Abstract
Integrating vectors can lead to the dysregulation of nearby chromosomal genes, with important consequences for clinical trials and cellular engineering. This includes the retroviral and lentiviral vectors commonly used for deriving induced pluripotent stem cells (iPSCs). We previously used integrating foamy virus (FV) vectors expressing OCT4, SOX2, MYC, and KLF4 to reprogram osteogenesis imperfecta mesenchymal stem cells (MSCs). Here we have studied the effects of 10 FV vector proviruses on neighboring gene expression in four iPSC lines and their corresponding iPSC-derived mesenchymal stem cells (iMSCs). Gene expression profiles in these iPSC lines showed that none of the 38 genes within 300 kb up- or downstream of integrated proviruses had a significant difference in mRNA levels, including 5 genes with proviruses in their transcription units. In the iMSCs derived from these iPSCs, the same type of analysis showed a single dysregulated transcript out of 46 genes found near proviruses. This frequency of dysregulation was similar to that of genes lacking nearby proviruses, so it may have been due to interclonal variation and/or measurement inaccuracies. While the number of integration sites examined in this paper is limited, our results suggest that integrated FV proviruses do not impact the expression of chromosomal genes in pluripotent human stem cells or their differentiated derivatives. This interpretation is consistent with previous reports that FV vectors have minimal genotoxicity, even when integrating near or within genes.
Keywords: Foamy virus vectors, retroviral integration, genotoxicity, cellular reprogramming
Introduction
Insertional mutagenesis due to vector integration is a major concern for gene and cell therapy. Integrated proviruses containing strong promoters and enhancers have the capacity to increase expression of nearby chromosomal genes, and activation of adjacent proto-oncogenes by gammaretroviral vectors can lead to clonal dominance and malignancies in human clinical trials1–4. Changes in vector design such as long terminal repeat (LTR) deletions (self-inactivating or SIN vectors) that lack enhancer activity and the use of internal promoters have been developed and may improve safety5–7. Nevertheless, integration of SIN vectors still results in the dysregulation of adjacent genes in hematopoietic stem cells and induced pluripotent stem cells (iPSCs)8, 9. SIN lentiviral (LV) vectors are less genotoxic than gammaretroviral vectors because in comparison they integrate away from transcription start sites and regulatory elements10–12. However, their integration within transcription units can still lead to dysregulation13, 14. In addition, it has been shown that the genotoxicity of SIN LV vector integration alone can give rise to iPSCs without reprogramming factors15.
Foamy virus (FV) vectors are an alternative type of retroviral vector with a large packaging capacity16, wide host range17, and a cDNA genome18, 19. FV vectors may be less oncogenic than gammaretroviral and LV vectors because they do not preferentially integrate within genes or active transcription units20, have less transcriptional read-through activity21, and their deleted LTRs lack enhancer activity21. Also, the integration of FV vectors in canine hematopoietic stem cells did not lead to clonal expansion or malignancies after transplantation22, even when near proto-oncogenes23. While these studies suggest that FV vectors may have minimal genotoxicity, the effects of integrated FV vector proviruses on neighboring gene expression have not been determined previously.
We have shown that FV vectors can be used to reprogram mesenchymal stem cells (MSCs) with efficiencies similar to other vector systems, especially when the reprogramming genes were expressed from an internal Moloney murine leukemia virus (MLV) LTR promoter24. FV-derived iPSCs were pluripotent and could be differentiated into bone-forming MSC-like cells (iMSCs). Here we examine the effects of these reprogramming vectors on chromosomal gene expression by determining their chromosomal locations and performing a global transcriptional analysis of FV-derived iPSC clones and their differentiated iMSC derivatives.
