Abstract
Background
Influenza-like illnesses (ILI) are estimated to cause millions of deaths annually. Despite this disease burden, the etiologic causes of ILI are poorly described for many geographical regions.
Methods
Beginning in April 2010, we conducted an observational cohort study at five hospitals in Mexico City, enrolling subjects who met the criteria for ILI. Evaluations were conducted at enrollment and on day 28, with the collection of clinical data and a nasopharyngeal swab (or nasal aspirate in children). Swabs were tested by multiplex PCR for 15 viral pathogens and real-time PCR for influenza.
Results
During the first year, 1065 subjects were enrolled in this study, 55% of whom were hospitalized; 24% of all subjects were children. One or more pathogens were detected by PCR in 64% of subjects, most commonly rhinovirus (25% of all isolates) and influenza (24% of isolates). Six percent of subjects died, and of those, 54% had no pathogen identified. Rhinovirus was the most common pathogen among those who died, although it did not have the highest case fatality rate.
Conclusions
Multiple respiratory viruses beyond influenza are associated with significant morbidity and mortality among adults and children in Mexico City. Detection of these agents could be useful for the adjustment of antibiotic treatment in severe cases.
Keywords: Respiratory viral pathogens, Epidemiology, Hospital burden of disease, Influenza, Respiratory syncytial virus, Rhinovirus, Coronavirus
1. Introduction
In April 2009, an outbreak of respiratory disease in young people was reported in Mexico City,1 and the etiologic agent was identified as influenza A virus H1N1pdm09.2 This outbreak and its consequences compelled the Ministry of Health of Mexico and the US National Institute of Allergy and Infectious Diseases (NIAID) to collaborate in establishing a Mexican Emerging Infectious Diseases Clinical Research Network (La Red).
Acute respiratory infections are estimated to cause 3.9 million deaths annually,3 many of which are influenza-like illnesses (ILI). The etiology of ILI in Mexican individuals seeking medical care is not well-defined. It has been reported in other populations that multiple respiratory viruses such as parainfluenza virus, rhinovirus, adenovirus, metapneumovirus, respiratory syncytial virus (RSV), and coronavirus can cause ILI,4 but the virologic etiology varies greatly among geographic locales, age groups, seasons, and years.5, 6, 7, 8, 9, 10, 11, 12, 13 Some studies have suggested that specific symptoms could be used for the clinical prediction of the etiology of ILI,14, 15, 16, 17, 18 though most of these studies only compared influenza to non-influenza etiology.
Although the outcomes of subjects infected with various influenza subtypes have been well documented,19, 20, 21 there is relatively little description of outcomes from other viral etiologies of ILI. Shlomai et al. compared outcomes of those hospitalized with an ILI, and found mortality of 4.6% in influenza patients vs. 7% in influenza-negative ILI patients.18 As further demonstration of the burden that non-influenza etiological agents contribute to ILIs, Widmer et al. reported that RSV hospitalization rates exceeded those of influenza (15.01 and 11.81 per 10 000, respectively).22
It is important to know the characteristics and seasonal behavior of the different types of virus causing ILI. Although current clinical diagnostic tests of all viruses are high in cost, improved diagnosis would allow a more judicious use of empirical wide-spectrum antibiotic treatment, which in many cases could be suspended, and thus reduce the cost of hospitalization.
Recognizing the need to investigate both influenza and non-influenza ILIs in the Mexican population, La Red implemented a study with the objective of describing the etiology, symptoms, and outcomes of subjects presenting with ILI in Mexico City.
2. Methods
2.1. Study design and sites
Beginning in April 2010, this hospital-based observational cohort study was conducted at five hospitals in Mexico City. These hospitals are located in the south of Mexico City, and include a general hospital, a tertiary care hospital that serves mainly those with respiratory problems, a tertiary care hospital that serves the metabolic/surgical population, and two pediatric centers. These centers were chosen given their capacity to conduct clinical research.
2.2. Case definition and study population
The study population included subjects aged >3 months who presented with an ILI. ILI was defined by the presence of at least one respiratory symptom (e.g., shortness of breath, postnasal drip, and cough) and one of the two following criteria: (1) fever (≥38 °C by any method) on examination, or participant-reported fever (≥38 °C), or feverishness in the past 24 h; (2) one or more non-respiratory symptoms (e.g., malaise, headache, myalgia, and chest pain). The subjects included were those who sought medical attention at our centers and agreed to participate in the study.
2.3. Study procedures
Demographic data were collected at enrollment. A nasopharyngeal swab (Copan, Brescia, Italy), or a nasal aspirate in children, was obtained for PCR detection of respiratory pathogens and sample storage. The samples were placed in transport medium, maintained at 4 °C, and sent within 72 h to a central facility (Molecular Biology Laboratory, Infectious Diseases Department, INNSZ, Mexico City), where they were stored at −70 °C. Clinical laboratory tests were carried out at enrollment. If available, the results of tests taken for standard clinical care (chest X-ray, bacteriological cultures, arterial blood gases, and liver tests) were also abstracted from the medical records. Subjects were seen again on day 28, and follow-up information was also obtained by phone on days 14 and 60. At each visit, clinical information (symptoms, chronic medical conditions, previous treatment, impact on daily function, hospitalizations, and death) was assessed.
