Abstract
Background
There is now compelling evidence that epigenetic modifications link adult disease susceptibility to environmental exposures during specific life stages, including pre-pubertal development. Animal studies indicate that bisphenol A (BPA), the monomer used in epoxy resins and polycarbonate plastics, may impact health through epigenetic mechanisms, and epidemiological data associate BPA levels with metabolic disorders, behavior changes, and reproductive effects. Thus, we conducted an environmental epidemiology study of BPA exposure and CpG methylation in pre-adolescent girls from Gharbiah, Egypt hypothesizing that methylation profiles exhibit exposure-dependent trends.
Methods
Urinary concentrations of total (free plus conjugated) species of BPA in spot samples were quantified for 60 girls aged 10 to 13. Genome-wide CpG methylation was concurrently measured in bisulfite-converted saliva DNA using the Infinium HumanMethylation27 BeadChip (N = 46). CpG sites from four candidate genes were validated via quantitative bisulfite pyrosequencing.
Results
CpG methylation varied widely among girls, and higher urinary BPA concentrations were generally associated with less genomic methylation. Based on pathway analyses, genes exhibiting reduced methylation with increasing urinary BPA were involved in immune function, transport activity, metabolism, and caspase activity. In particular, hypomethylation of CpG targets on chromosome X was associated with higher urinary BPA. Using the Comparative Toxicogenomics Database, we identified a number of candidate genes in our sample that previously have been associated with BPA-related expression change.
Conclusions
These data indicate that BPA may affect human health through specific epigenomic modification of genes in relevant pathways. Thus, epigenetic epidemiology holds promise for the identification of biomarkers from previous exposures and the development of epigenetic-based diagnostic strategies.
Keywords: Bead array, Bisphenol A, Egypt, Epigenetics, DNA methylation
Background
The developmental origins of health and disease (DOHaD) hypothesis postulates that environmental and nutritional factors influence developmental plasticity during critical periods of development, thereby altering susceptibility to diseases later in life [1-3]. Increasingly, this emerging field of research points to the epigenome as an important modifier of disease susceptibility. Although the epigenome is vulnerable to environmental perturbations during multiple points in life, it is particularly susceptible during embryogenesis, neonatal development, and adolescence. For example, similar to the rapid synthetic rate of DNA in early development, during the pre-pubertal period, mammary growth is highly proliferative and tightly regulated [4-8], making pre-adolescent females especially vulnerable to environmental insults acting via an epigenetic mechanism.
Accumulating work suggests that exposure to endocrine-active compounds (EACs), including bisphenol A (BPA), increases susceptibility for adverse phenotypic outcomes via epigenetic mechanisms. Methylation studies conducted by Li et al. on the estrogenic pharmaceutical agent diethylstilbestrol (DES) observed hypomethylation in two critical DNA control regions in mice exposed in utero or in the perinatal period [9,10]. Exposure to the anti-androgenic fungicide vinclozolin, the estrogenic pesticide methoxychlor, or a mixture of plastic compounds during reprogramming of the germ line has been linked with epigenetic alterations that are inherited transgenerationally even in the absence of continued exposure [11-15]. Further, BPA, a high-production volume monomer used in the manufacture of polycarbonate plastic and epoxy resin, has been associated with epigenetic alterations following developmental exposure [16-19]. Using a rat model, Ho et al. observed multiple changes in gene-specific DNA methylation patterns in the adult male prostate, including hypomethylation of the phosphodiesterase type 4 variant 4 (Pde4d4) gene following neonatal exposure to low BPA concentration (10 μg/kg of body weight BPA) [16]. Using the Avy mouse model, we have shown that maternal dietary exposure to physiologically relevant concentrations of BPA results in altered DNA methylation at two metastable loci [17,19]. Moreover, restoration of normal methylation patterns occurs with maternal supplementation of genistein or methyl donors including folate, choline, betaine, and vitamin B12[17,20].
Recent toxicological and epidemiological data warrant the identification of epigenetic biomarkers associated with BPA exposure. For example, rodent studies have associated environmentally relevant pre- or perinatal BPA exposure with higher body weight, increased breast and prostate cancer susceptibility, altered reproductive function, and other chronic health effects [21]. In 2008, the first comprehensive human epidemiological study associated higher urinary BPA concentrations with increased risk for cardiovascular disease, type 2 diabetes, and liver enzyme abnormalities [22], and recent data correlated urinary BPA concentrations and obesity in U.S. children [23]. An analysis of 2003–04 National Health and Nutrition Examination Survey (NHANES) data found that the half life of BPA in the human body is longer than would be expected from acute exposure studies, suggesting that BPA either bioaccumulates in the body or BPA exposure occurs through non-dietary sources or both [24]. Hence, it is now hypothesized that life course exposure to environmental pollutants, such as BPA, are linked to adult disease, such as breast cancer [25].
Building upon ongoing epidemiological and exposure studies in Gharbiah, Egypt [26-30], we correlated urinary concentrations of total (free plus conjugated) species of BPA in spot samples with genome-wide CpG methylation levels in concurrently collected saliva DNA samples. We conducted an environmental epidemiology study of BPA exposure and CpG methylation in pre-adolescent girls hypothesizing that methylation profiles exhibit exposure-dependent trends.
Utilizing a semi-unbiased, quantitative genome-wide methylation array, which evaluates 27,543 individual CpG sites targeting 14,475 genes, we identified 57 agnostic and 35 biologically prioritized candidate CpG sites as well as gene pathways differentially methylated with respect to BPA concentrations. Four candidate genes were further validated using bisulfite pyrosequencing, a sensitive and quantitative method of DNA methylation detection. Using the Comparative Toxicogenomics Database (CTD) [31-33], we identified previously published candidate genes with up-regulated expression associated with BPA and found that many of the targets had BPA-associated hypomethylation in our sample set, and vice versa.
Methods
Study population
This study originated from a pilot study addressing environmental exposures in a developing country, and our study population was previously described in Nahar et al.[27]. Briefly we recruited healthy females 10–13 years of age living in either rural (N = 30) or urban (N = 30) areas of the Gharbiah province of Egypt, located 90 kilometers north of Cairo. We obtained written informed consent from the mother of each participant, and approval from the Institutional Review Boards of the University of Michigan and the Gharbiah Cancer Society were obtained before starting the study. The involvement of the Centers for Disease Control and Prevention (CDC) laboratory was limited and determined not to constitute engagement in human subject research. The average age of menarche in Egypt is 12.44 ± 1.3, and all study participants were pre-menstrual, as ascertained by asking the each mother about any prior menstrual cycles of her daughter [34]. To reduce variability arising from various exposures throughout the day, the participants provided one spot urine sample during the time period between noon to 4 pm and were measured for height, weight, waist, and hip circumference by trained nurses, using standardized techniques as previously described and validated [35,36]. In brief, each anthropometric measurement was taken twice. If the difference in the height measurements was > 0.5 centimeters (cm); if the difference in weight was > 0.2 kilograms; if the difference in waist circumference was > 0.5 cm; or if the difference in hip circumference was > 0.5 cm, then a third measurement was taken. We discarded the most discrepant value and averaged the other two. The Division of Laboratory Sciences of the National Center for Environmental Health at the CDC determined the urinary concentrations of total (free plus conjugated species) BPA as previously described [27,37]. An interviewer administered a lifestyle and diet questionnaire in Arabic in order to assess potential routes of BPA exposure, and field-collected anthropometric measurements were acquired. The questionnaire, entitled “Comparison of Xenoestrogen Levels Among Prepubertal Females in Urban and Rural Gharbiah, Egypt,” contained questions addressing residential history, personal care product usage, family history of cancer, use of canned foods, breast feeding status, and food preparation and storage behaviors.
Saliva DNA was concurrently collected using the Oragene DNA Self-Collection Kit (DNA Genotek, Ontario, Canada). The Oragene kit allows for long-term DNA storage at room temperature with little or no DNA degradation. Total genomic DNA was isolated from saliva samples using Oragene-DNA Self-Collecting kit standard protocols. Bisulfite conversion was carried out on 600 ng of genomic DNA using the manufacturer’s protocol (EZ DNA Methylation-Direct Kit, Zymo Research Corp., Orange, CA) for genome-wide methylation assessment using the Illumina Infinium HumanMethylation27 BeadChip platform (Illumina, San Diego, CA). For pyrosequencing validation 600 ng of DNA was bisulfite converted using the Qiagen EpiTect Bisulfite Conversion kit (Qiagen, Valencia, CA).