Results & Discussion
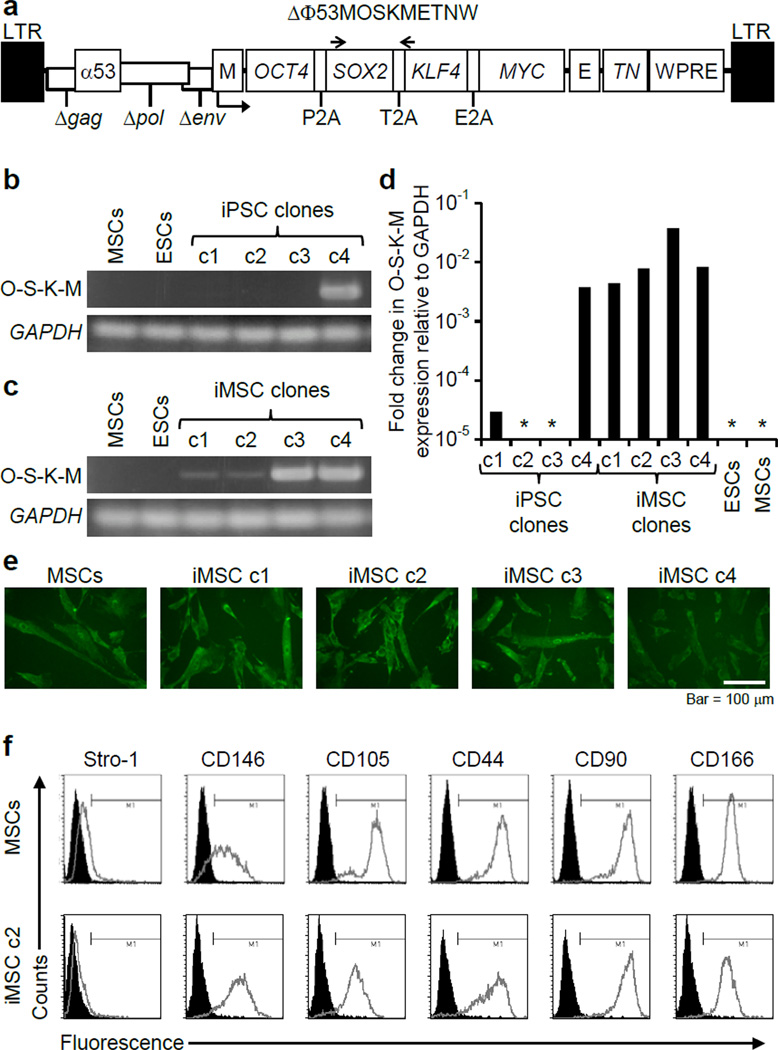
Four independent iPSC lines were chosen for this study. iPSC clones c1, c2, c3, and c4 were derived by transducing bone marrow MSCs from osteogenesis imperfecta patients with the polycistronic FV reprogramming vector ΔΦ53MOSKMETNW, which expresses OCT4, SOX2, KLF4 and MYC as a single transcript separated by peptide cleavage signals under the control of an internal MLV LTR promoter (Figure 1a). These iPSC lines were shown to express pluripotency genes and had trilineage developmental potential by teratoma assays24. Cytogenetic analysis of two of these 4 iPSC lines showed that they had normal karyotypes24. Expression of the vector-encoded, polycistronic transgene cassette was assessed by RT-PCR and qRT-PCR and found to be absent in iPSC clones c2 and c3, at a low level in iPSC clone c1, and persistent in iPSC clone c4 (Figure 1b and 1d). The MLV LTR promoter present in the vector is known to be frequently silenced in pluripotent cells25.
Figure 1. iPSC derivation and differentiation.