2.4. Virology
All samples were tested by real-time reverse transcriptase PCR for influenza A following the Centers for Disease Control and Prevention (CDC) protocol (CDC 2009), as described previously.23
For multi-pathogen detection, the samples were tested with the RespiFinder19 kit (previously RespiFinder Plus, from PathoFinder BV, Maastricht, The Netherlands), as described previously.24 This multiplex PCR test can detect and differentiate 15 viruses (coronavirus NL63, OC43, and 229E, human metapneumovirus, influenza A, influenza A H5N1, influenza B, parainfluenza virus types 1 to 4, RSV A and B, rhinovirus, and adenovirus), as well as four bacteria (Bordetella pertussis, Chlamydophila pneumoniae, Legionella pneumophila, and Mycoplasma pneumoniae). The analytical sensitivity (reported by the manufacturer) varies between 5 and 50 copies per reaction for most targets.25
For the samples with discordant real-time reverse transcriptase PCR results and multiplex PCR for influenza, the samples were further tested with a third method, based on a final-point nested PCR protocol.26 For these samples, the result of this test was considered the final result.
2.5. Statistical analysis
Data were analyzed with the Statistical Analysis System (SAS Institute, Cary, NC, USA). Median (range) and percentage were used to summarize quantitative and qualitative variables, respectively. To discard the role of chance, the Mann–Whitney test and the Chi-square or Fisher's exact test were used as appropriate. Univariate logistic regression analyses were performed to evaluate the effect of each baseline covariate on the risk of death. Odds ratios (OR) and the corresponding 95% confidence intervals (CI) were reported based on the fitted logistic regression models. All p-values were two-sided, with no correction for multiple comparisons; p < 0.05 was considered statistically significant.
3. Results
3.1. Patients
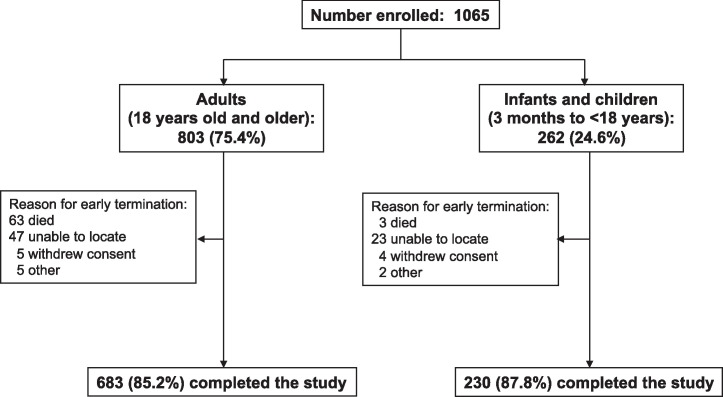
From April 11, 2010 through April 10, 2011, 1065 subjects were enrolled in this study. A total of 86 withdrew or were withdrawn from the study (Figure 1 ). Characteristics of the study participants are listed in Table 1 . Just under a quarter of the study subjects were children and 58.8% were female. Four hundred thirty-six (40.9%) subjects were hospitalized. Asthma and cardiovascular disease were the most common chronic medical conditions, though neither condition accounted for more than 17% of subjects.
Figure 1.
Enrollment and follow-up.
Table 1.
Baseline characteristics
| Demographics | Total, N = 1065 | Children, n = 262 | Adults, n = 803 |
|---|---|---|---|
| Age, years | |||
| Median | 33 | ||
| Range | 0–96 | ||
| ≤18 | 262 (24.6%) | ||
| 19–59 | 614 (57.6%) | ||
| ≥60 | 189 (17.8%) | ||
| Sex | |||
| Male | 439 (41.2%) | 132 (50.4%) | 307 (38.2%) |
| Female | 626 (58.8%) | 130 (49.6%) | 496 (61.8%) |
| Pregnant | 4 | 4 | |
| Ethnicity | |||
| Mixed | 1026 (96.4%) | 252 (96.2%) | 775 (96.5%) |
| White | 33 (3.1%) | 10 (3.8%) | 23 (2.9%) |
| Other | 5 (0.5%) | 5 (0.6%) | |
| Medical history | |||
| Influenza vaccination | 504 (47.3%) | 121 (46.2%) | 383 (47.7%) |
| Asthma | 151 (14.2%) | 16 (6.1%) | 135 (16.8%) |
| CVD | 147 (13.8%) | 17 (6.5%) | 130 (16.2%) |
| Diabetes | 99 (9.3%) | 99 (12.3%) | |
| Immunodeficiency | 48 (4.5%) | 8 (3.1%) | 40 (5.0%) |
| Renal disorder | 42 (3.9%) | 4 (1.5%) | 38 (4.8%) |
| COPD | 36 (3.4%) | 36 (4.5%) | |
| BMI categorya | |||
| Obese | 207 (19.9%) | 23 (9.2%) | 184 (23.3%) |
| Overweight | 313 (30.2%) | 42 (16.1%) | 273 (34.6%) |
| Normal | 417 (40.2%) | 121 (48.6%) | 296 (37.5%) |
| Underweight | 101 (9.7%) | 65 (26.1%) | 36 (4.6%) |
| Median days since symptoms onset | 1 | ||
| Inpatient at baseline | 436 (40.9%) | 113 (43.1%) | 323 (40.2%) |
| Outpatient at baseline | 629 (59.1%) | 149 (56.9%) | 480 (59.8) |
BMI, body mass index; COPD, chronic obstructive pulmonary disease; CVD, cardiovascular disease; SD, standard deviation.
BMI categories: underweight = BMI < 18.5 kg/m2 (or <1 SD for children); normal weight = 18.5–24.9 kg/m2; overweight = 25–29.9 kg/m2 (>1SD for children); obese = BMI of ≥30 kg/m2 (>2 SD for children).
3.2. Etiology
A total of 821 etiologic agents were identified among the 678 (64% of total) subjects from whom a pathogen was isolated. Rhinovirus was the most frequently isolated pathogen, infecting 15.3% of enrolled subjects and accounting for 25.3% of all isolates. Rhinovirus was followed by influenza, which was detected in 14.3% of subjects, and 24% of isolates. Adenovirus, coronavirus, metapneumovirus, and RSV all had an isolate frequency of 9% or greater. No pathogen was detected for 35.5% of subjects (Table 2 ).
