Abstract
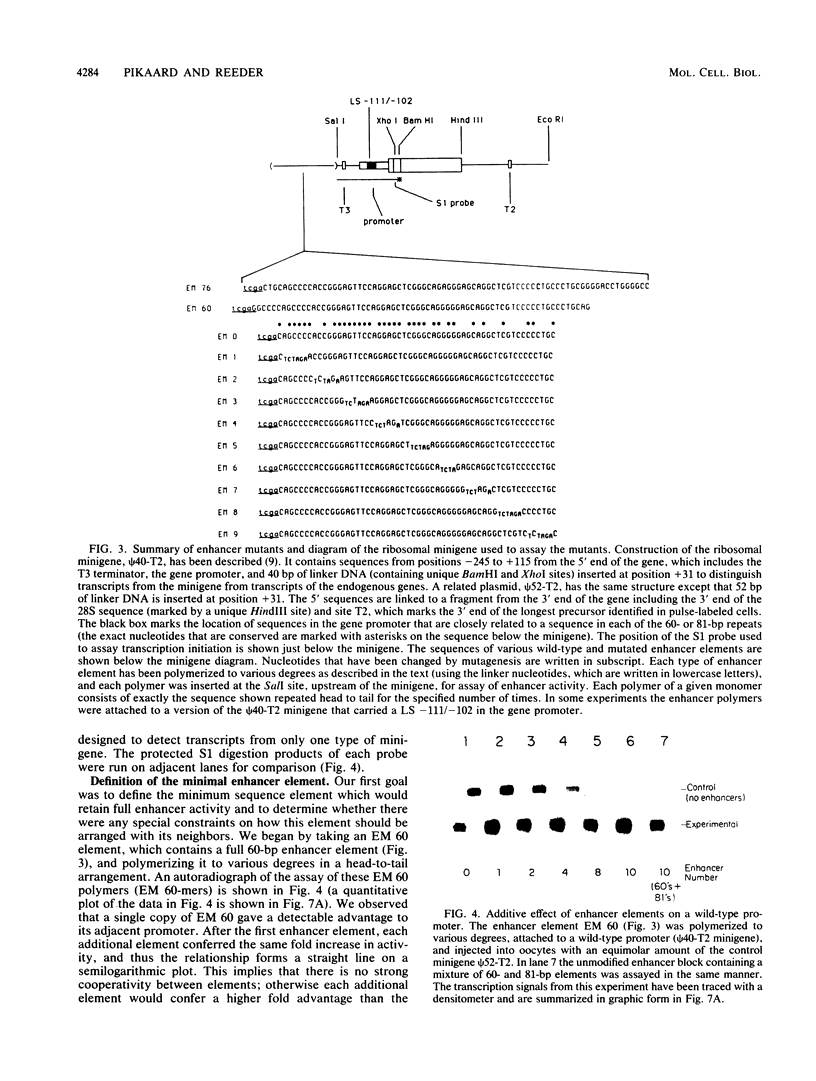
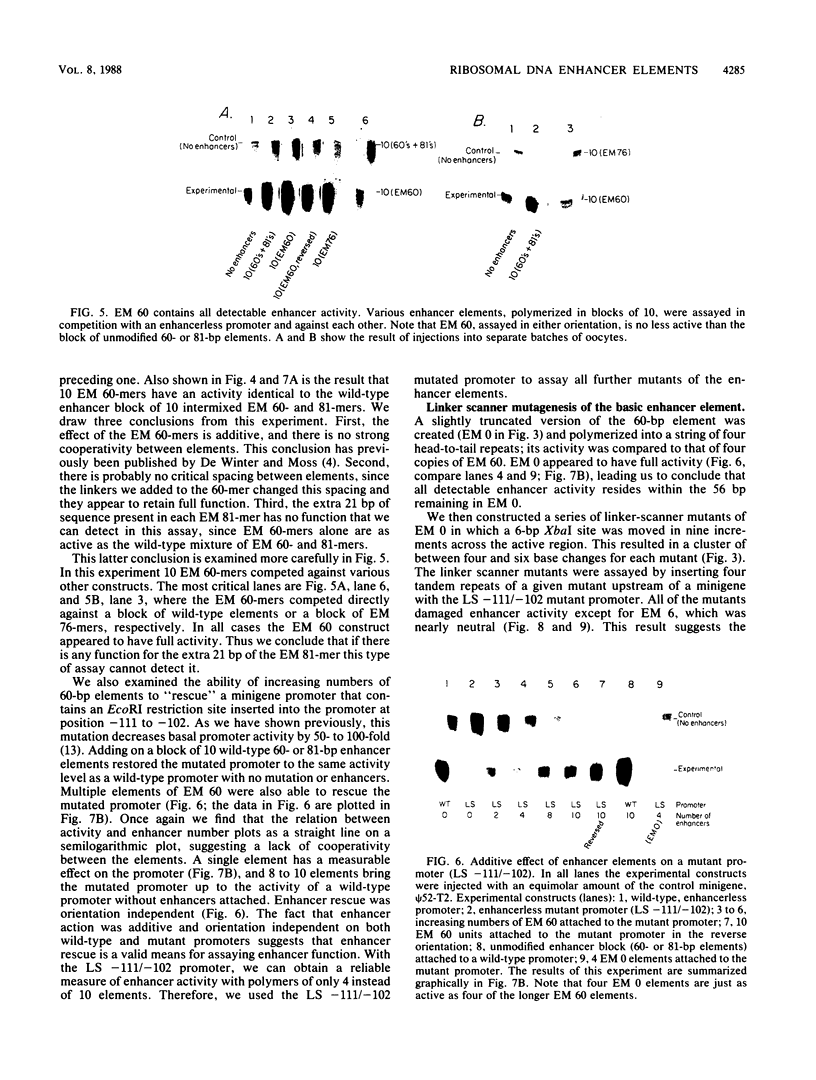
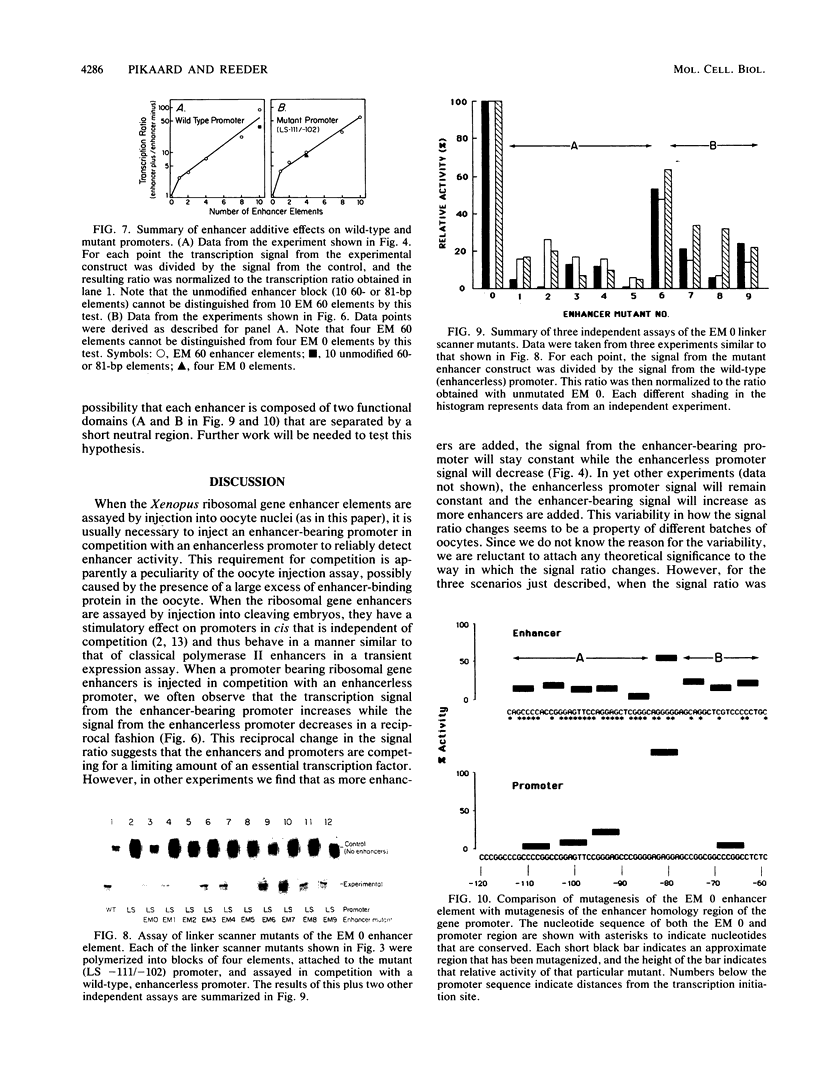
The intergenic spacer region of the Xenopus laevis ribosomal DNA contains multiple elements which are either 60 or 81 base pairs long. Clusters of these elements have previously been shown to act as position- and distance-independent enhancers on an RNA polymerase I promoter when located in cis. By a combination of deletion and linker scanner mutagenesis we show that the sequences essential for enhancer function are located within a 56-base-pair region that is present in both the 60- and 81-base-pair repeats. Within the 56-base-pair region one linker scanner mutation was found to be relatively neutral, suggesting that each enhancer element may be composed of two smaller domains. Each 56-base-pair region appears to be an independent enhancer with multiple enhancers being additive in effect. We review the current evidence concerning the mechanism of action of these enhancers.