mol Biol. 1983 Nov 5;170(3):699–722. doi: 10.1016/s0022-2836(83)80128-4. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim U. J., Kayne P., Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Humphries S. E., Young D., Carroll D. Chromatin structure of the 5S ribonucleic acid genes of Xenopus laevis. Biochemistry. 1979 Jul 24;18(15):3223–3231. doi: 10.1021/bi00582a006. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Franke W. W. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982 Jul;29(3):799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Seiter A. Identification of domains involved in nuclear uptake and histone binding of protein N1 of Xenopus laevis. EMBO J. 1988 Jun;7(6):1605–1614. doi: 10.1002/j.1460-2075.1988.tb02986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Razvi F., Worcel A. The role of DNA-mediated transfer of TFIIIA in the concerted gyration and differential activation of the Xenopus 5S RNA genes. Cell. 1986 Apr 25;45(2):209–218. doi: 10.1016/0092-8674(86)90385-5. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Ryoji M., Worcel A. Gyration is required for 5S RNA transcription from a chromatin template. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1305–1309. doi: 10.1073/pnas.83.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene induces a 5S DNA-specific gyration in Xenopus oocyte extracts. Cell. 1985 Jul;41(3):945–953. doi: 10.1016/s0092-8674(85)80075-1. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene proteolyses during direct exchange between 5S DNA sites. J Cell Biol. 1986 Sep;103(3):673–681. doi: 10.1083/jcb.103.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezetic J. A., Luse D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986 Apr 11;45(1):95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Earnshaw W. C. Nucleosome assembly. Nature. 1980 Aug 21;286(5775):763–767. doi: 10.1038/286763a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Morris N. R., Wyllie A. H., Mertz J. E., De Roberts E. M., Gurdon J. B. Chromatin assembly and transcription in eggs and oocytes of Xenopus laevis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):171–178. doi: 10.1101/sqb.1978.042.01.019. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Hamer D. H., Roeder R. G. Stable transcription complex on a class III gene in a minichromosome. Mol Cell Biol. 1985 Jan;5(1):40–45. doi: 10.1128/mcb.5.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lyon M. F. Mechanisms and evolutionary origins of variable X-chromosome activity in mammals. Proc R Soc Lond B Biol Sci. 1974 Nov 5;187(1088):243–268. doi: 10.1098/rspb.1974.0073. [DOI] [PubMed] [Google Scholar]
- McConkey G. A., Bogenhagen D. F. TFIIIA binds with equal affinity to somatic and major oocyte 5S RNA genes. Genes Dev. 1988 Feb;2(2):205–214. doi: 10.1101/gad.2.2.205. [DOI] [PubMed] [Google Scholar]
- Mertz J. E. Linear DNA does not form chromatin containing regularly spaced nucleosomes. Mol Cell Biol. 1982 Dec;2(12):1608–1618. doi: 10.1128/mcb.2.12.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Wiegand R., Brutlag D. Ribonucleic acid and other polyanions facilitate chromatin assembly in vitro. Biochemistry. 1981 Apr 28;20(9):2594–2601. doi: 10.1021/bi00512a035. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Millstein L., Eversole-Cire P., Gottesfeld J. M., Varshavsky A. Transcriptionally inactive oocyte-type 5S RNA genes of Xenopus laevis are complexed with TFIIIA in vitro. Mol Cell Biol. 1987 Oct;7(10):3503–3510. doi: 10.1128/mcb.7.10.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razvi F., Gargiulo G., Worcel A. A simple procedure for parallel sequence analysis of both strands of 5'-labeled DNA. Gene. 1983 Aug;23(2):175–183. doi: 10.1016/0378-1119(83)90049-5. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti I., Worcel A. Mechanism of chromatin assembly in Xenopus oocytes. J Mol Biol. 1986 Jun 5;189(3):457–476. doi: 10.1016/0022-2836(86)90317-7. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Structure of the two distinct types of minichromosomes that are assembled on DNA injected in Xenopus oocytes. Cell. 1985 Apr;40(4):923–932. doi: 10.1016/0092-8674(85)90352-6. [DOI] [PubMed] [Google Scholar]
- Schlissel M. S., Brown D. D. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984 Jul;37(3):903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- Senshu T., Fukuda M., Ohashi M. Preferential association of newly synthesized H3 and H4 histones with newly replicated DNA. J Biochem. 1978 Oct;84(4):985–988. doi: 10.1093/oxfordjournals.jbchem.a132213. [DOI] [PubMed] [Google Scholar]
- Shang Z. G., Windsor W. T., Liao Y. D., Wu C. W. Purification of Xenopus transcription factor IIIA and 5 S RNA from 7 S ribonucleoprotein particle by ammonium sulfate precipitation. Anal Biochem. 1988 Jan;168(1):156–163. doi: 10.1016/0003-2697(88)90023-1. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina M. A model chromatin assembly system. Factors affecting nucleosome spacing. J Mol Biol. 1984 Sep 15;178(2):341–363. doi: 10.1016/0022-2836(84)90148-7. [DOI] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Closely spaced nucleosome cores in reconstituted histone.DNA complexes and histone-H1-depleted chromatin. Eur J Biochem. 1978 Feb;83(2):615–628. doi: 10.1111/j.1432-1033.1978.tb12131.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Tremethick D. J., Molloy P. L. High mobility group proteins 1 and 2 stimulate transcription in vitro by RNA polymerases II and III. J Biol Chem. 1986 May 25;261(15):6986–6992. [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H. High-resolution mapping of S1- and DNase I-hypersensitive sites in chromatin. Mol Cell Biol. 1985 Jun;5(6):1538–1539. doi: 10.1128/mcb.5.6.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Andrews M. T., Crawford E., Losa R., Brown D. D. Negative supercoiling is not required for 5S RNA transcription in vitro. Cell. 1987 May 8;49(3):301–303. doi: 10.1016/0092-8674(87)90279-0. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Woodland H. R. The modification of stored histones H3 and H4 during the oogenesis and early development of Xenopus laevis. Dev Biol. 1979 Feb;68(2):360–370. doi: 10.1016/0012-1606(79)90210-0. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H., Laskey R. A., Finch J., Gurdon J. B. Selective DNA conservation and chromatin assembly after injection of SV40 DNA into Xenopus oocytes. Dev Biol. 1978 May;64(1):178–188. doi: 10.1016/0012-1606(78)90069-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]