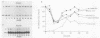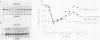Abstract
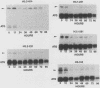
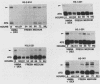
We investigated the expression characteristics of the fully replication-dependent (FRD) and the partially replication-dependent (PRD) histone gene variants by measuring changes in steady-state mRNA levels during hexamethylene bisacetamide (HMBA)-induced differentiation of murine erythroleukemia (MEL) cells. Between 24 and 60 h after induction, there was a dramatic switch in histone gene expression, such that the ratio of PRD to FRD transcripts increased severalfold over that found in uninduced MEL cells. We demonstrated that this gene switching was not simply a partial or complete uncoupling of PRD gene expression from DNA synthesis. PRD and FRD transcript levels were regulated coordinately upon treatment of uninduced or induced MEL cells with inhibitors of DNA synthesis, protein synthesis, or both. Using several criteria, we were unable to detect any difference in PRD and FRD gene expression under any conditions except in cells undergoing differentiation. MEL cells were arrested at a precommitment stage of differentiation by induction with HMBA in the presence of dexamethasone (DEX). If DEX was subsequently removed, DNA synthesis resumed, the cells underwent commitment, and histone gene switching was observed. In contrast, if both DEX and HMBA were removed, DNA synthesis still resumed, but commitment did not occur and no gene switching was observed. These results imply that histone gene switching is intimately related to the differentiation process.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I. Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol Cell Biol. 1984 Jan;4(1):123–132. doi: 10.1128/mcb.4.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artishevsky A., Delegeane A. M., Lee A. S. Use of a cell cycle mutant to delineate the critical period for the control of histone mRNA levels in the mammalian cell cycle. Mol Cell Biol. 1984 Nov;4(11):2364–2369. doi: 10.1128/mcb.4.11.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A., Superti-Furga G., Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987 Jul 31;50(3):347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Biard-Roche J., Gorka C., Lawrence J. J. The structural role of histone H1: properties of reconstituted chromatin with various H1 subfractions (H1-1, H1-2, and H1o). EMBO J. 1982;1(12):1487–1492. doi: 10.1002/j.1460-2075.1982.tb01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