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- Busby S. J., Reeder R. H. Spacer sequences regulate transcription of ribosomal gene plasmids injected into Xenopus embryos. Cell. 1983 Oct;34(3):989–996. doi: 10.1016/0092-8674(83)90556-1. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. A complex array of sequences enhances ribosomal transcription in Xenopus laevis. J Mol Biol. 1987 Aug 20;196(4):813–827. doi: 10.1016/0022-2836(87)90407-4. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell. 1986 Jan 31;44(2):313–318. doi: 10.1016/0092-8674(86)90765-8. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A 12-base-pair sequence is an essential element of the ribosomal gene terminator in Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1900–1905. doi: 10.1128/mcb.7.5.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. DNA sequences for typical ribosomal gene spacers from Xenopus laevis and Xenopus borealis. Nucleic Acids Res. 1987 Apr 24;15(8):3623–3624. doi: 10.1093/nar/15.8.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Xenopus ribosomal gene enhancers function when inserted inside the gene they enhance. Nucleic Acids Res. 1985 Dec 20;13(24):8999–9009. doi: 10.1093/nar/13.24.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Mechanisms of nucleolar dominance in animals and plants. J Cell Biol. 1985 Nov;101(5 Pt 1):2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Pennock D., McStay B., Roan J., Tolentino E., Walker P. Linker scanner mutagenesis of the Xenopus laevis ribosomal gene promoter. Nucleic Acids Res. 1987 Sep 25;15(18):7429–7441. doi: 10.1093/nar/15.18.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G. The mechanism of nucleolar dominance in Xenopus hybrids. Cell. 1984 Aug;38(1):38–44. doi: 10.1016/0092-8674(84)90524-5. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Searle P. F., Stuart G. W., Palmiter R. D. Building a metal-responsive promoter with synthetic regulatory elements. Mol Cell Biol. 1985 Jun;5(6):1480–1489. doi: 10.1128/mcb.5.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Toohey M. G., Morley K. L., Peterson D. O. Multiple hormone-inducible enhancers as mediators of differential transcription. Mol Cell Biol. 1986 Dec;6(12):4526–4538. doi: 10.1128/mcb.6.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]