Genome-wide DNA methylation discovery
Genome-wide DNA methylation was assessed with the Infinium HumanMethylation27 BeadChip (Illumina) performed at the University of Michigan DNA Sequencing Core facility in accordance with manufacturer’s instructions as previously published [38]. Samples exhibiting various BPA concentrations (ranging from non-detect to 12 ng/mL) were randomly applied to each of the 12-sample arrays to avoid bias by batch effect. The Illumina BeadStudio Software provided estimates, termed beta values or β-scores, of the percent methylation for each probe site based on the Cy3 and Cy5 intensities. We utilized a previously described normalization strategy that improves the correlation among replicate samples and tends to increase the level of significance when the methylation percentages are compared between groups [39].
In order to maximize power to detect methylation differences specific to higher BPA concentrations, we performed analysis both categorizing the samples into three groups: BPA-low (n = 22), BPA-intermediate (n = 14), and BPA-high (n = 10) and treating BPA as a continuous variable. First, in the categorization strategy, samples were classified as BPA-low if the BPA measurement was less than 1 ng/mL; BPA-high if BPA measurement was greater than 2 ng/mL; and BPA-intermediate for remaining samples. The observed BPA concentrations within our Egyptian cohort were relatively low, ranging from below the limit of detection (LOD) to 12 ng/mL. To identify differential methylation associated with high BPA concentrations, we performed the categorical analysis focusing on the tails of the BPA distribution. We used an empirical Bayesian moderated t-test to assess significance between the percent methylation in BPA-high versus BPA-low samples, which improves results for studies of small sample size [40]. Those with p-value < 0.05 with average ß-score difference greater than 5% were reported as candidate sites. To identify differential methylation associated with the highest concentrations of BPA in this cohort, the analysis described above was repeated using a more stringent cutoff for the BPA-high group (n = 5, where urinary BPA concentrations are greater than 2.60 ng/mL, the median concentration of age and gender matched individuals from NHANES 2009–2010) (Additional file 1: Table S1). The list of significant CpG sites at candidate genes from the above analysis was compared with BPA-interacting genes acquired from CTD and used for the pathway enrichment analysis.
Bioinformatics analyses
All statistical analyses were performed with R Statistical Software (version 2.10.1). CpG sites that failed on 10% of samples were not included in subsequent analyses. Using the LIMMA package (Version 3.4.4) in R, BPA-dependent differential methylation regions among Egyptian girls were examined using covariates including hybridization date, age, breast-fed status, body mass index (BMI), and specific gravity to adjust for urinary dilutions, and log-transformed urinary concentrations of BPA. As is standard in microarray analyses, empirical Bayesian variance methods were incorporated to perform site-specific moderated t-tests [40]. The linear model used for each individual CpG site was as follows:
RPMM clustering analysis
The Recursively Partitioned Mixture Model (RPMM) clustering analysis in R software program was performed on the top 200 most variable CpG sites using the glcTree function, which performs Gaussian latent class modeling. The only non-default parameter used during the analysis was for the level of verbosity, which was set at 0 (default is at 2). The glcTree clustering analysis separated 46 samples into 3 leaf classes (clusters 1, 2, and 3).
Logistic regression-based pathway enrichment analysis
An enrichment analysis was performed using logistic regression-based pathway enrichment analysis tool (LRpath) available at http://lrpath.ncibi.org[41,42]. The data representing 27,543 sites generated from the R statistical analysis described above was reformatted to contain Entrez gene IDs, p-values, and fold-changes in tab-delimited text file format. Gene Ontology (GO) terms and pathway concept types were selected (Biocarta pathway, Edinburgh Human Metabolic Network (EHMN) metabolic pathway, GO biological process, cellular component, and molecular function, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Panther pathway) for enrichment analysis in LRpath web-application. For both dichotomous comparison of low- versus high-BPA sample groups and the continuous linear models, LRpath was performed using the directional option with default settings, to distinguish pathways enriched with hyper- versus hypo-methylated sites.
Gene set enrichment analysis
Positional gene set enrichment analysis was performed using Gene Set Enrichment Analysis (GSEA) to determine statistical over-representation of BPA-concentration specific epigenetic marks within chromosomal cytogenic bands. The differences in average β-score of each probe (N = 27,543) between BPA-low and BPA-high groups were pre-ranked and subjected to GseaPreranked analysis tool. After collapsing features into gene symbols, a total of 14,474 genes were represented, and the enrichment among 240 cytogenic band sets were tested with default setting.
Comparative toxicogenomics database (CTD)
The publicly available Comparative Toxicogenomics Database (CTD) combines manually curated information from peer-reviewed articles on environmental chemicals, interacting genes, and known associated health effects in human as well as in animal models [31-33]. The tool allows investigators to quickly query their epigenomics-derived gene list with curated genes associated with a particular exposure or phenotypic endpoint of interest. A list of 903 BPA-interacting genes in humans was acquired using a chemical-gene interaction query using Bisphenol-A as a search term, and after mapping the list to the genes present on the BeadChip, 657 unique genes represented by 1,355 probes remained for downstream analysis.
Validation by pyrosequencing
CpG methylation at specific loci of the biologically-prioritized and agnostic candidates was validated by pyrosequencing using 2 ul bisulfite converted DNA. PCR and sequencing primers were designed in-house using PyroMark software are listed in Additional file 1: Table S2. The PCR products were run on a 1.5% agarose gel to ensure PCR quality, correct product length, and lack of contamination. Pyrosequencing was performed following the manufacturer’s protocol on a PyroMark MD (Qiagen).
Results
Characterization of DNA methylation across Egyptian girls
High-throughput analysis of DNA methylation in a cohort of Egyptian girls was performed using the Infinium HumanMethylation27 BeadChip (Illumina) containing 27,543 CpG sites targeting 14,475 gene promoters, including > 200 cancer-related and imprinted genes and 110 microRNAs [43-45]. To demonstrate experimental consistency in our research setting, we selected four samples with the most abundant genomic DNA and profiled them on the bead array in duplicate. All duplicates were shown to be highly correlated with a median correlation coefficient value of 0.98 (data not shown).
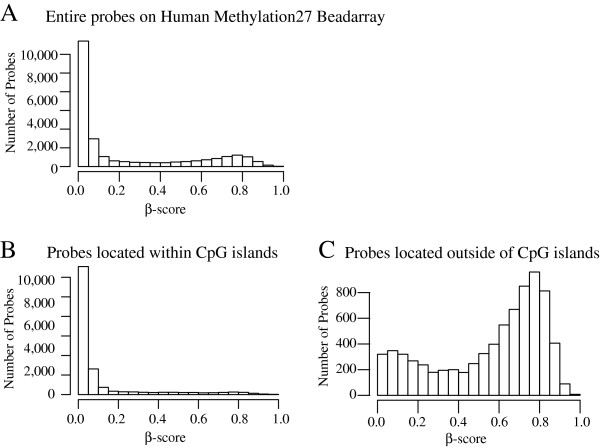
We evaluated the overall distribution of the β-score, which indicates the level of methylation at CpG sites (Figure 1A). Among the 27,543 probes present in all 46 samples with a significant p-value (p < 0.05), 19,985 (73%) probes were located within annotated CpG islands, while the remaining 7,558 (27%) probes were not. More than half of the probes (average n = 11,700 with s.d. of 1,786) located within CpG islands had a β-score less than 0.05 showing highly shifted distribution (Figure 1B). Probes located outside of CpG islands displayed skewed bimodal distributions of β-scores (Figure 1C). The secondary peak observed on the long right tail of the distribution in Figure 1A is likely associated with probes from CpG island depleted regions.
Figure 1.
β-score distribution in the HumanMethylation27 BeadChip in the presence and absence of CpG islands in 1 randomly chosen sample. All samples display similar distributions as shown above. (A) The first histogram represents the β-score frequency of all probes. (B) The β-score frequency of the probes located within CpG islands is graphed. (C) The β-score frequency of the probes located outside of CpG islands is graphed.