Table 2.
Frequency distribution of viral isolates
| Pathogen | Age <18a, n (%) | Age 18–59a, n (%) | Age ≥60a, n (%) | Totala, n (%) | Totalb, n (%) |
|---|---|---|---|---|---|
| Adenovirus | 17 (6.6) | 15 (2.5) | 3 (1.6) | 35 (3.3) | 74 (9.0) |
| Bordetella pertussis | 2 (0.8) | 3 (0.5) | 0 (0) | 5 (0.5) | 9 (1.1) |
| Coronavirus | 12 (4.7) | 50 (8.2) | 15 (8.2) | 77 (7.3) | 118 (14.4) |
| 229E | 0 | 11 | 2 | 13 | 22 |
| NL63 | 2 | 8 | 1 | 11 | 18 |
| OC43 | 10 | 31 | 12 | 53 | 78 |
| Influenza | 43 (16.7) | 89 (14.6) | 18 (9.8) | 150 (14.3) | 197 (24.0) |
| A | 3 | 5 | 0 | 8 | 11 |
| A H1N1 2009 | 0 | 5 | 1 | 6 | 7 |
| A H3N2 | 16 | 46 | 11 | 73 | 91 |
| B | 24 | 33 | 6 | 63 | 88 |
| Metapneumovirus | 11 (4.3) | 22 (3.6) | 9 (4.9) | 42 (4.0) | 78 (9.5) |
| Mycoplasma pneumoniae | 1 (0.4) | 4 (0.7) | 1 (0.5) | 6 (0.6) | 10 (1.2) |
| Parainfluenza virus | 6 (2.3) | 12 (1.9) | 2 (1.1) | 20 (1.9) | 42 (5.1) |
| 1 | 0 | 0 | 0 | 0 | 3 |
| 2 | 0 | 0 | 0 | 0 | 3 |
| 3 | 6 | 7 | 1 | 14 | 24 |
| 4 | 0 | 5 | 1 | 6 | 12 |
| Rhinovirus | 29 (11.2) | 100 (16.4) | 32 (17.4) | 161 (15.3) | 208 (25.3) |
| RSV | 34 (13.2) | 15 (2.5) | 8 (4.3) | 57 (5.4) | 85 (10.3) |
| A | 33 | 15 | 6 | 54 | 72 |
| B | 1 | 0 | 2 | 3 | 13 |
| Mixed infections | 51 (19.8) | 60 (9.9) | 14 (7.6) | 125 (11.9) | |
| Negative cases | 52 (20.2) | 238 (39.1) | 83 (45.1) | 373 (35.5) | |
| Total | 258 (100) | 609 (100) | 184 (100) | 1051 (100)c | 821 (100) |
RSV, respiratory syncytial virus.
Frequency by subject. Infections with more than one virus are counted as a single category (mixed infection).
Frequency by isolates. Excludes negative cases.
Fourteen subjects did not have a baseline sample for virologic testing.
Among the 125 subjects (11.9%) with mixed virus infections, 62 unique combinations were recorded. The most common combinations were adenovirus/rhinovirus in nine subjects (7.2% of combinations), adenovirus/metapneumovirus in eight subjects (6.4%), and influenza B/metapneumovirus in eight subjects (6.4%). Influenza was the most common virus involved in co-infections (when considering all subtypes), present in 45/125 (36%) combinations. Most combinations were limited to two viruses.
3.3. Seasonal distribution of viruses
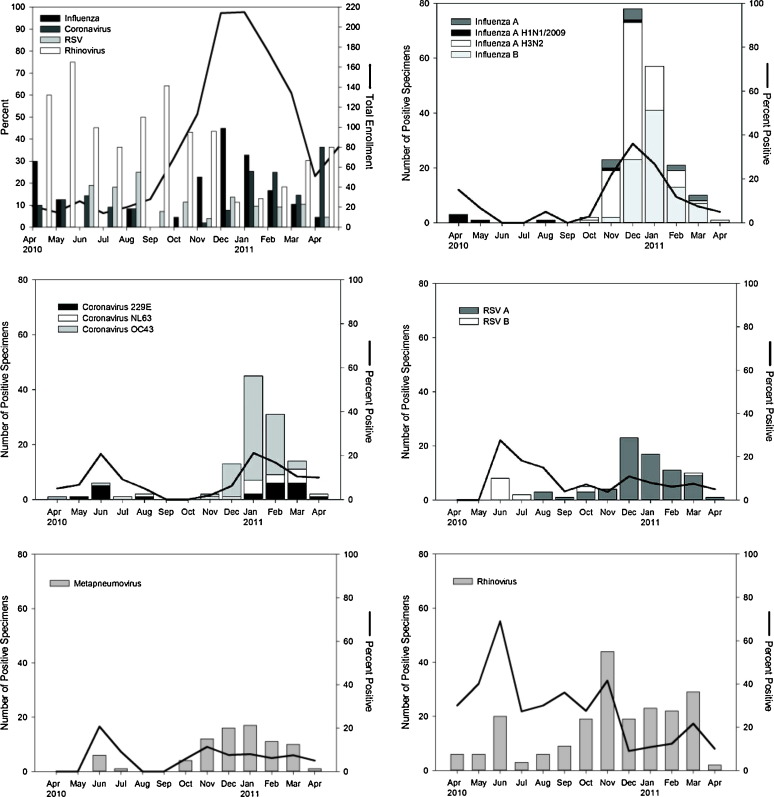
The four most common pathogens (rhinovirus, influenza, coronavirus, and RSV) all had seasonal distributions with more isolates observed during cooler months (December–March). The seasonal variation of rhinovirus and RSV, however, was less pronounced than that of coronavirus and influenza (Figure 2 ). During April – November, rhinovirus accounted for 20–60% of all ILI. However, between December and March – a period during which the total number of ILI cases increased – the relative contribution of rhinovirus to ILI was only 10–20%.