(a) The FV vector ΔΦ53MOSKMETNW is shown containing a polycistronic 2A peptide-linked reprogramming cassette with OCT4, SOX2, KLF4, and MYC open reading frames. E, EF1α promoter; M, MLV promoter; TN, Thymidine kinase-neomycin fusion protein gene; α53, anti-p53 shRNA; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. The locations of primers Foamy-f and Foamy-r are indicated by arrows. (b) RT-PCR analysis showed silencing of the FV polycistronic transcript in three of the four iPSC clones during reprogramming, with GAPDH transcript controls. O-S-K-M, reprogramming vector transcript. (c) RT-PCR showing expression of the reprogramming vector after differentiation of iPSCs into iMSCs. (d) mRNA levels of the FV polycistronic transcript (O-S-K-M) as determined by qRT-PCR and shown as fold change relative to GAPDH. *No transcript detected. (e) Collagen expression detected by immunohistochemistry with anti-human α2 Type I procollagen antibody in MSCs and iMSCs. Bar = 100 µm. (f) Representative flow cytometry analysis of MSC surface markers produced by MSCs and iMSC c2.
Each iPSC clone was differentiated into iMSCs by embyroid body formation, plating on gelatin-coated dishes, and serial passaging in the presence of fetal bovine serum as described26. Because reactivation of the viral transgenes can occur with differentiation, we evaluated the expression of the FV reprogramming cassette in these iMSC cultures, and observed reactivation in clones c1 – c3, and persistent expression in c4 (Figure 1c and 1d). These iMSCs expressed type I collagen and mesenchymal cell surface markers (Figure 1e and 1f), and formed bone in vitro and in vivo24.
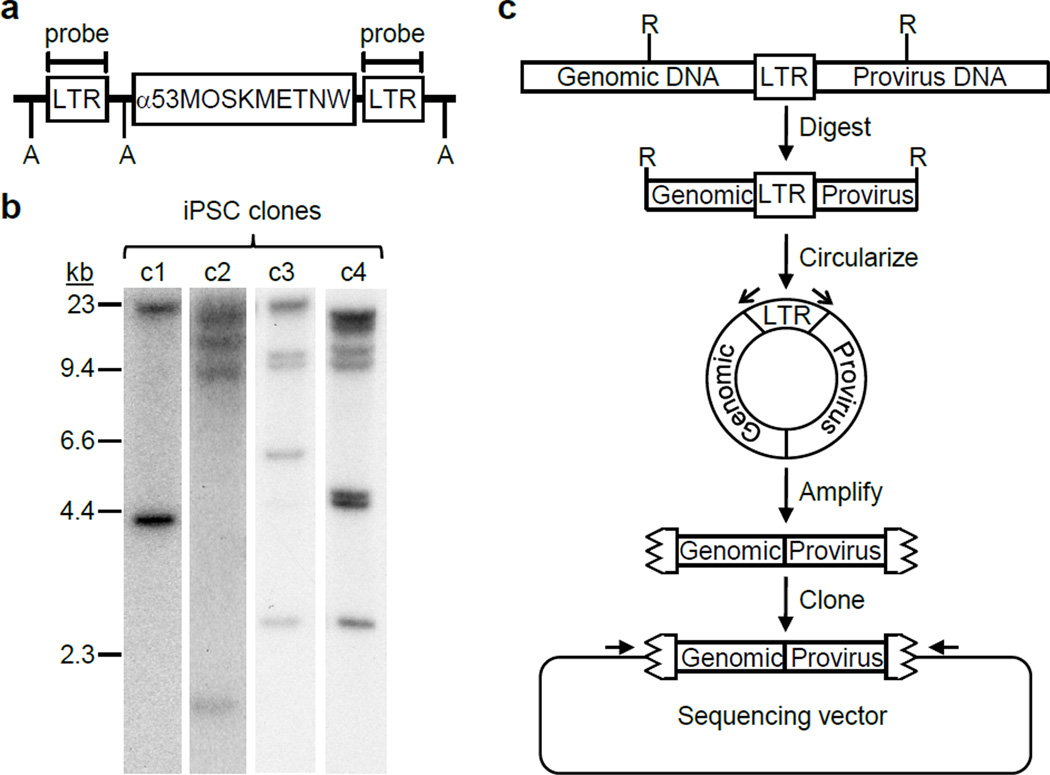
We identified a total of 10 FV integration sites in the four iPSC lines by Southern blot analysis (Figure 2a–b). Each of these was mapped unambiguously by inverse PCR (Figure 2c) to a unique location in the human genome that agreed with the restriction digest fragment sizes observed on Southern blots (Table 1). Six of the integrants were located inside transcription units and four were outside of genes (Figure 3). Two of these integrants were within 300 kb of the EPHA5 or ERBB4 proto-oncogenes associated with lung cancer27 and multiple types of human tumors28–30 respectively. Overall, the 10 integrants were located within 300 kb of 46 different measurable transcripts.