Urinary BPA concentration affects CpG methylation profiles
As described previously, urinary BPA concentrations and variability were low in the Egyptian girls cohort compared to age-matched females in the United States as reported in 2009–2010 National Health and Nutrition Examination Survey (NHANES) [27]. Concentrations of urinary BPA in Egyptian girls ranged from below the limit of detection (LOD = 0.4 ng/ml) to 12 ng/mL (total median unadjusted: 0.70 ng/mL) compared to age-matched American girls ranging from below the LOD to 16.1 ng/mL (median unadjusted: 2.60 ng/mL). Comparing samples with BPA-low and BPA-high concentrations as a dichotomous trait resulted in 1,439 differentially methylated sites (Figure 2A) with p-value less than 0.05 and 171 sites with a p-value less than 0.01, compared to 190 differentially methylated sites by age and 444 differentially methylated sites by BMI. The overall difference in average β-score between the two BPA exposure groups was not significant (0.177 vs. 0.180); however, when we limited the analysis to those sites that are differentially methylated, there were a greater number of hypo- (178 with at least 5% change in β-score) compared to hyper-methylated (54 with at least 5% change in β-score) sites in the BPA-high group compared to the BPA-low group (Additional file 2: Figure S1). The top 57 agnostic candidate genes with average β-score difference greater than 5% (ranges from −0.11 to 0.12) and a p-value < 0.05 are listed in Additional file 1: Table S3.
Figure 2.
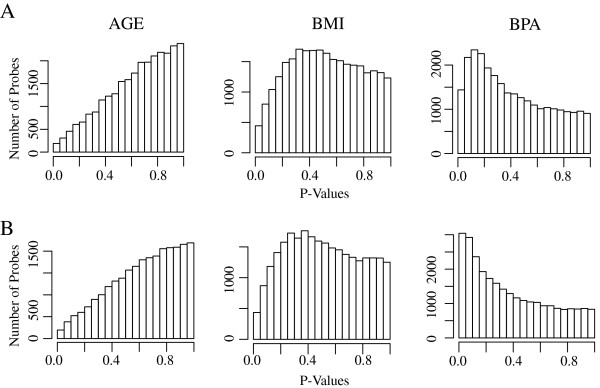
Histogram of p-values (bin = 0.1) in methylation analysis using covariates including age, BMI, and log BPA in empirical Bayes modeling using LIMMA package in R software with BPA as a (A) dichotomous or (B) continuous variable.
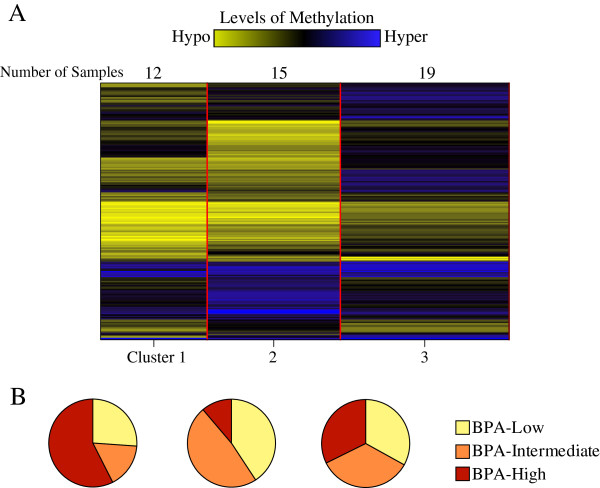
Using Recursively Partitioned Mixture Model (RPMM) clustering analysis within the R software program, we examined the patterns of differential methylation using the top 200 most variable CpG sites (Figure 3). The resulting cluster was divided into 3 major leaf nodes: clusters 1, 2, and 3. Cluster 1 displayed a hypomethyation signature, and more than half of BPA-high samples were in this cluster (Figure 3A and B). While the least number of BPA-high samples were categorized into the cluster 2, it contained the greatest number of BPA-low and -intermediate samples. In general, BPA-low and -intermediate samples showed similar methylation profiles in all resulting leaves, indicating similarity in CpG methylation between these two groups. Lastly, cluster 3, which exhibited a high methylation signature was evenly composed of samples from all three groups (Figure 3). The tree plot of mean-centered β-scores of these top 200 most variable CpG sites is provided in Additional file 2: Figure S2.
Figure 3.
The 200 most variable CpG sites from 46 samples were clustered using Recursively Partitioned Mixture Model (RPMM) for Beta and Gaussian Mixtures from R package, a model-based clustering algorithm that returns a hierarchy of classes, similar to hierarchical clustering and finite mixture models. (A) RPMM clustering divided 46 samples into 3 clusters (1, 2, and 3), containing 12, 15, and 19 samples, respectively. (B) More than half of the BPA-high samples were clustered within cluster 1, while BPA-low and –intermediate samples were more evenly distributed across all 3 clusters.
An enrichment analysis on the dichotomous model candidate sites was performed using LRpath with resulting p-value output from R statistical analysis [42]. Highly enriched GO molecular functions included various receptor and receptor binding activities of G-protein-coupled receptor (p-value < 1.91e-5 and FDR < 2.00e-3), chemokine (p-value < 2.48e-5 and FDR < 2.02e-3), G-protein chemoattractant, and other sensory-related receptors (Table 1 and Additional file 1: Table S4). The cytokine-related concepts were further supported by a top-ranked related KEGG pathway concept (p-value < 3.06e-6 and FDR < 5.69e-4). Other top KEGG pathways were involved with metabolism and biosynthesis activities (Additional file 1: Table S4), and GO terms were enriched with immune and inflammatory responses. Interestingly, these concepts were enriched among the hypomethylated gene sets, indicating possible induced activity with higher BPA concentrations. The enrichment analysis using a stringent BPA-high group (N = 5), whose BPA measurements were above the mean of age-matched American girls, yielded similar findings, with immune-related and metabolic concepts enriched among hypomethylated genes.
Table 1.
LRpath analysis reveals that cytokine and hormone activity, immune response, and metabolic processes among the top enriched concepts within differentially methylated genes
|
Dichotomous model - pathway (FDR <0.2, P-value <0.02) | |||||||
|---|---|---|---|---|---|---|---|
| Name | Concept type | #Genes | Coeff | Odds ratio | P-Value | FDR | Enriched |
| Cytokine-cytokine receptor interaction |
KEGG Pathway |
229 |
−0.200253028 |
0.288086617 |
3.06E-06 |
5.69E-04 |
Down |
| Metabolism of xenobiotics by cytochrome P450 |
KEGG Pathway |
61 |
−0.287221818 |
0.167801921 |
1.58E-04 |
0.014739788 |
Down |
| Vitamin A (retinol) metabolism |
EHMN Pathway Gene |
25 |
−0.428286702 |
0.069834019 |
3.14E-04 |
0.015401391 |
Down |
| N-Glycan biosynthesis |
KEGG Pathway |
44 |
0.359631955 |
9.346216996 |
0.001289861 |
0.079971357 |
Up |
| Proteogylcan biosynthesis |
EHMN Pathway Gene |
21 |
−0.40681778 |
0.079801493 |
0.001359963 |
0.0333191 |
Down |
| Oxidative phosphorylation |
KEGG Pathway |
101 |
0.206370232 |
3.605679051 |
0.004079267 |
0.176257686 |
Up |
| C21-Steroid hormone metabolism |
KEGG Pathway |
12 |
−0.42731737 |
0.07025597 |
0.00473811 |
0.176257686 |
Down |
| Proteasome |
KEGG Pathway |
21 |
0.442939628 |
15.68486443 |
0.006686061 |
0.18372292 |
Up |
| Ribosome |
KEGG Pathway |
59 |
0.255415299 |
4.890555948 |
0.006914303 |
0.18372292 |
Up |
| Natural killer cell mediated cytotoxicity |
KEGG Pathway |
116 |
−0.157029295 |
0.376862256 |
0.008222671 |
0.188057588 |
Down |
|
Dichotomous model - GO (FDR <0.01, P-value <5e-4, Odds ratio >5) | |||||||
|
Name |
Concept type |
#Genes |
Coeff |
Odds ratio |
P-value |
FDR |
Enriched |
| Epidermis development |
GO Biological Process |
120 |
−0.288785876 |
0.166178787 |
7.22E-08 |
2.83E-05 |
Down |
| Ectoderm development |
GO Biological Process |
130 |
−0.27220051 |
0.184220938 |
1.69E-07 |
4.41E-05 |
Down |
| Sensory perception of chemical stimulus |
GO Biological Process |
81 |
−0.326598184 |
0.131377584 |
2.59E-07 |
5.80E-05 |
Down |