Figure 2.
Monthly distribution of viral isolates. Total monthly enrollment (line), and the four most prevalent viruses (A). Monthly isolates by virus subtype (bars) and percent positive are demonstrated for the five most common viruses: influenza (B), coronavirus (C), respiratory syncytial virus (D), metapneumovirus (E), and rhinovirus (F).
3.4. Vaccination
Previous influenza vaccination was reported for 46.2% of the children and 47.7% of the adults. We found no difference in the proportion of vaccinated and unvaccinated subjects in the persons with virus or without virus detection, neither for individual agents nor as a group.
3.5. Symptoms
Evaluation across the four most common pathogens showed that children were more likely to have fever, productive cough, and nausea, whereas adults were more likely to have fatigue, headache, and sore throat (Table 3 ). Respiratory symptoms, such as dyspnea, were more common among those with rhinovirus or RSV, and gastrointestinal symptoms, such as nausea or diarrhea, were more common among those with influenza.
Table 3.
Demographics and symptoms by age and pathogen
| Children |
Adults |
|||||||
|---|---|---|---|---|---|---|---|---|
| Coronavirus | Influenza | Rhinovirus | RSV | Coronavirus | Influenza | Rhinovirus | RSV | |
| Number enrolled | 12 | 43 | 29 | 34 | 65 | 107 | 132 | 23 |
| Any antiviral | 0% | 9% | 7% | 6% | 28% | 33% | 27% | 35% |
| Chronic medical condition | 42% | 28% | 38% | 26% | 42% | 44% | 63% | 74% |
| Asthma | 8% | 7% | 10% | 3% | 11% | 21% | 19% | 26% |
| CVD | 0% | 0% | 10% | 6% | 15% | 10% | 20% | 17% |
| COPD | 0% | 0% | 0% | 0% | 5% | 6% | 2% | 13% |
| DM | 0% | 0% | 0% | 0% | 8% | 6% | 19% | 13% |
| Antibiotics | 50% | 44% | 52% | 71% | 40% | 48% | 66% | 65% |
| Inhaled steroids | 8% | 9% | 21% | 24% | 18% | 20% | 14% | 35% |
| Systemic steroids | 25% | 9% | 17% | 21% | 22% | 24% | 28% | 43% |
| Pregnant | 0% | 0% | 0% | 0% | 0% | 1% | 0% | 0% |
| Non-smoker | 0% | 0% | 0% | 0% | 72% | 51% | 60% | 65% |
| Current/former smoker | 0% | 0% | 0% | 0% | 28% | 49% | 40% | 35% |
| Seasonal flu vaccine | 50% | 47% | 45% | 38% | 60% | 39% | 39% | 52% |
| Outpatient | 67% | 81% | 38% | 35% | 68% | 67% | 52% | 57% |
| Hospitalized | 33% | 19% | 62% | 65% | 32% | 33% | 48% | 43% |
| Symptoms | ||||||||
| Rales/crepitations | 33% | 16% | 55% | 79% | 25% | 29% | 39% | 26% |
| Wheezing | 25% | 16% | 21% | 29% | 12% | 22% | 25% | 35% |
| Fever | 67% | 95% | 69% | 100% | 45% | 79% | 65% | 70% |
| Productive cough | 75% | 65% | 83% | 88% | 52% | 59% | 58% | 57% |
| Diarrhea | 0% | 9% | 0% | 12% | 6% | 11% | 4% | 4% |
| Dyspnea | 0% | 16% | 31% | 38% | 22% | 33% | 39% | 43% |
| Fatigue | 0% | 58% | 34% | 32% | 71% | 64% | 41% | 57% |
| Headache | 0% | 51% | 21% | 12% | 74% | 75% | 55% | 39% |
| Nausea | 17% | 42% | 14% | 24% | 15% | 21% | 9% | 22% |
| Red eyes | 0% | 49% | 17% | 9% | 37% | 27% | 13% | 17% |
| Sore throat | 25% | 60% | 14% | 15% | 65% | 61% | 47% | 52% |
COPD, chronic obstructive pulmonary disease; CVD, cardiovascular disease; DM, diabetes mellitus; RSV, respiratory syncytial virus.
3.6. Outcomes
Hospitalization was required for 585 subjects (54.9%) (Table 4 ). A lower percentage of subjects infected with coronavirus and influenza were hospitalized (32% and 29%) than those infected with rhinovirus and RSV (51% and 56%, respectively) (Table 3). In the pediatric population, RSV infections had the highest rate of hospitalization (76.5%), as well as the highest absolute number of hospitalizations (n = 26, 24.4% of all hospitalized pediatric subjects). Mixed infections (n = 25, 20.7%), rhinovirus (n = 17, 14.0%), and influenza (n = 11, 9.1%) also contributed significantly to hospitalization in the pediatric cohort. Rhinovirus was also the most common pathogen isolated from adults and the elderly who were hospitalized, followed by mixed infections, influenza, and metapneumovirus.
Table 4.