Figure 2. Identification of FV integration sites.
(a) Diagram of an integrated reprogramming vector with locations of the LTR probe shown. A, Avr II sites. (b) Southern blot analysis of Avr II-digested genomic DNAs to determine the number of FV vector integration sites in each iPSC clone. Each integrant produces 2 LTR-hybridizing fragments. (c) Inverse PCR strategy for identifying chromosome-provirus junctions. R, restriction enzymes sites; open arrows, LTR-specific PCR primers; jagged box, LTR remnant; closed arrow, sequencing primers.
Table 1.
Predicted LTR fragment sizes after restriction digestion.
| iPSC clone |
Number of proviruses |
LTR Fragment |
Junction 1 |
Junction 2 |
Junction 3 |
Junction 4 |
|---|---|---|---|---|---|---|
| c1 | 1 | 1 | 18874 | |||
| 2 | 4048 | |||||
| c2 | 2 | 1 | 24771 | 12496 | ||
| 2 | 8676 | 1276 | ||||
| c3 | 3 | 1 | 13996 | 15656 | 18215 | |
| 2 | 6063 | 2713 | 9014 | |||
| c4 | 4 | 1 | 12431 | 28874 | 11901 | 21449 |
| 2 | 3078 | 9216 | 4915 | 5555 |
Fragment sizes are in base pairs and based on the February 2009 freeze of the human (hg19) genome.
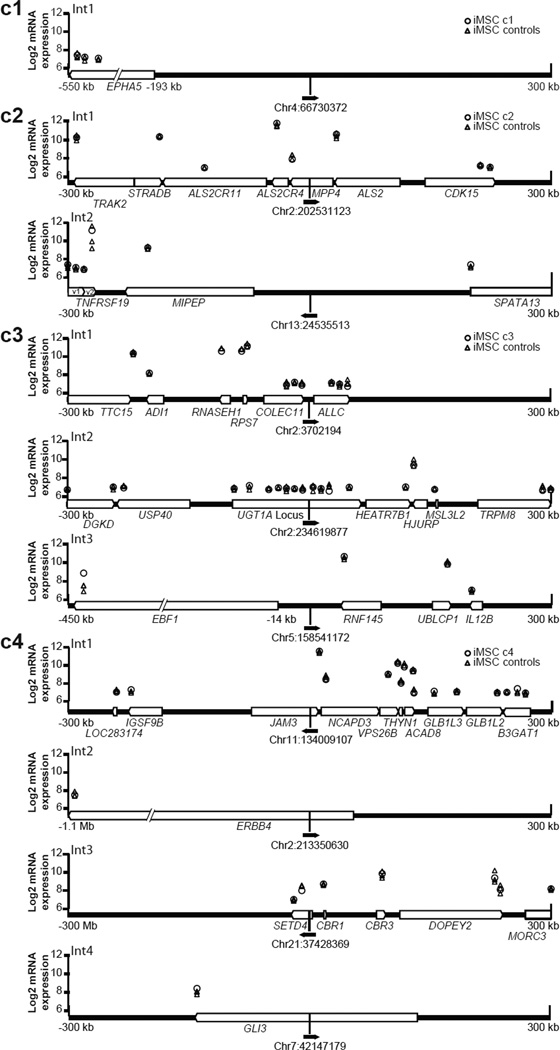
Figure 3. Provirus locations and neighboring gene expression in iMSCs.