| Digestion |
GO Biological Process |
85 |
−0.314243619 |
0.141861941 |
4.72E-07 |
8.22E-05 |
Down |
| Ribonucleoprotein complex biogenesis and assembly |
GO Biological Process |
161 |
0.282212002 |
5.776725456 |
5.25E-07 |
8.23E-05 |
Up |
| Regulation of homeostatic process |
GO Biological Process |
18 |
−0.534094322 |
0.03618224 |
2.19E-06 |
3.12E-04 |
Down |
| G-protein-couples receptor binding |
GO Molecular Function |
69 |
−0.298251857 |
0.156684914 |
1.91E-05 |
0.001995771 |
Down |
| Chemokine binding |
GO Molecular Function |
26 |
−0.431856927 |
0.068301636 |
2.48E-05 |
0.002021324 |
Down |
| Sensory perception of smell |
GO Biological Process |
56 |
−0.318324204 |
0.138309662 |
2.90E-05 |
0.002387889 |
Down |
| Serine-type endopeptidase inhibitor activity |
GO Molecular Function |
73 |
−0.285841884 |
0.169247136 |
2.90E-05 |
0.002021324 |
Down |
|
Continuous model - pathway (FDR <0.2, P-value <0.02) | |||||||
|
Name |
Concept type |
#Genes |
Coeff |
Odds ratio |
P-Value |
FDR |
Enriched |
| Cytokine-cytokine receptor interation |
KEGG Pathway |
229 |
−0.205604496 |
0.278663207 |
9.68E-09 |
1.80E-06 |
Down |
| Metabolism of xenobiotics by cytochrome P450 |
KEGG Pathway |
61 |
−0.2927432 |
0.162141761 |
4.33E-06 |
4.03E-04 |
Down |
| Complement and coagulation cascades |
KEGG Pathway |
66 |
0.25256689 |
0.208127535 |
5.11E-05 |
0.003167145 |
Down |
| Plaminogen activating cascade |
Panther Pathway |
13 |
−0.497645229 |
0.045380623 |
6.32E-05 |
0.114613557 |
Down |
| Cytokines and Inflammatory Response |
Biocarta Pathway |
29 |
−0.33471697 |
0.124913374 |
195E-04 |
0.035217055 |
Down |
| Ribosome |
KEGG Pathway |
59 |
0.287062223 |
5.953499593 |
2.21E-04 |
0.010264183 |
Up |
| Vitamin A (retinol) metabolism |
EHMN Pathway Gene |
25 |
−0.385600917 |
0.091048898 |
2.51E-04 |
0.012317017 |
Down |
| Autoimmune thyroid disease |
KEGG Pathway |
47 |
−0.26174305 |
0.196590877 |
3.38E-04 |
0.012575822 |
Down |
| Oxidative phosphorylation |
KEGG Pathway |
101 |
0.209255664 |
3.670918657 |
4.30E-04 |
0.013316379 |
Up |
| Asthma |
KEGG Pathway |
26 |
−0.321259111 |
0.135809858 |
6.79E-04 |
0.015209508 |
Down |
|
Continuous model - GO (FDR <0.01, P-value <5e-4, Odds ratio >5) | |||||||
|
Name |
Concept type |
#Genes |
Coeff |
Odds ratio |
P-Value |
FDR |
Enriched |
| Sensory perception of chemical stimulus |
GO Biological Process |
81 |
−0.378994195 |
0.094865006 |
8.35E-14 |
6.54E-11 |
Down |
| Digestion |
GO Biological Process |
85 |
−0.332578826 |
0.126584265 |
6.03E-11 |
2.10E-08 |
Down |
| Epidermis development |
GO Biological Process |
120 |
−0.281910372 |
0.17343325 |
2.06E-10 |
5.38E-08 |
Down |
| Ectoderm development |
GO Biological Process |
130 |
−0.26538019 |
0.192197099 |
7.20E-10 |
1.34E-07 |
Down |
| Sensory perception of smell |
GO Biological Process |
56 |
−0.372825845 |
0.09857215 |
7.72E-10 |
1.34E-07 |
Down |
| Acute inflammatory response |
GO Biological Process |
69 |
−0.321398071 |
0.135692626 |
1.28E-08 |
1.55E-06 |
Down |
| Serine-type endopeptidase inhibitor activity |
GO Molecular Function |
73 |
−0.304550109 |
0.150670556 |
3.09E-08 |
2.71E-06 |
Down |
| olfactory receptor activity |
GO Molecular Function |
42 |
−0.377710874 |
0.095624611 |
3.89E-08 |
2.71E-06 |
Down |
| Immunoglobulin mediated immune response |
GO Biological Process |
50 |
−0.352071918 |
0.11214204 |
5.47E-08 |
5.72E-06 |
Down |
| B cell mediated immunity |
GO Biological Process |
51 |
−0.34665743 |
0.115979703 |
7.23E-08 |
7.08E-06 |
Down |
|
Continuous model - metabolite | |||||||
| Progesterone |
Metabolite |
34 |
−0.456431331 |
0.058628061 |
2.25E-07 |
1.04E-04 |
Down |
| Testerone |
Metabolite |
53 |
−0.364806957 |
0.103608877 |
5.41E-07 |
1.25E-04 |
Down |
| Trichloroethanol |
Metabolite |
46 |
−0.365998732 |
0.102844341 |
2.20E-06 |
2.59E-04 |
Down |
| Estradiol-17beta | Metabolite | 47 | −0.362340859 | 0.105208999 | 2.25E-06 | 2.59E-04 | Down |
This table depicts the top 10 GO and pathway concepts in both the dichotomous and continuous analyses.
As a second approach, modeling BPA as a continuous linear model utilizing all 46 samples identified 3,042 differentially methylated sites with a p-value < 0.05 and 523 sites with a p-value < 0.01 (Figure 2B and Additional file 1: Table S3). Seventy-six percent of the continuous model sites overlap with the initial categorical modeling hits. We uploaded significant CpG sites from the continuous approach to LRpath for gene set enrichment testing, and observed a high resemblance between dichotomous and continuous LRpath pathways. For example, highly enriched GO molecular functions and pathways included receptor binding activities such as G-protein, chemokine, and sensory-related, as well as immune, and inflammatory responses (Table 1 and Additional file 1: Table S4). Significantly enriched GO and pathway terms among hypermethylated sites were ribosome-related, such as structural constituent of ribosome, rRNA metabolic process, rRNA processing, and ribosome biogenesis and assembly. On the other hand, enriched concepts among hypomethylated sites included metabolite concepts such as progesterone (p-value < 2.25e-7 and FDR < 1.04e-4), testosterone (p-value < 5.41e-7 and FDR < 1.25e-4), and estradiol-17beta (p-value < 2.25e-6 and FDR < 2.59e-4). The ratio of the odds of an event was 17, 9.6, and 9.5 respectively.
Higher BPA concentration is associated with hypomethylation of X-chromosome CpG sites
Using the dichotomous categorical model, we investigated the chromosomal location of the differentially methylated CpG sites. The differences in mean β-score of each probe between BPA-low and BPA-high groups were pre-ranked and subjected to the GseaPreranked analysis [46]. Several cytogenic regions on chromosome X, including both the p and q arms (Xq21, Xp11, Xq24, etc.) were significantly enriched (p-value < 0.05) with hypomethylated probes in BPA-high vs. BPA-low samples (Figure 4 and Additional file 1: Table S5). No regions from chromosome X were enriched with hypermethylated genes in BPA-high vs. BPA-low groups, and none of the autosomes were significantly enriched in either direction. Chromosome X enrichment is also observed, even when BPA-intermediate group is added to the analysis and combined with the BPA-low group (data not shown). However, when the BPA-intermediate group is combined with BPA-high group to be tested against BPA-low group, the enrichment within chromosome X is no longer significant.
Figure 4.
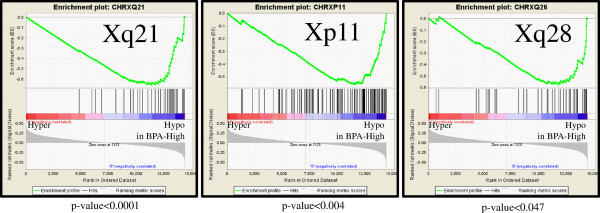
In the dichotomous model analysis, multiple regions in chromosome X were enriched with hypomethylated genes in BPA-high vs. BPA-low groups.