Hospitalizations and deaths by pathogen
| Pathogen | Hospitalized % (n) |
Death % (n) |
||||
|---|---|---|---|---|---|---|
| Age <18 | Age 18–59 | Age ≥60 | Age <18 | Age 18–59 | Age ≥60 | |
| Adenovirus | 41.8% (7) | 66.7% (10) | 100% (3) | 6.7% (1) | ||
| Bordetella pertussis | 100% (2) | 33.3% (1) | ||||
| Coronavirus | 33.3% (4) | 24% (12) | 73.3% (11) | 2% (1) | 13.3% (2) | |
| 229E | 45.5% (5) | 100% (2) | ||||
| NL63 | 25% (2) | 100% (1) | ||||
| OC43 | 40% (4) | 16.1% (5) | 66.7% (8) | 3.2% (1) | 16.7% (2) | |
| Influenza | 25.6% (11) | 43.8% (39) | 72.2% (13) | 11.1% (2) | ||
| A | 60% (3) | |||||
| A H1N1 2009 | 80% (4) | |||||
| A H3N2 | 25% (4) | 50% (23) | 72.7% (8) | 9.1% (1) | ||
| B | 29.2% (7) | 27.3% (9) | 83.3% (5) | 16.7% (1) | ||
| Metapneumovirus | 63.6% (7) | 50% (11) | 88.9% (8) | 22.2% (2) | ||
| Mycoplasma pneumoniae | 100% (4) | 100% (1) | 100% (1) | |||
| Parainfluenza virus | 66.7% (4) | 41.7% (5) | 100% (2) | 16.7% (1) | ||
| 1 | ||||||
| 2 | ||||||
| 3 | 66.7% (4) | 42.9% (3) | 100% (1) | 16.7% (1) | ||
| 4 | 40% (2) | 100% (1) | ||||
| Rhinovirus | 58.6% (17) | 59% (59) | 81.3% (26) | 7% (7) | 12.5% (4) | |
| Respiratory syncytial virus | 76.5% (26) | 53.3% (8) | 100% (8) | 5.9% (2) | 25% (2) | |
| A | 75.8% (25) | 53.3% (8) | 100% (6) | 6.1% (2) | 33.3% (2) | |
| B | 100% (1) | 100% (2) | ||||
| Mixed infections | 49% (25) | 56.7% (34) | 100% (14) | 10% (6) | 7.1% (1) | |
| Negative cases | 34.6% (18) | 51.7% (123) | 88% (73) | 4.6% (11) | 27.7% (23) | |
| Total | 46.2% (121) | 49.8% (306) | 84.1% (159) | 1.5% (4) | 4.2% (26) | 19% (36) |
Of the 1065 patients enrolled, 66 died (6.2%). Of the patients who died, 54% had no pathogen identified. Rhinovirus was the most common etiologic agent identified in subjects who died (37.9% of deaths with a pathogen identified, and 17.5% of all deaths). Only four pediatric subjects died: two with RSV, one with parainfluenza, and one with Mycoplasma. Subjects ≥60 years of age infected with coronavirus, influenza, metapneumovirus, rhinovirus, or RSV all had mortality rates exceeding 10%. The highest mortality rate (27.7%, n = 23) was seen in the cohort of patients ≥60 years old with no identified pathogen, followed by rhinovirus. Mixed viral infections did not have an appreciably larger rate of hospitalization or death.
Subjects who died were more likely to have elevated creatinine, lactate dehydrogenase, and C-reactive protein, lower lymphocyte counts, and higher neutrophil counts (Table 5 ). Subjects who died were more likely to have underlying medical conditions (except asthma). Subjects on systemic steroids prior to enrollment were also more likely to die. Headaches and sore throats were seen more commonly in non-fatal cases, whereas rales (representing lower respiratory tract disease) were more likely in fatal cases. A productive cough did not seem to differentiate fatal from non-fatal cases.
Table 5.
Outcomes
| Variable at baseline | Patients who survived | Patients who died | Odds ratio | p-Value |
|---|---|---|---|---|
| Median (range), n = 999 | Median (range), n = 66 | Estimate (95% CI) | ||
| Creatinine phosphokinase (U/l) | 95 (9–7160) | 90.5 (10–1840) | 1.04 (1.00, 1.08) | 0.066 |
| Creatinine (mg/dl) | 0.6 (0–14) | 1.1 (0.1–9.7) | 1.37 (1.19, 1.58) | <0.001 |
| C-reactive protein (mg/l) | 1.4 (0–36) | 14.25 (1.6–34.1) | 1.12 (1.09, 1.16) | <0.001 |
| Hematocrit (%) | 41.7 (14.3–76.1) | 34.9 (13.2–57.5) | 0.73 (0.66, 0.8) | <0.001 |
| Hemoglobin (g/dl) | 14.1 (4.1–25.4) | 11.7 (4.7–19) | 0.73 (0.66, 0.8) | <0.001 |
| Lactate dehydrogenase (IU/l) | 224 (26–4003) | 338 (31–5175) | 1.19 (1.08, 1.32) | 0.001 |
| Lymph (%) | 18 (0–92) | 7 (1–35) | 0.92 (0.90, 0.95) | <0.001 |
| Lymph absolute (×109/l) | 1.4 (0–35.1) | 0.7 (0.02–6.8) | 0.92 (0.88, 0.95) | <0.001 |
| Neutrophil (%) | 70 (0–97) | 87 (16–98) | 1.06 (1.04, 1.08) | <0.001 |
| Neutrophil absolute (×109/l) | 5.7 (0–26.9) | 8.6 (0.27–27.2) | 1.01 (1.00, 1.01) | 0.001a |
| Platelets (×109/l) | 231 (10–744) | 140 (3–551) | 0.91 (0.88, 0.94) | <0.001 |
| White blood count (×109/l) | 8.5 (0.1–54) | 10.4 (0.3–29.2) | 1.03 (0.99, 1.07) | 0.179 |
| Patients who survived | Patients who died | Odds ratio | p-Value | |
|---|---|---|---|---|
| Percent | Percent | Estimate (95% CI) | ||
| Any antiviral medications | 21.2 | 39.4 | 2.41 (1.44, 4.05) | <0.001a |
| Inhaled steroids | 18.4 | 18.2 | 1.00 (0.52, 1.91) | 1.00 |
| Systemic steroids | 23.7 | 36.4 | 1.88 (1.11, 3.1) | 0.02 |
| History of smoking | 30.3 | 50.0 | 2.30 (1.39, 3.79) | <0.001a |
| Medical history | ||||
| Asthma | 15.0 | 1.5 | 0.09 (0.01, 0.63) | 0.02a |
| Cardiovascular disorder | 12.7 | 30.3 | 2.99 (1.71, 5.21) | <0.001a |
| COPD | 3.0 | 9.1 | 3.23 (1.29, 8.06) | 0.01a |
| Diabetes | 8.7 | 18.2 | 2.33 (1.20, 4.52) | 0.01 |
| Immunodeficiency | 3.7 | 16.7 | 5.20 (2.52, 10.75) | <0.001 |
| Renal disorder | 3.5 | 10.6 | 3.27 (1.39, 7.67) | 0.01a |
| Signs and symptoms | ||||