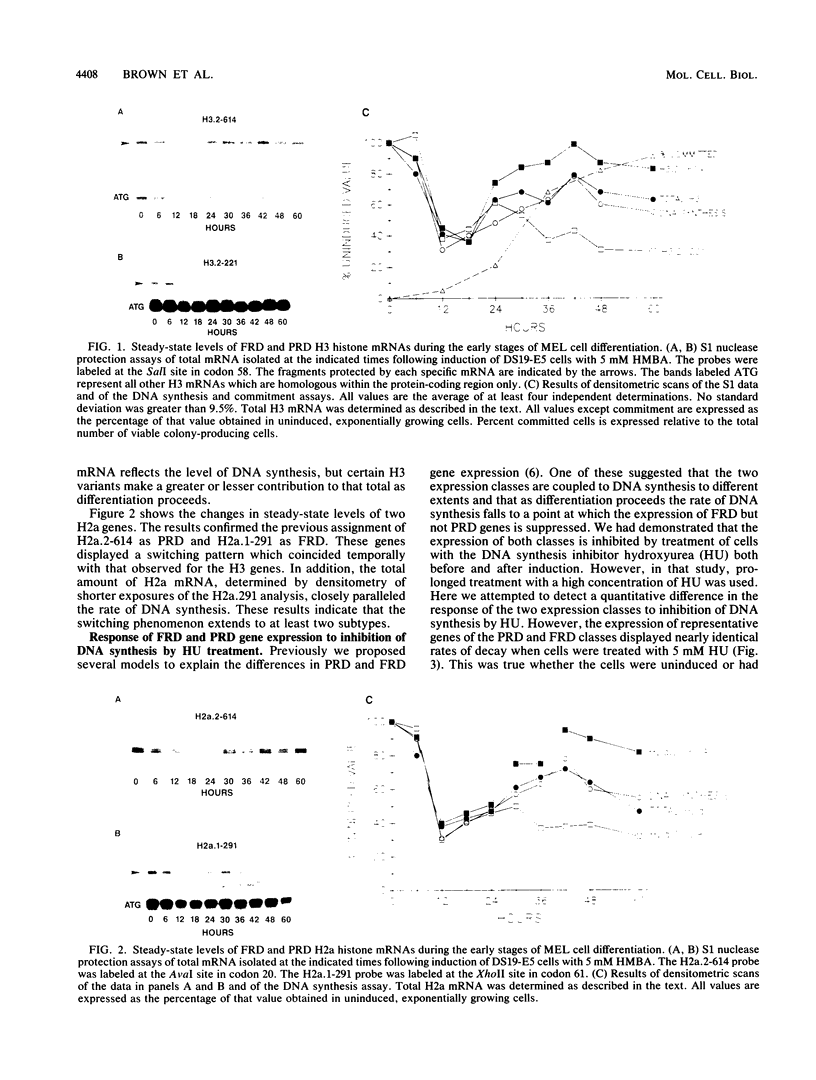
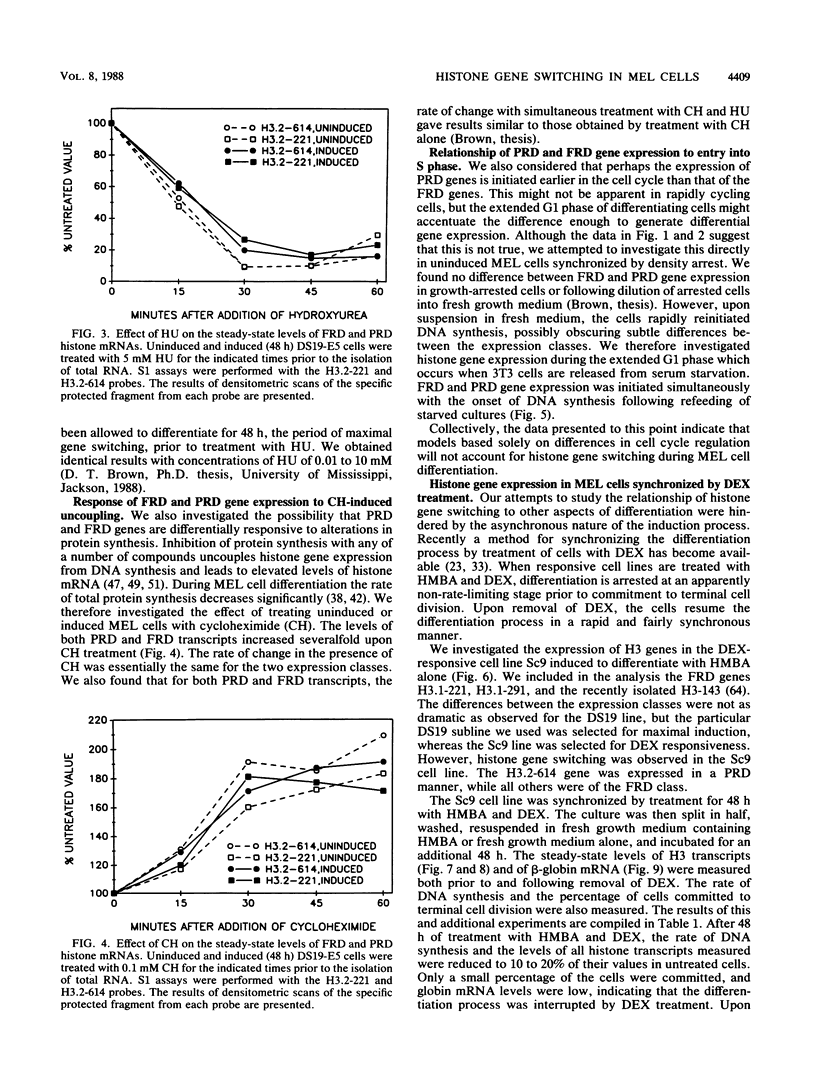
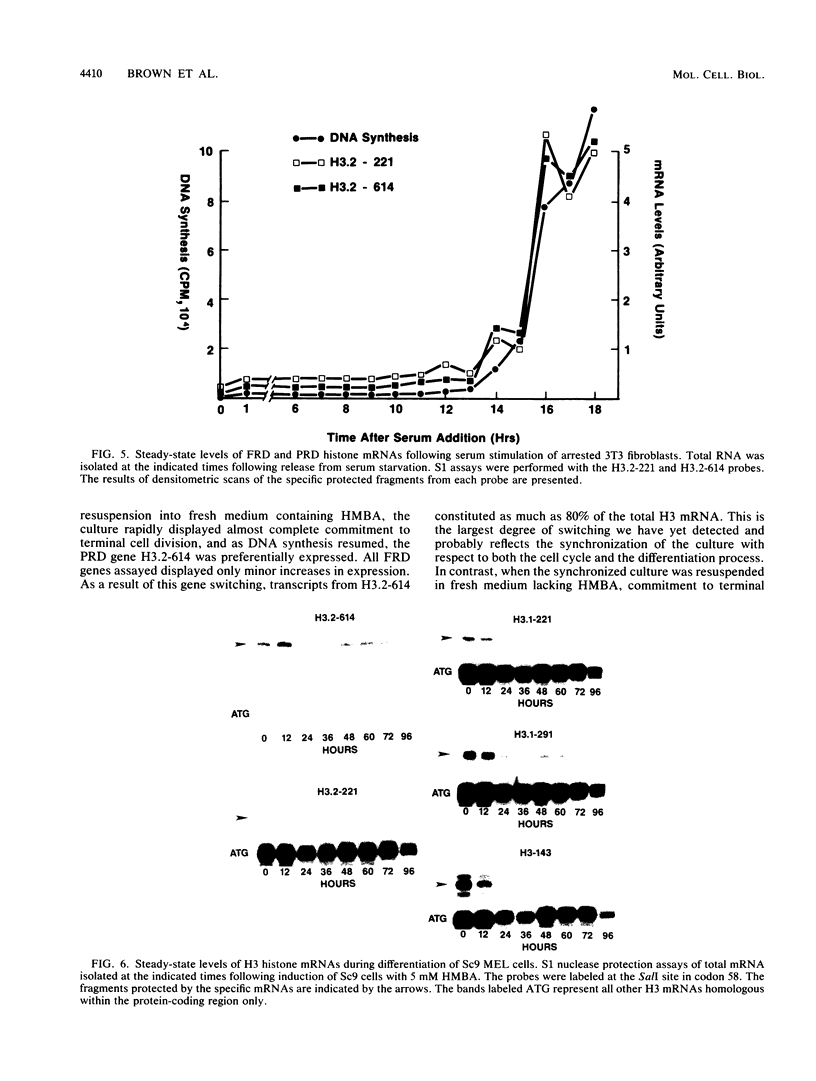
- Brown D. T., Wellman S. E., Sittman D. B. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol Cell Biol. 1985 Nov;5(11):2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R. D. Microheterogeneity in H1 histones and its consequences. Int J Pept Protein Res. 1987 Oct;30(4):433–449. doi: 10.1111/j.1399-3011.1987.tb03352.x. [DOI] [PubMed] [Google Scholar]
- Dailey L., Hanly S. M., Roeder R. G., Heintz N. Distinct transcription factors bind specifically to two regions of the human histone H4 promoter. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7241–7245. doi: 10.1073/pnas.83.19.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisle A. J., Graves R. A., Marzluff W. F., Johnson L. F. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol Cell Biol. 1983 Nov;3(11):1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabar J. M. Nonrandom location of H1-H1 degree histones on chromatin of mouse liver and brain. J Biol Chem. 1985 Oct 15;260(23):12622–12628. [PubMed] [Google Scholar]
- Fletcher C., Heintz N., Roeder R. G. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987 Dec 4;51(5):773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Garrett C., Kredich N. M. Induction of hemoglobin synthesis by xylosyladenine in murine erythroleukemia cells. Metabolism of xylosyladenine and effects on transmethylation. J Biol Chem. 1981 Dec 25;256(24):12705–12709. [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Wellman S. E., Chiu I. M., Marzluff W. F. Differential expression of two clusters of mouse histone genes. J Mol Biol. 1985 May 25;183(2):179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- Grove G. W., Zweidler A. Regulation of nucleosomal core histone variant levels in differentiating murine erythroleukemia cells. Biochemistry. 1984 Sep 11;23(19):4436–4443. doi: 10.1021/bi00314a030. [DOI] [PubMed] [Google Scholar]
- Hall J. M., Cole R. D. Mechanisms of H1o accumulation in mouse neuroblastoma cells differ with different treatments. J Biol Chem. 1986 Apr 15;261(11):5168–5174. [PubMed] [Google Scholar]
- Hanly S. M., Bleecker G. C., Heintz N. Identification of promoter elements necessary for transcriptional regulation of a human histone H4 gene in vitro. Mol Cell Biol. 1985 Feb;5(2):380–389. doi: 10.1128/mcb.5.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Jacob E. Histone-gene reiteration in the genome of mouse. Eur J Biochem. 1976 May 17;65(1):275–284. doi: 10.1111/j.1432-1033.1976.tb10415.x. [DOI] [PubMed] [Google Scholar]