Imprinted gene methylation is not broadly affected by BPA concentrations
The methylation patterns observed in differentially methylated regions (DMRs) of imprinted regions in the human genome play an important role in parent-of-origin monoallelic gene expression. We compared the methylation level of imprinted genes obtained from the catalogue of parent-of-origin effects (http://igc.otago.ac.nz/home.html). The methylation differences of 34 unique imprinted genes measured by 233 probes on BeadChip in BPA-low and BPA-high samples were evaluated, and none were found to be differentially methylated by BPA exposure group (Additional file 2: Figure S3A) among Egyptian pre-adolescent girls, with the exception of Necdin (NDN). Among six probes (999, 776, 80, and 77 bps downstream, and 308 and 170 bps upstream of TSS) targeting the promoter of NDN, one CpG site (308 bp upstream from TSS) was significantly less methylated in BPA-high samples (p-value < 0.02) with average methylation difference greater than 5% (Additional file 2: Figure S3B). Neighboring CpG sites (77 bp downstream and 170 bp upstream of TSS) were not differentially methylated.
Differential methylation is associated with known BPA-interaction genes
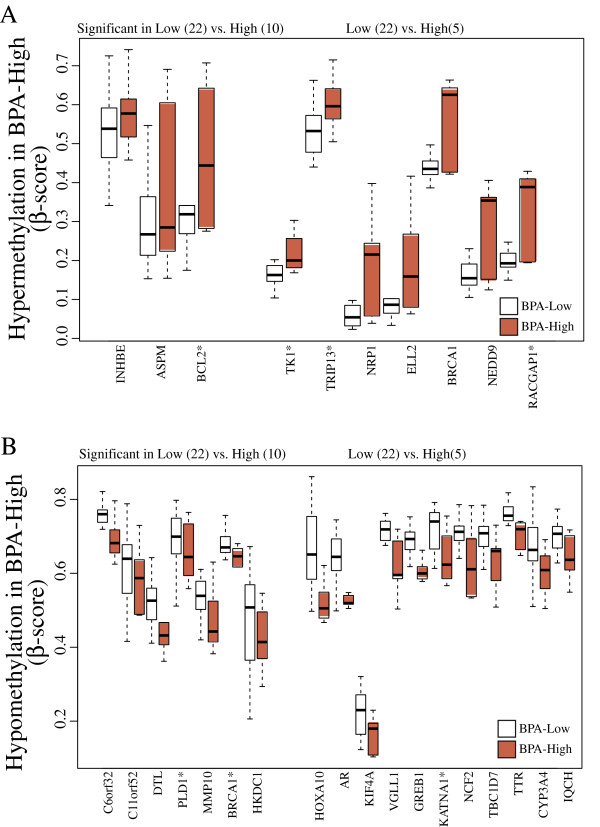
The publicly available Comparative Toxicogenomics Database (CTD) provides manually curated information on environmental chemicals, interacting genes, and associated health effects in human as well as animal models [31-33]. A list of 903 genes in humans whose expression levels are known to be affected by BPA was acquired using a chemical-gene interaction query, yielding 333 genes with increased and 257 genes with decreased level of expression upon BPA exposure. The directionality of the expression changes in remaining 313 genes was not available. A total of 657 unique genes (from 1,355 probes) from this list were represented on the BeadChip, and the methylation levels were compared to identify “biologically-prioritized” candidate genes with previously characterized altered expression by the CTD. Overall, 293 out of 1,355 sites (21.6%) were differentially methylated (p-value < 0.05) with BPA exposure in either the dichotomous or continuous model described in the Methods. The top biologically-prioritized candidates with an average β-score difference more than 5% are shown in Figure 5 and Additional file 1: Table S6 (n = 35). In general, hypomethylated candidates exhibit high level β-scores (above 0.5), while hypermethylated candidates tend to have basal or low level methylation (Figure 5).
Figure 5.
Differential methylation observed in genes known to be interacting with BPA from the Comparative Toxicogenomics Database (CTD). (A) Top candidate genes that undergo hypermethylation in BPA-high vs. BPA-low groups. (B) Top candidate genes that undergo hypomethylation in BPA-high vs. BPA-low groups. The reported expression change in genes with the asterisk did not inversely correlate with the methylation status.
Several hyper- and hypo-methylated genes in BPA-high compared to BPA-low samples corresponded with expression change, adhering to the general agreement that methylation and expression often exhibit inverse directionality. For example, the hypomethylated probes mapping to C11orf52, HOXA10, KIF4A, DTL, GREB1, and NCF2 were identified in CTD to have increased expression, while the hypermethylated probes mapping to NCK1, CSDA, and BRCA1 were reported by CTD to have decreased expression with BPA exposure. The directionality of the expression change in hypomethylated genes including C6orf32, MMP10, HKDC1, SLC3A1, VGLL1, and IQCH, and hypermethylated genes including INHBE, ASPM, NRP1, ELL2, and NEDD9 was not provided in the CTD. The expression change of the hypomethylated genes, PLD1, AR, KATNA1, and CYP3A4, and the hypermethylated genes, BCL2, TK1, TRIP13, and RACGAP1, was not inversely correlated with the methylation status.
Candidate gene validation
The high ranking CpG sites from four candidate genes, characterized as either A) agnostic based on p-value alone (BEX2 and cXorf23; Additional file 1: Table S3) or B) biologically prioritized based on a previously established association with BPA (HOXA10 and DTL; Additional file 1: Table S6) were validated by quantitative bisulfite pyrosequencing (Additional file 1: Table S2). DTL with two CpG sites and HOXA10 with a single CpG site were run in duplicate on all samples with sufficient DNA, resulting in high correlation between replicates (N = 59, including the original 46 profiled on the BeadChip). The regions encoding BEX2 with 3 CpG sites and cXorf23 with a single CpG site were each run in a single sample well due to DNA quantity limitations (N = 59 including the original 46 profiled on BeadChip).
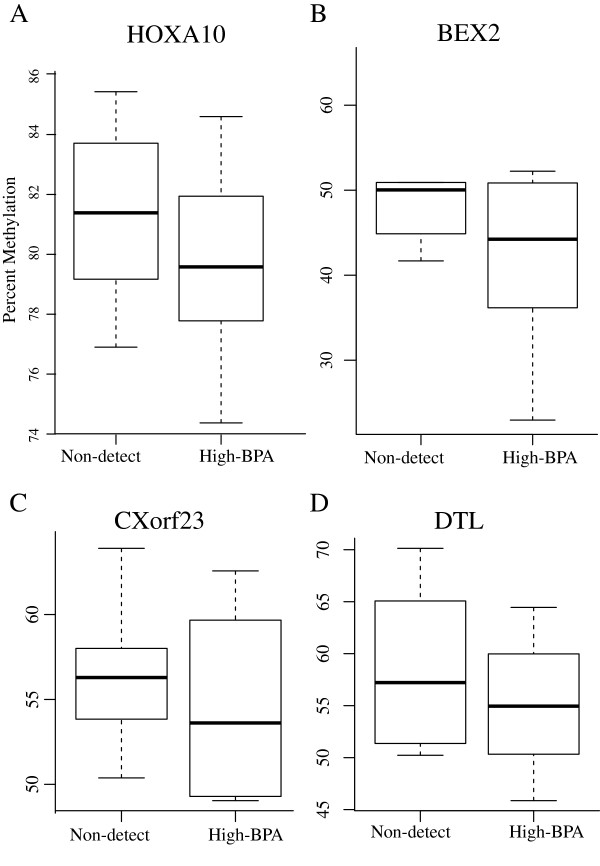
When methylation levels were compared between girls classified as BPA-low (<0.7 ng/mL, N = 22) to girls classified as BPA-high (>2 ng/mL, N = 13), no significant differences were observed at either the agnostic or biologically prioritized CpG sites (data not shown). However, when the analysis was restricted to girls with urinary concentrations of BPA below the limit of detection (N = 12) compared to girls characterized as BPA-high (N = 13), a decreasing, yet statistically non-significant, trend in DNA methylation in all candidate CpG sites was observed in all 4 genes (Figure 6A-D). Interestingly, further dichotomization of exposure categorization to girls with non-detectable concentrations of BPA (N = 12) and those with BPA concentrations 2 s.d. above the mean (>4 ng/mL, N = 3) revealed differential methylation with statistical significance (p-value < 0.024) at the HOXA10 biologically-prioritized candidate gene (Additional file 2: Figure S4).
Figure 6.
Quantitative levels of methylation for HOXA10, BEX2, CXorf23, and DTL (N = 12 for Non-detect and 13 for High-BPA for HOXA10, CXorf23, and DTL, and N = 9 for Non-detect and N = 9 for High-BPA for BEX2).