| Productive cough | 62.9 | 51.5 | 0.63 (0.38, 1.03) | 0.07 |
| Diarrhea | 7.5 | 3.0 | 0.39 (0.09, 1.60) | 0.19 |
| Dyspnea | 32.5 | 39.4 | 1.35 (0.81, 2.25) | 0.25 |
| Fatigue | 51.8 | 42.4 | 0.69 (0.42, 1.14) | 0.14 |
| Fever | 70.9 | 75.8 | 1.28 (0.72, 2.29) | 0.40 |
| Headache | 53.2 | 21.2 | 0.24 (0.13, 0.43) | <0.001 |
| Nausea | 18.9 | 1.5 | 0.07 (0.01, 0.48) | 0.01a |
| Rales/crepitations | 36.7 | 83.3 | 8.61 (4.45, 16.66) | <0.001 |
| Red eyes | 21.6 | 4.5 | 0.17 (0.05, 0.56) | <0.001a |
| Sore throat | 46.4 | 21.2 | 0.31 (0.17, 0.57) | <0.001 |
| Wheezing | 23.2 | 9.1 | 0.33 (0.14, 0.78) | 0.01a |
CI, confidence interval; COPD, chronic obstructive pulmonary disease.
When analysis for deaths attributed to ILI, the p-value was no longer <0.05.
4. Discussion
This study presents the most thorough description of the causes of ILI in Mexico City to date. Using a multiplex PCR test, one or more pathogens were identified in 65% of enrolled subjects. The most common pathogen identified in the study was rhinovirus (25% of all isolates), and influenza was the second most common pathogen (24% of all isolates). This is consistent with other series of adult ILI in various geographic regions, where rhinovirus has been reported to account for 25–50% of adult ILI isolates, and influenza for 14–23%.10, 13, 27, 28 Coronavirus – most commonly OC43, but also NL63 and 2229E – was also a significant contributor to the burden of ILI (14% of all isolates). Recent studies have reported coronavirus to be the third to fifth most common pathogen.10, 13
Among children in this study, the most common pathogen was influenza, followed by RSV and rhinovirus. Similar significant contributions of RSV and influenza have been noted in prior studies.29, 30, 31 Rhinovirus, however, was rarely tested in previous studies. In one other prospective cohort study of children with lower respiratory tract infections, rhinovirus was reported as second only to RSV in causing hospitalizations.32 The frequency at which it was isolated in our hospitalized children (14%, 17/121) suggests that rhinovirus is a previously under-appreciated pathogen for this age group.
Rhinovirus was a cause of significant morbidity in all age groups of our study population, such that 60–80% of individuals infected with rhinovirus required hospitalization. Rhinovirus was also the most common isolate from subjects who died (isolated from 11/66 subjects). Given the study design, a clear attribution to the cause of hospitalization or death is not possible.
Coronavirus species made up 14% of all isolates in our population. Coronavirus can present with a similar syndrome to influenza.33 Twenty-two percent of all subjects with isolated coronavirus were hospitalized, but in the population aged ≥60 years this increased to 73%. While our study design precludes the determination of exact hospitalization rates as a result of coronavirus infections, the hospitalization rates were similar to those for influenza.
No pathogen was identified in 35% of subjects. This was more common in adults (no pathogen detected in 41% (321/793) of all adult subjects aged ≥18 years) than in children (no pathogen detected in 20% of subjects). It is unclear if these subjects had bacterial infections, pathogens not tested for in our assay, or non-infectious etiologies of their ILI. Unfortunately the lack of paired sputum limited our ability to determine etiology. In a cohort in New Caledonia, paired sputum samples were required to determine the etiologic agent in 23% of subjects with lower respiratory tract infections.32 In a population of hospitalized children in Mexico with lower respiratory tract infections, viral etiologies were determined in only 47%.34 This study found a virologic agent in 80% of children. This improved detection is likely in part due to the extended panel of viruses that were tested with the multiplex PCR.
More than one virus was isolated in 11.9% of subjects. It is not possible to determine the contribution of each virus to the clinical syndrome, nor is it possible to determine if some viruses that were detected were not causing disease. While PCR significantly improves the sensitivity of detecting respiratory viruses, the detection of viral nucleic acids may not always represent causation. In prior case–control studies, virus nucleic acids could be isolated from 20–27% of asymptomatic subjects.35, 36 This might be supported by the data showing that among those with mixed infections, the outcomes were not significantly worse than those for subjects with single infections. While all subjects enrolled were symptomatic, and this would increase the likelihood that isolated pathogens were causative, cohort studies such as ours are not able to directly determine causation. With 62 unique combinations in 125 different subjects, the assessment of outcomes lacks statistical power to determine differences between the various combinations of mixed infections.