The vector proviruses found in each clone are shown with chromosomal location (February 2009 freeze of the human (hg19) genome) and a solid black arrow in the direction of vector transcription. Cellular genes within a 300 kb window up- or downstream of each provirus are shown as white block arrows in the direction of transcription. Micorarray probe signal levels are shown above their chromosomal positions as log2 values. Probe signals from each iMSC clone containing the integrant (○) were compared to the remaining three iMSC clones as controls (△). Int1–Int4 identifies distinct integration sites within each clone.
We determined the effects of FV proviruses on chromosomal gene expression by performing a global transcriptional analysis on undifferentiated cultures of iPSC clones c1, c3, and c4, as well as differentiated iMSC cultures of clones c1–c4 (undifferentiated iPSC clone c2 was not available for analysis). The Illumina Bead Chip v3 was used to measure the expression levels of 25,000 genes with 48,801 probes, and the levels obtained from each clone were compared to the average of the other clones for both undifferentiated iPSCs and differentiated iMSCs. The percentage of > 2-fold up- or down-regulated probe signals ranged from 0.02–0.51% between the iPSC clones and 0.17–0.68% between their iMSC cultures (Table 2). These findings demonstrate that nearly 1% of all probe signals may vary >2-fold between clones of iPSCs or iMSCs, and this represents the background variation level in our expression profiles.
Table 2.
mRNA probe signals in iPSCs and iMSCs.
| Cell type | No. of probe ignals with >2-fold up or down variation |
Percent of all probe signals with >2-fold differences |
No. of probes for genes within 300 kb of an integrated provirus |
No. of probe signals for genes within 300 kb of proviruses with >2-fold differences |
|---|---|---|---|---|
| iPSC c1 | 248 | 0.51 | 3 | 0 |
| iPSC c3 | 12 | 0.02 | 35 | 0 |
| iPSC c4 | 64 | 0.13 | 23 | 0 |
| iMSC c1 | 81 | 0.17 | 3 | 0 |
| iMSC c2 | 332 | 0.68 | 14 | 0 |
| iMSC c3 | 219 | 0.45 | 34 | 1 |
| iMSC c4 | 281 | 0.58 | 23 | 0 |
We next evaluated the signal level of every microarray probe that detected a transcript located within a 300 kb window up- or downstream of an FV integration site. In the three iPSC clones studied, none of the 61 probes within this window had > 2-fold signal differences when comparing cells that contained the nearby provirus to those lacking the provirus (Supplemental Figure 1). These iPSC data must be interpreted cautiously, since the reprogramming vectors were silenced or expressed at a low level in 2 of these clones (c1 and c3).
In the four iMSC cultures studied, all of which expressed the reprogramming vector transcript, one of 74 probes assayed (1.4%) had a >2-fold signal increase (3-fold). This single probe detects the EBF1 transcript. In clone c3, integrant number 3 is 14 kb away from the EBF1 transcription start site and transcribed from the opposite strand (Figure 3). Although this places the probe 415 kb from the integration site and outside the ±300 kb window, it was included because it was the only probe that detected the EBF1 gene. We confirmed these microarray data by quantitative RT-PCR of EBF1 mRNA which showed a 5, 12, and 210-fold increase in clone c3 iMSCs when compared to clone c2, c1 and c4 iMSCs respectively. While it is possible that the enhancer activity of the internal MLV promoter increased EBF1 transcription, this observation may also have been unrelated to the provirus. The 1.4% of probes found near integration sites that were dysregulated (Table 2) was not statistically different than the 0.45% percent of all probes dysregulated in iMSC c3 (Fisher exact test, p value = 0.284). In addition, a power analysis showed that at least 2 neighboring probes would have to be dysregulated to achieve significance. This probe signal was also particularly inconsistent among iMSCs, since it varied 42-fold in the 3 iMSC cultures that lacked a nearby provirus.