Discussion
The effect of environmental exposures on epigenetic regulation in human populations remains poorly understood. The consequences of maternal exposure to EACs, including BPA, on the epigenetic landscape of offspring has been studied using animal models, and this includes the discovery of altered DNA methylation at candidate metastable epialleles such as CDK5 activator-binding protein (Cabp) and viable yellow Agouti (Avy) alleles [17,19]. The effects of postnatal BPA exposure on the human epigenome, however, remain largely unexplored. To date, the majority of environmental epidemiology studies that incorporate epigenetics have been conducted in the context of a cohort study, where both environmental exposures and epigenetic outcomes are quantified in healthy populations [47-49]. The majority of these studies have looked at single genes or global methylation profiles. Increasingly, epigenome-wide approaches are useful in identifying a broader constellation of epigenetic changes associated with environmental exposures [50]. Here we measured DNA methylation levels genome-wide in 46 pre-adolescent Egyptian girls with varying concentrations of BPA exposure. The study population was homogeneous with respect to age and genetic population stratification was limited, thereby increasing our ability to identify exposure based methylation profiles.
While the overall levels of urinary BPA concentrations of Egyptian girls were low and less variable compared to age-matched American girls [27], we identified several CpG sites showing differential methylation upon exposure and biological pathways of interest using a genome-wide approach. This provides support for the hypothesis that even low concentrations of BPA exposure may be associated with health effects via epigenetic modification, and studies of relevant ranges of BPA exposure in human populations are warranted. These results should be interpreted cautiously as the use of spot urine for measuring BPA exposure is a limitation of our study. For example, interpersonal variation in amount of water consumed may affect urine output. Thus, we adjusted urine BPA concentrations using specific gravity. Similarly, to reduce variability arising from exposures throughout the day, the participants provided spot urine sample during a specified time period between noon to 4 pm. Additionally, previous studies on various populations have shown that individuals are exposed to mixtures of chemicals, potentially with similar mechanisms of action as BPA, such as phthalates, pesticides, and heavy metals. Future studies using more comprehensive methods of exposure assessment, including the measurement of chemical mixtures, are warranted [51].
Of interest, many previously identified pathways associated with BPA exposure were also identified in our analysis as top differentially methylated pathways. LRpath enrichment analysis identified BPA-induced hypomethylation among genes involved in metabolism and steroid hormones, including progesterone, testosterone, and estradiol. BPA is one of several widely known EACs or xenoestrogens [52,53], and steroid hormones are known to influence metabolism and inflammation including various immune responses. As such, several immune-response concepts were enriched with hypomethylated CpG sites in girls in the BPA-high group. In previous studies, higher cytokine activity was observed in BPA-exposed rodents [54]. In addition, the autoimmune thyroid disease pathway was significantly enriched with hypomethylated genes in girls categorized in the BPA-high group, and BPA is a known thyroid receptor antagonist [55,56]. Genes involved in the N-glycan biosynthesis pathway were also found to be differentially methylated based on BPA exposure, supporting previous work linking BPA concentrations with insulin resistance and type II diabetes [57], For example, N-glycans are key players mediating cell-to-cell communication, interacting with glycan-binding proteins [58], and can be attached to human insulin receptor (IR) and interfere with the IR interaction with antibodies or binding proteins by masking the surface of IR dimers [59].
Prenatal BPA exposure has been associated with altered DNA methylation and expression of imprinted genes in mouse embryos [60,61]. A methylation analysis of oocytes from CD-1 mice perinatally exposed to 20 or 40 ug BPA/kg diet identified decreased methylation among imprinted genes, including insulin like growth factor 2 receptor (IGF2R) and paternally expressed 3 (PEG3) [60]. When pregnant JF1 mice were exposed to 200 ug BPA/kg diet, embryos showed a 10% difference in Slc22a18 expression compared with controls [61]. When we examined DNA methylation of 34 imprinted genes (from 233 sites) in these subjects, BPA-associated changes were not observed in the imprinted genes, with the exception of a single CpG site in the NDN promoter (Additional file 2: Figure S3B). The imprinted gene, NDN, is located within the Prader-Willi/Angelman syndrome (PWS) chromosomal region [62]. Loss of function of this gene has been associated with post-natal lethality [63]; thus NDN has a critical role during development. Thus, methylation profiles at imprinted genes, while labile to nutritional and environmental influences early in development [64-66], may remain stable and relatively resistant to environmental factors over the life course.
DNA methylation of CpG islands acting in concert with other epigenetic mechanisms, are known to be important in female mammalian X chromosome inactivation and maintenance [67]. As with genomic imprinting, chromosome X inactivation may thus be potentially vulnerable to the effects of environmentally-induced epigenetic reprogramming [68]. In our data, we observed an enrichment of hypomethylated sites on chromosome X with increasing BPA concentrations (Figure 4 and Additional file 1: Table S5). One of the significantly hypomethylated cytobands in chromosome X was Xq13 (p-value < 0.02), where the X Inactivation Center (XIC) is located. The XIC is known to play a role in Xist expression, a major regulator of X inactivation. Two of the four sites validated by quantitative bisulfite pyrosequencing were CXorf23 (Xp22.12) and BEX2 (Xq22.1) from chromosome X. Although we were unable to achieve robust statistical significance as seen in the BeadChip data, we did observe a slight decrease in DNA methylation in BPA-high samples (Figure 6). As the girls observed in our study had low urinary BPA concentrations compared to age-matched American girls [27], it will be important to further evaluate the chromosome X findings in other exposed populations.
A functional validation of DNA methylation targets using gene expression was not performed in this cohort due to the unavailability of field-collected RNA. To overcome this limitation, we extracted gene expression information from multiple previously conducted studies available in CTD. The increasing wealth of data in the public arena now allows for the comparison of our genome-wide signatures with those from other studies, efficiently providing additional insight into key biologically-relevant pathways. When the curated genes known to interact or harbor differential expression upon BPA exposure in the CTD were compared with the list of differentially methylated genes in our study, more than 20% of BPA-associated genes were found to overlap. The biologically-prioritized candidate gene, BEX2, is associated with estrogen, cell cycle, and apoptosis pathways [69] and over-expressed in Ishikawa cells, a well-differentiated human endometrial adenocarcinoma cells, treated with BPA [70]. In addition, the biologically-prioritized candidate, HOXA10, has previously been associated with decreased DNA methylation among in utero BPA-exposed mouse samples [71]. Altered methylation at HOXA10 was confirmed by bisulfite pyrosequencing in our Egyptian girls, despite relatively low BPA concentrations with small variation compared to US populations [27]. Although the validation did not result in the same depth of significance as seen in the array data, it showed a clear trend toward differential methylation (Additional file 2: Figure S4). Importantly, some biologically-prioritized candidate genes are known to target the major disease pathways, such as DNA repair, cell cycle, and development. Thus, future epigenetic epidemiology studies with increased sample size and increased exposure ranges are needed to further elucidate the consequences of genome-wide altered methylation upon BPA exposure, including BRCA1, BEX2, and HOXA10.
Conclusions
BPA exposure levels in Egyptian pre-adolescent girls were relatively low with less variability compared to age-matched American girls; however, using a genome-wide methylation approach, we identified alterations in saliva DNA methylation associated with BPA concentrations. Moreover, a number of identified genes are important regulators in developmental processes and/or disease susceptibility. Pathways involved in metabolism, steroid hormone, thyroid disease, and immune response were also enriched among differentially methylated genes. The integrative analysis with data obtained from the CTD database identified candidate genes, where epigenetic modifications may be associated with concomitant transcriptomic changes observed upon BPA exposure. Although methylation is more susceptible to environmental exposures during early development, these data indicate that BPA may affect human health through specific epigenomic modification of genes in relevant pathways even throughout pre-adolescent development.
Abbreviations
Avy: Viable yellow agouti; BPA: Bisphenol A; BMI: Body mass index; Cabp: CDK5 activator-binding protein; CTD: Comparative Toxicogenomics Database; DES: Diethylstilbestrol; DMRs: Differentially methylated regions; DOHaD: Developmental origins of health and disease; EACs: Endocrine-active compounds; EHMN: Edinburgh Human Metabolic Network; GO: Gene ontology; GSEA: Gene set enrichment analysis; IGF2: Insuline-like growth factor 2; IGF2R: Insulin like growth factor 2 receptor; IR: Insulin receptor; KEGG: Kyoto Encyclopedia of Genes and Genomes; LIMMA: Linear models for microarray data; LOD: Limit of detection; LRpath: Logistic regression based pathway analysis tool; NDN: Necdin; NHANES: National Health and Nutrition Examination Survey; Pde4d4: Phosphodiesterase type 4 variant 4; PEG3: Paternally expressed 3; PWS: Prader-Willi/Angelman Syndrome; RPMM: Recursively partitioned mixture model.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AH, ASS, DCD, IAS, and LSR conceived of this project and designed the study protocols. AH, ASS, and IAS supervised and conducted field collection, including subject recruitment. AMC was responsible for laboratory analysis of BPA. JHK, MSN, and MAS performed statistical analysis. JHK, JAC, and CW carried out pyrosequencing analysis. JHK, DCD, and LSR drafted the manuscript, and all authors edited and approved the final draft of the manuscript.