- Kaneda T., Murate T., Sheffery M., Brown K., Rifkind R. A., Marks P. A. Gene expression during terminal differentiation: dexamethasone suppression of inducer-mediated alpha 1- and beta maj-globin gene expression. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5020–5024. doi: 10.1073/pnas.82.15.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Bella F., Zhong R., Heintz N. Cell type-specific expression of a human histone H1 gene. J Biol Chem. 1988 Feb 15;263(5):2115–2118. [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. J., Liu L., Marzluff W. F. Mouse histone H2A and H2B genes: four functional genes and a pseudogene undergoing gene conversion with a closely linked functional gene. Nucleic Acids Res. 1987 Apr 10;15(7):3023–3039. doi: 10.1093/nar/15.7.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C., Roche J., Roux B., Gorka C. Differences in the condensation of chromatin by individual subfractions of histone H1: implications for the role of H1(0) in the structural organization of chromatin. Biochemistry. 1985 Nov 5;24(23):6328–6335. doi: 10.1021/bi00344a002. [DOI] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Childs G., Kedes L. Distinct organizations and patterns of expression of early and late histone gene sets in the sea urchin. Nature. 1983 Jan 13;301(5896):120–125. doi: 10.1038/301120a0. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Murate T., Kaneda T., Rifkind R. A., Marks P. A. Inducer-mediated commitment of murine erythroleukemia cells to terminal cell division: the expression of commitment. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3394–3398. doi: 10.1073/pnas.81.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newrock K. M., Alfageme C. R., Nardi R. V., Cohen L. H. Histone changes during chromatin remodeling in embryogenesis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):421–431. doi: 10.1101/sqb.1978.042.01.045. [DOI] [PubMed] [Google Scholar]
- Ohta Y., Tanaka M., Terada M., Miller O. J., Bank A., Marks P., Rifkind R. A. Erythroid cell differentiation: murine erythroleukemia cell variant with unique pattern of induction by polar compounds. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1232–1236. doi: 10.1073/pnas.73.4.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne H. B., Chabanas A. Kinetics of histone H10 accumulation and commitment to differentiation in murine erythroleukemia cells. Exp Cell Res. 1984 Jun;152(2):449–458. doi: 10.1016/0014-4827(84)90646-3. [DOI] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Housman D. Regulation of protein synthesis and accumulation during murine erythroleukemia cell differentiation. J Biol Chem. 1985 Jan 10;260(1):604–609. [PubMed] [Google Scholar]