The determination of non-influenza respiratory viruses in ILI has several implications. First, increasing knowledge that acute respiratory illness and its associated morbidity are often caused by respiratory viruses may stimulate the development of more affordable rapid diagnostic tests as well as therapeutics for these viruses. With the consideration of increasing antibiotic resistance, the diagnosis of respiratory viruses could decrease the quantities of empiric broad-spectrum antibiotics used.
The aim of this study was not the epidemiological surveillance of respiratory diseases. We included only patients who by the severity of their illnesses sought medical attention. The attending physicians determined the need for hospitalization or outpatient treatment. This limited our population to a particular group of patients with clinical manifestations of the disease. With this strategy we seek to find the more serious cases. From this we believe we can more easily determine the factors associated with severe disease. It is a limitation of this study, however, that we lose a part of the population with mild or asymptomatic disease.
In conclusion, viruses that were traditionally believed to produce minor disease have the capacity, in certain subjects, to cause severe cases, as suggested by the high proportion of adult patients with rhinovirus infection who required in-hospital treatment. We believe that these findings mark the need for specific studies that will help us to better understand the behavior of these viruses, and the factors associated with a more serious presentation.
Acknowledgements
La Red is funded by the Mexico Ministry of Health and the U.S. National Institute of Allergy and Infectious Diseases. This project has been funded in part by funding provided by: CONACYT through FONSEC SSA/IMSS/ISSSTE No. 71260 and No. 127088. Intramural Research Programs of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) through a contract with Westat, Inc., Contract Number: HHSN2722009000031, Task Order Number: HHSN27200002. And through the National Cancer Institute, National Institutes of Health, 318 under Contract No. HHSN261200800001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, or Westat, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Ethical approval: The Ethics committee of each institution reviewed the study, and the study was conducted following the principles of the ICH GCP, Declaration of Helsinki, and the Mexican General Health Law. All subjects provided informed consent. All data was de-identified during data collection. The project was registered on clinicaltrials.gov (NCT01418287).
Conflict of interest: The authors have no competing interests to declare.
Corresponding Editor: Eskild Petersen, Skejby, Denmark
References
- 1.Perez-Padilla R., de la Rosa-Zamboni D., Ponce de Leon S., Hernandez M., Quinones-Falconi F., Bautista E. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;361:680–689. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- 2.Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization . WHO; Geneva: 2002. World health report 2002. [Google Scholar]
- 4.Monto A.S. Epidemiology of viral respiratory infections. Am J Med. 2002;112(Suppl 6A):4S–12S. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 5.Kammerer P.E., Montiel S., Kriner P., Bojorquez I., Ramirez V.B., Vazquez-Erlbeck M. Influenza-like illness surveillance on the California–Mexico border, 2004–2009. Influenza Other Respi Viruses. 2012;6:358–366. doi: 10.1111/j.1750-2659.2011.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed J.A., Katz M.A., Auko E., Njenga M.K., Weinberg M., Kapella B.K. Epidemiology of respiratory viral infections in two long-term refugee camps in Kenya, 2007–2010. BMC Infect Dis. 2012;12:7. doi: 10.1186/1471-2334-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe A.S., Carraro E., Candeias J.M., Donalisio M.R., Leal E., Granato C.F. Viral etiology among the elderly presenting acute respiratory infection during the influenza season. Rev Soc Bras Med Trop. 2011;44:18–21. doi: 10.1590/s0037-86822011000100005. [DOI] [PubMed] [Google Scholar]
- 8.Juven T., Mertsola J., Waris M., Leinonen M., Meurman O., Roivainen M. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J. 2000;19:293–298. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Laguna-Torres V.A., Gomez J., Ocana V., Aguilar P., Saldarriaga T., Chavez E. Influenza-like illness sentinel surveillance in Peru. PLoS One. 2009;4:e6118. doi: 10.1371/journal.pone.0006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellei N., Carraro E., Perosa A., Watanabe A., Arruda E., Granato C. Acute respiratory infection and influenza-like illness viral etiologies in Brazilian adults. J Med Virol. 2008;80:1824–1827. doi: 10.1002/jmv.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiberville S.D., Ninove L., Vu Hai V., Botelho-Nevers E., Gazin C., Thirion L. The viral etiology of an influenza-like illness during the 2009 pandemic. J Med Virol. 2012;84:1071–1079. doi: 10.1002/jmv.23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren L., Gonzalez R., Wang Z., Xiang Z., Wang Y., Zhou H. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005–2007. Clin Microbiol Infect. 2009;15:1146–1153. doi: 10.1111/j.1469-0691.2009.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razanajatovo N.H., Richard V., Hoffmann J., Reynes J.M., Razafitrimo G.M., Randremanana R.V. Viral etiology of influenza-like illnesses in Antananarivo, Madagascar, July 2008 to June 2009. PLoS One. 2011;6:e17579. doi: 10.1371/journal.pone.0017579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boivin G., Hardy I., Tellier G., Maziade J. Predicting influenza infections during epidemics with use of a clinical case definition. Clin Infect Dis. 2000;31:1166–1169. doi: 10.1086/317425. [DOI] [PubMed] [Google Scholar]