Supplementary Material
Distribution of urinary BPA concentration (ng/ml). Table S2. Pyrosequencing Primer sequences. S3. Candidate Genes. Table S4. Gene ontology enrichment analysis using LRpath. Table S5. Cytoband enrichment analysis using GSEA. Table S6. Differential methylation in BPA interacting genes.
Average percent change in differentially methylated probes (p-value < 0.05) between BPA-low and BPA-high samples (N=1,439). Figure S2. The tree plot of mean-centered β-scores of top 200 most variable CpG sites. Figure S3. The levels of methylation observed among imprinted genes in Egyptian cohort (A) Averaged β-score of 34 unique genes from 233 probes is displayed in heatmap. Red color indicates higher levels of methylation. (B) The β-score boxplots of a CpG site located 308 bp upstream of NDN promoter from BPA-Low and BPA-High groups. Figure S4. Quantitative levels of HOXA10 methylation in the Egyptian girls cohort.
Contributor Information
Jung H Kim, Email: junghk@umich.edu.
Laura S Rozek, Email: rozekl@umich.edu.
Amr S Soliman, Email: amr.soliman@unmc.edu.
Maureen A Sartor, Email: sartorma@umich.edu.
Ahmed Hablas, Email: ahablas@hotmail.com.
Ibrahim A Seifeldin, Email: ieldein@hotmail.com.
Justin A Colacino, Email: colacino@umich.edu.
Caren Weinhouse, Email: carenw@umich.edu.
Muna S Nahar, Email: msnahar@umich.edu.
Dana C Dolinoy, Email: ddolinoy@umich.edu.
Acknowledgements
This work was supported by the University of Michigan NIEHS Center of excellence P30 ES017885 and by NIH grant ES017524. Support for CW, JAC, and MSN was provided by NIEHS Institutional Training Grant T32 ES007062. Support for JAC was also provided by NHGRI Institutional Training Grant T32 HG00040. We would like to acknowledge Dr. Antonia Calafat and her team at the National Center for Environmental Health, Centers for Disease Control and Prevention, Atlanta, GA for their part in the analysis of BPA concentrations. We would also like to acknowledge Ms. Stacy Endres for her effort in part of the data collection in Egypt and the Cancer Epidemiology Education in Special Population Program Grant R25 CA112383. The authors have no conflicts of interest and declare no competing financial interests.
References
- Barker DJ. Intrauterine programming of coronary heart disease and stroke. Acta Paediatra Suppl. 1997;423:178–182. doi: 10.1111/j.1651-2227.1997.tb18408.x. [DOI] [PubMed] [Google Scholar]
- Barker DJP. In: Mothers, Babies, and Disease in Later Life. 2. Barker DJP, editor. Edinburgh: New York: Churchill Livingstone; 1998. Programming the baby; pp. 13–42. [Google Scholar]
- Wojtyla A. Application of the hypothesis of Developmental Origin of Health and Diseases (DOHaD) in epidemiological studies of women at reproductive age and pregnant women in Poland. Ann Agric Environ Med. 2011;18:355–364. [PubMed] [Google Scholar]
- Elston CW, Ellis IO. In: The breast: systemic pathology. Volume 13. 3. Elston CW, Ellis IO, editor. Churchill Livingston: Edinburgh; 1998. Normal structure and developmental abnormalities; pp. 1–19. [Google Scholar]
- Russo J, Russo IH. In: The mammary gland: development, regulation, and function. Volume 1. Neville MC, Daniel CW, editor. New York: Springer; 1987. Development of the human mammary gland; pp. 67–93. [Google Scholar]
- Daniel CW, Silberstein GB. In: The mammary gland: development, regulation, and function. Volume 1. Neville MC, Daniel CW, editor. New York: Springer-Verlag; 1987. Postnatal development of the rodent mammary gland. [Google Scholar]
- McNally S, Martin F. Molecular regulators of pubertal mammary gland development. Ann Med. 2011;43:212–234. doi: 10.3109/07853890.2011.554425. [DOI] [PubMed] [Google Scholar]
- Russo J, Russo IH. Development of the human breast. Maturitas. 2004;49:2–15. doi: 10.1016/j.maturitas.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog. 2003;38:78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- Li S, Washburn K, Moore R, Uno T, Teng C, Newbold R, McLachlan J, Negishi M. Developmental exposure to diethylstilbestrol elicits demethylation of estrogen-responsive lactoferrin gene in mouse uterus. Cancer Res. 1997;57:4356–4359. [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H-S, Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147:5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci USA. 2012;109:9143–9148. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova MI, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS One. 2012;7:e36129. doi: 10.1371/journal.pone.0036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S, Tang W, Belmonte de Frausto J, Prins G. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Anderson O, Nahar M, Faulk C, Jones T, Liao C, Kannan K, Weinhouse C, Rozek L, Dolinoy D. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen. 2012;53:334–342. doi: 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Wiedman J, Waterland R, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini MV, Rubin BS, Sonnenschein C, Soto AM. Endocrine disruptors and reproductive health: the case of bisphenol-A. Mol Cell Endocrinol. 2006;254–255:179–186. doi: 10.1016/j.mce.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Lang IA, Galloway TS, Scarlett A, Henley WE, Depledge M, Wallace RB, Melzer D. Association of urinary bisphenol A concentration with medical disorders and laboratory abnormalities in adults. JAMA. 2008;300:1303–1310. doi: 10.1001/jama.300.11.1303. [DOI] [PubMed] [Google Scholar]
- Trasande L, Attina TM, Blustein J. Association between urinary bisphenol A concentration and obesity prevalence in children and adolescents. JAMA. 2012;308:1113–1121. doi: 10.1001/2012.jama.11461. [DOI] [PubMed] [Google Scholar]
- Stahlhut RW, Welshons WV, Swan SH. Bisphenol A data in NHANES suggest longer than expected half-life, substantial nonfood exposure, or both. Environ Health Perspect. 2009;117:784–789. doi: 10.1289/ehp.0800376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody JG, Rudel RA, Michels KB, Moysich KB, Bernstein L, Attfield KR, Gray S. Environmental pollutants, diet, physical activity, body size, and breast cancer: where do we stand in research to identify opportunities for prevention? Cancer. 2007;109:2627–2634. doi: 10.1002/cncr.22656. [DOI] [PubMed] [Google Scholar]
- Colacino JA, Soliman AS, Calafat AM, Nahar MS, Van Zomeren-Dohm A, Hablas A, Seifeldin IA, Rozek LS, Dolinoy DC. Exposure to phthalates among premenstrual girls from rural and urban Gharbiah, Egypt: a pilot exposure assessment study. Environ Health. 2011;10:40. doi: 10.1186/1476-069X-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar MS, Soliman AS, Colacino JA, Calafat AM, Battige K, Hablas A, Seifeldin IA, Dolinoy DC, Rozek LS. Urinary bisphenol A concentrations in girls from rural and urban Egypt: a pilot study. Environ Health. 2012;11:20. doi: 10.1186/1476-069X-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Zhang Z, Hablas A, Seifeldein IA, Ramadan M, El-Hamzawy H, Soliman AS. Geographic patterns of cancer in the population-based registry of Egypt: Possible links to environmental exposures. Cancer Epidemiol. 2011;35:254–264. doi: 10.1016/j.canep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Dey S, Soliman AS, Hablas A, Seifeldein IA, Ismail K, Ramadan M, El-Hamzawy H, Wilson ML, Banerjee M, Boffetta P. Urban–rural differences in breast cancer incidence in Egypt (1999–2006) Breast. 2010;19:417–423. doi: 10.1016/j.breast.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel AM, Soliman AS, Zhang Q, El-Ghawalby N, Ezzat F, Soultan A, Abdel-Wahab M, Fathy O, Ebidi G, Bassiouni N. Serum cadmium levels in pancreatic cancer patients from the East Nile Delta region of Egypt. Environ Health Perspect. 2006;114:113–119. doi: 10.1289/ehp.114-a113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, Wiegers T, Mattingly CJ. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Murphy CG, Saraceni-Richards CA, Rosenstein MC, Wiegers TC, Mattingly CJ. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009;37:D786–792. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Murphy CG, Rosenstein MC, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database facilitates identification and understanding of chemical-gene-disease associations: arsenic as a case study. BMC Med Genomics. 2008;1:48. doi: 10.1186/1755-8794-1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaly I, Hussein FH, Abdelghaffar S, Anwar G, Seirvogel RM. Optimal age of sexual maturation in Egyptian children. East Mediterr Health J. 2008;14:1391–1399. [PubMed] [Google Scholar]