- Ross J., Peltz S. W., Kobs G., Brewer G. Histone mRNA degradation in vivo: the first detectable step occurs at or near the 3' terminus. Mol Cell Biol. 1986 Dec;6(12):4362–4371. doi: 10.1128/mcb.6.12.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeon F., Mach B. Cloning and amplification of alpha and beta mouse globin gene sequences synthesised in vitro. Gene. 1977 May;1(3-4):229–239. doi: 10.1016/0378-1119(77)90047-6. [DOI] [PubMed] [Google Scholar]
- STEDMAN E. Cell specificity of histones. Nature. 1950 Nov 4;166(4227):780–781. doi: 10.1038/166780a0. [DOI] [PubMed] [Google Scholar]
- Seyedin S. M., Cole R. D., Kistler W. S. H1 histones from mammalian testes. The widespread occurrence of H1t. Exp Cell Res. 1981 Dec;136(2):399–405. doi: 10.1016/0014-4827(81)90019-7. [DOI] [PubMed] [Google Scholar]
- Sherton C. C., Kabat D. Changes in RNA and protein metabolism preceding onset of hemoglobin synthesis in cultured Friend leukemia cells. Dev Biol. 1976 Jan;48(1):118–131. doi: 10.1016/0012-1606(76)90051-8. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Modulation of nucleosome structure by histone subtypes in sea urchin embryos. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6803–6807. doi: 10.1073/pnas.78.11.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Chiu I. M., Pan C. J., Cohn R. H., Kedes L. H., Marzluff W. F. Isolation of two clusters of mouse histone genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4078–4082. doi: 10.1073/pnas.78.7.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983 Oct 11;11(19):6679–6697. doi: 10.1093/nar/11.19.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Heintz N., Roeder R. G. Multiple sequence elements are required for maximal in vitro transcription of a human histone H2B gene. Mol Cell Biol. 1986 Oct;6(10):3329–3340. doi: 10.1128/mcb.6.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Heintz N., Roeder R. G. Regulation of human histone gene expression during the HeLa cell cycle requires protein synthesis. Mol Cell Biol. 1984 Dec;4(12):2723–2734. doi: 10.1128/mcb.4.12.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl H., Gallwitz D. Fate of histone messenger RNA in synchronized HeLa cells in the absence of initiation of protein synthesis. Eur J Biochem. 1977 Jan;72(2):385–392. doi: 10.1111/j.1432-1033.1977.tb11263.x. [DOI] [PubMed] [Google Scholar]