- 15.Peltola V., Reunanen T., Ziegler T., Silvennoinen H., Heikkinen T. Accuracy of clinical diagnosis of influenza in outpatient children. Clin Infect Dis. 2005;41:1198–1200. doi: 10.1086/444508. [DOI] [PubMed] [Google Scholar]
- 16.Nakanishi M., Yoshida Y., Takeda N., Hirana H., Horita T., Shimizu K. Community-acquired pneumonia distinguished from influenza infection based on clinical signs and symptoms during a novel (swine) influenza A/H1N1 pandemic. Prim Care Respir J. 2011;20:421–426. doi: 10.4104/pcrj.2011.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee V.J., Yap J., Cook A.R., Tan C.H., Loh J.P., Koh W.H. A clinical diagnostic model for predicting influenza among young adult military personnel with febrile respiratory illness in Singapore. PLoS One. 2011;6:e17468. doi: 10.1371/journal.pone.0017468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlomai A., Nutman A., Kotlovsky T., Schechner V., Carmeli Y., Guzner-Gur H. Predictors of pandemic (H1N1) 2009 virus positivity and adverse outcomes among hospitalized patients with a compatible syndrome. Isr Med Assoc J. 2010;12:622–627. [PubMed] [Google Scholar]
- 19.Dominguez-Cherit G., Lapinsky S.E., Macias A.E., Pinto R., Espinosa-Perez L., de la Torre A. Critically Ill patients with 2009 influenza A(H1N1) in Mexico. JAMA. 2009;302:1880–1887. doi: 10.1001/jama.2009.1536. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A., Zarychanski R., Pinto R., Cook D.J., Marshall J., Lacroix J. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA. 2009;302:1872–1879. doi: 10.1001/jama.2009.1496. [DOI] [PubMed] [Google Scholar]
- 21.Webb S.A., Pettila V., Seppelt I., Bellomo R., Bailey M., Cooper D.J. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009;361:1925–1934. doi: 10.1056/NEJMoa0908481. [DOI] [PubMed] [Google Scholar]
- 22.Widmer K., Zhu Y., Williams J.V., Griffin M.R., Edwards K.M., Talbot H.K. Rates of hospitalizations for respiratory syncytial virus, human metapneumovirus, and influenza virus in older adults. J Infect Dis. 2012;206:56–62. doi: 10.1093/infdis/jis309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. CDC protocol of realtime RTPCR for swine influenza A (H1N1). 28 April 2009; revision 1 (30 April 2009). Atlanta, GA: WHO/CDC. Available at: http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090428.pdf [accessed 29.06.09].
- 24.Reijans M., Dingemans G., Klaassen C.H., Meis J.F., Keijdener J., Mulders B. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol. 2008;46:1232–1240. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loens K., van Loon A.M., Coenjaerts F., van Aarle Y., Goossens H., Wallace P. Performance of different mono- and multiplex nucleic acid amplification tests on a multipathogen external quality assessment panel. J Clin Microbiol. 2012;50:977–987. doi: 10.1128/JCM.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam W.Y., Yeung A.C., Tang J.W., Ip M., Chan E.W., Hui M. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007;45:3631–3640. doi: 10.1128/JCM.00280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson K.G., Kent J., Hammersley V., Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ. 1997;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boivin G., Osterhaus A.D., Gaudreau A., Jackson H.C., Groen J., Ward P. Role of picornaviruses in flu-like illnesses of adults enrolled in an oseltamivir treatment study who had no evidence of influenza virus infection. J Clin Microbiol. 2002;40:330–334. doi: 10.1128/JCM.40.2.330-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carballal G., Videla C.M., Espinosa M.A., Savy V., Uez O., Sequeira M.D. Multicentered study of viral acute lower respiratory infections in children from four cities of Argentina, 1993–1994. J Med Virol. 2001;64:167–174. doi: 10.1002/jmv.1032. [DOI] [PubMed] [Google Scholar]
- 30.Berkley J.A., Munywoki P., Ngama M., Kazungu S., Abwao J., Bett A. Viral etiology of severe pneumonia among Kenyan infants and children. JAMA. 2010;303:2051–2057. doi: 10.1001/jama.2010.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathisen M., Basnet S., Sharma A., Shrestha P.S., Sharma B.N., Valentiner-Branth P. RNA viruses in young Nepalese children hospitalized with severe pneumonia. Pediatr Infect Dis J. 2011;30:1032–1036. doi: 10.1097/INF.0b013e31822f845f. [DOI] [PubMed] [Google Scholar]
- 32.Mermond S., Zurawski V., D’Ortenzio E., Driscoll A.J., DeLuca A.N., Deloria-Knoll M. Lower respiratory infections among hospitalized children in New Caledonia: a pilot study for the Pneumonia Etiology Research for Child Health project. Clin Infect Dis. 2012;54(Suppl 2):S180–S189. doi: 10.1093/cid/cir1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabeca T.K., Bellei N. Human coronavirus NL-63 infection in a Brazilian patient suspected of H1N1 2009 influenza infection: description of a fatal case. J Clin Virol. 2012;53:82–84. doi: 10.1016/j.jcv.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noyola D.E., Rodriguez-Moreno G., Sanchez-Alvarado J., Martinez-Wagner R., Ochoa-Zavala J.R. Viral etiology of lower respiratory tract infections in hospitalized children in Mexico. Pediatr Infect Dis J. 2004;23:118–123. doi: 10.1097/01.inf.0000110269.46528.a5. [DOI] [PubMed] [Google Scholar]
- 35.Jansen R.R., Wieringa J., Koekkoek S.M., Visser C.E., Pajkrt D., Molenkamp R. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanghavi S.K., Bullotta A., Husain S., Rinaldo C.R. Clinical evaluation of multiplex real-time PCR panels for rapid detection of respiratory viral infections. J Med Virol. 2012;84:162–169. doi: 10.1002/jmv.22186. [DOI] [PMC free article] [PubMed] [Google Scholar]