- Soliman AS, Schairer C. Considerations in setting up and conducting epidemiologic studies of cancer in middle- and low-income countries: the experience of a case–control study of inflammatory breast cancer in North Africa in the past 10 years. Cancer Med. 2012;1:338–349. doi: 10.1002/cam4.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schairer C, Soliman AS, Omar S, Khaled H, Eissa S, Ben Ayed F, Khalafallah S, Ben Ayoub W, Kantor ED, Merajver S. Assessment of diagnosis of inflammatory breast cancer cases at two cancer centers in Egypt and Tunisia. Cancer Med. 2013;2:178–184. doi: 10.1002/cam4.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Kuklenyik Z, Needham LL, Calafat AM. Quantification of urinary conjugates of bisphenol A, 2,5-dichlorophenol, and 2-hydroxy-4-methoxybenzophenone in humans by online solid phase extraction-high performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2005;383:638–644. doi: 10.1007/s00216-005-0019-4. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou LX, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium (R) assay. Epigenomics-Uk. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, Rozek LS. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6(6):777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Kim JH, Karnovsky A, Mahavisno V, Weymouth T, Pande M, Dolinoy DC, Rozek LS, Sartor MA. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics. 2012;13:526. doi: 10.1186/1471-2164-13-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirlwell C, Eymard M, Feber A, Teschendorff A, Pearce K, Lechner M, Widschwendter M, Beck S. Genome-wide DNA methylation analysis of archival formalin-fixed paraffin-embedded tissue using the Illumina Infinium HumanMethylation27 BeadChip. Methods. 2010;52:248–254. doi: 10.1016/j.ymeth.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Cotton AM, Lam L, Affleck JG, Wilson IM, Penaherrera MS, McFadden DE, Kobor MS, Lam WL, Robinson WP, Brown CJ. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 2011;130:187–201. doi: 10.1007/s00439-011-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics. 2011;6:920–927. doi: 10.4161/epi.6.7.16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, Jacobs DR, Steffes M, Lee DH. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect. 2010;118:370–374. doi: 10.1289/ehp.0901131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA. 450 K Epigenome-Wide Scan Identifies Differential DNA Methylation in Newborns Related to Maternal Smoking During Pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff TJ, Zota AR, Schwartz JM. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ Health Perspect. 2011;119:878–885. doi: 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Tokunaga T, Liu X, Takayanagi S, Matsushima A, Shimohigashi Y. Direct evidence revealing structural elements essential for the high binding ability of bisphenol A to human estrogen-related receptor-gamma. Environ Health Perspect. 2008;116:32–38. doi: 10.1289/ehp.116-a32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Saal FS, Myers JP. Bisphenol A and risk of metabolic disorders. JAMA. 2008;300:1353–1355. doi: 10.1001/jama.300.11.1353. [DOI] [PubMed] [Google Scholar]
- Yoshino S, Yamaki K, Li X, Sai T, Yanagisawa R, Takano H, Taneda S, Hayashi H, Mori Y. Prenatal exposure to bisphenol A up-regulates immune responses, including T helper 1 and T helper 2 responses, in mice. Immunology. 2004;112:489–495. doi: 10.1111/j.1365-2567.2004.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Tagami T, Akamizu T, Usui T, Saijo M, Kanamoto N, Hataya Y, Shimatsu A, Kuzuya H, Nakao K. Thyroid hormone action is disrupted by bisphenol A as an antagonist. J Clin Endocrinol Metab. 2002;87:5185–5190. doi: 10.1210/jc.2002-020209. [DOI] [PubMed] [Google Scholar]
- Zoeller RT, Bansal R, Parris C. Bisphenol-A, an environmental contaminant that acts as a thyroid hormone receptor antagonist in vitro, increases serum thyroxine, and alters RC3/neurogranin expression in the developing rat brain. Endocrinology. 2005;146:607–612. doi: 10.1210/en.2004-1018. [DOI] [PubMed] [Google Scholar]
- Thayer KA, Heindel JJ, Bucher JR, Gallo MA. Role of environmental chemicals in diabetes and obesity: a National Toxicology Program workshop review. Environ Health Perspect. 2012;120:779–789. doi: 10.1289/ehp.1104597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulos A, North SJ, Haslam SM, Dell A. Glycosylation of mouse and human immune cells: insights emerging from N-glycomics analyses. Biochem Soc Trans. 2011;39:1334–1340. doi: 10.1042/BST0391334. [DOI] [PubMed] [Google Scholar]
- Sparrow LG, Lawrence MC, Gorman JJ, Strike PM, Robinson CP, McKern NM, Ward CW. N-linked glycans of the human insulin receptor and their distribution over the crystal structure. Proteins. 2008;71:426–439. doi: 10.1002/prot.21768. [DOI] [PubMed] [Google Scholar]
- Chao HH, Zhang XF, Chen B, Pan B, Zhang LJ, Li L, Sun XF, Shi QH, Shen W. Bisphenol A exposure modifies methylation of imprinted genes in mouse oocytes via the estrogen receptor signaling pathway. Histochem Cell Biol. 2012;137:249–259. doi: 10.1007/s00418-011-0894-z. [DOI] [PubMed] [Google Scholar]
- Kang ER, Iqbal K, Tran DA, Rivas GE, Singh P, Pfeifer GP, Szabo PE. Effects of endocrine disruptors on imprinted gene expression in the mouse embryo. Epigenetics. 2011;6:937–950. doi: 10.4161/epi.6.7.16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei MG, Malzac P, Roeckel N, Taviaux S, Lefranc JL, Cau P. The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- Gerard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Slagboom PE, van Dongen J, Kremer D, Stein AD, Putter H, Heijmans BT, Lumey LH. Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLoS One. 2012;7:e37933. doi: 10.1371/journal.pone.0037933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF. X chromosome inactivation, XIST, and pursuit of the X-inactivation center. Cell. 1996;86:5–7. doi: 10.1016/S0092-8674(00)80071-9. [DOI] [PubMed] [Google Scholar]
- Naderi A, Teschendorff AE, Beigel J, Cariati M, Ellis IO, Brenton JD, Caldas C. BEX2 is overexpressed in a subset of primary breast cancers and mediates nerve growth factor/nuclear factor-kappaB inhibition of apoptosis in breast cancer cell lines. Cancer Res. 2007;67:6725–6736. doi: 10.1158/0008-5472.CAN-06-4394. [DOI] [PubMed] [Google Scholar]
- Naciff JM, Khambatta ZS, Reichling TD, Carr GJ, Tiesman JP, Singleton DW, Khan SA, Daston GP. The genomic response of Ishikawa cells to bisphenol A exposure is dose- and time-dependent. Toxicology. 2010;270:137–149. doi: 10.1016/j.tox.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of urinary BPA concentration (ng/ml). Table S2. Pyrosequencing Primer sequences. S3. Candidate Genes. Table S4. Gene ontology enrichment analysis using LRpath. Table S5. Cytoband enrichment analysis using GSEA. Table S6. Differential methylation in BPA interacting genes.
Average percent change in differentially methylated probes (p-value < 0.05) between BPA-low and BPA-high samples (N=1,439). Figure S2. The tree plot of mean-centered β-scores of top 200 most variable CpG sites. Figure S3. The levels of methylation observed among imprinted genes in Egyptian cohort (A) Averaged β-score of 34 unique genes from 233 probes is displayed in heatmap. Red color indicates higher levels of methylation. (B) The β-score boxplots of a CpG site located 308 bp upstream of NDN promoter from BPA-Low and BPA-High groups. Figure S4. Quantitative levels of HOXA10 methylation in the Egyptian girls cohort.