- Stimac E., Groppi V. E., Jr, Coffino P. Inhibition of protein synthesis stabilizes histone mRNA. Mol Cell Biol. 1984 Oct;4(10):2082–2090. doi: 10.1128/mcb.4.10.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Wellman S. E., Marzluff W. F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J Mol Evol. 1986;23(3):242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]
- Terada M., Fried J., Nudel U., Rifkind R. A., Marks P. A. Transient inhibition of initiation of S-phase associated with dimethyl sulfoxide induction of murine erythroleukemia cells to erythroid differentiation. Proc Natl Acad Sci U S A. 1977 Jan;74(1):248–252. doi: 10.1073/pnas.74.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor C. D., Stamler S. J., Engel J. D. Erythroid-specific transcription of the chicken histone H5 gene is directed by a 3' enhancer. 1987 Aug 27-Sep 2Nature. 328(6133):827–830. doi: 10.1038/328827a0. [DOI] [PubMed] [Google Scholar]
- Trostle-Weige P. K., Meistrich M. L., Brock W. A., Nishioka K. Isolation and characterization of TH3, a germ cell-specific variant of histone 3 in rat testis. J Biol Chem. 1984 Jul 25;259(14):8769–8776. [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman S. E., Casano P. J., Pilch D. R., Marzluff W. F., Sittman D. B. Characterization of mouse H3.3-like histone genes. Gene. 1987;59(1):29–39. doi: 10.1016/0378-1119(87)90263-0. [DOI] [PubMed] [Google Scholar]
- Wells D. E. Compilation analysis of histones and histone genes. Nucleic Acids Res. 1986;14 (Suppl):r119–r149. doi: 10.1093/nar/14.suppl.r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley P. L., Wells J. R. H5 gene specific trans-activation by nuclear extracts from avian erythroid cells. Mol Cell Biol. 1987 Oct;7(10):3853–3856. doi: 10.1128/mcb.7.10.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E., Levy D., Gordon J. S. Changes in the H-1 histone complement during myogenesis. I. Establishment by differential coupling of H-1 species synthesis to DNA replication. J Cell Biol. 1985 Jul;101(1):167–174. doi: 10.1083/jcb.101.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Yang Y. S., Brown D. T., Wellman S. E., Sittman D. B. Isolation and characterization of a mouse fully replication-dependent H1 gene within a genomic cluster of core histone genes. J Biol Chem. 1987 Dec 15;262(35):17118–17125. [PubMed] [Google Scholar]