Six of the proviruses we studied were located within the introns of transcription units, and none of these transcripts were dysregulated by provirus integration. In two cases, intron-embedded proviruses were in the same orientation as the transcription unit, and the signals of downstream probes were still not significantly altered. These data confirm that FV vector proviruses do not result in read-through transcription, as suggested by prior transfection-based assays21. They also suggest that integration within an intron is not genotoxic, presumably due to splicing out of the vector provirus, since nonsense-mediated decay or premature termination would otherwise have decreased transcript levels.
In summary, our analysis of 10 independent FV vector proviruses failed to demonstrate any dysregulation of nearby genes, although we cannot rule out that a larger sample size might have included a small percentage of dysregulated loci. Nevertheless, these results stand in contrast to those obtained with gammaretroviral and LV vectors, where dysregulation was noted in 3.2–20% or 3–13% of genes near integration sites respectively14, 31–33. Also, a study analyzing only intragenic integration sites in LV-derived iPSC clones showed that 11% of these genes were dysregulated9, while in our study none of the genes containing FV vector proviruses were altered. Even when using SIN gammaretroviral and LV vectors, internal promoters with strong enhancer elements such as the MLV LTR increased the frequency of nearby gene dysregulation14. Notably, the FV vector used in our experiments contained the MLV promoter, but no dysregulation of nearby genes was observed. In addition, read-through transcription and aberrant splicing events from gammaretroviral and LV vectors can result in fusion transcripts that alter adjacent gene transcript levels13, 21, 34–37, which we did not observe with FV vectors, suggesting that fusion transcripts were not produced at significant levels. In conclusion, we have found that FV vectors have minimal genotoxicity, supporting their use in gene therapy and cellular reprogramming.
Materials & Methods
Cell Culture
Osteogenesis imperfecta MSCs were established from discarded bone fragments of affected individuals undergoing corrective surgery with Institutional Review Board approval38. Both MSCs and iMSCs were cultured in Dulbecco’s modified Eagle's medium (DMEM) with low glucose containing 10% characterized fetal bovine serum (HyClone Laboratories, Logan, UT), 100 U/ml penicillin, 100 µg/ml streptomycin, and supplemented with 2 mM L-glutamine. Human iPSCs were cultured on irradiated mouse embryonic fibroblasts (MEFs) derived from the progeny of DR-4 mice39 crossed with CF-1 mice (Charles River Laboratories, Wilmington, MA) in DMEM/F12 medium supplemented with 20% Knockout Serum Replacement (Invitrogen, Grand Island, NY), 1% nonessential amino acid solution, 1% sodium pyruvate, 0.1 mM β-mercaptoethanol, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 ng/ml basic fibroblast growth factor as described40, or on human embryonic stem cell (hESC)-qualified matrigel (BD Biosciences, Bedford, MA) in TeSR 2 medium (StemCell Technologies, Vancouver, BC) when expanded for RNA analysis. iPSCs were derived from MSCs with the FV vector ΔΦ53MOSKMETNW as described previously24, where iPSC clones c1–c4 corresponded to iPSC clones iPSCe2-FVc1, iPSCe2-FVc2, iPSCe2-FVc3, and iPSCe2-FVc4. iMSCs were derived from iPSCs as described for hESCs26.
Identification of FV integration sites
Genomic DNA was isolated from each iPSC line by using the Puregene DNA purification system (Gentra Systems, Minneapolis, MN). Southern blot analysis and restriction digests were performed according to standard protocols. Radiolabeled probes were synthesized by random priming using Rediprime II (GE Healthcare, Piscataway, NJ). For inverse PCR, two µg of genomic DNA was digested with 8 units of Nla III, Hae III, Aci I, Hha I, or Msp I restriction endonuclease (New England Biolabs, Beverly, MA) at 37°C for 2 hours, extracted with phenol/chloroform, and precipitated with ethanol. Nucleic acid pellets were resuspended in 355 µl of water, 40 µl of 10X ligase buffer, and 5 µl of T4 DNA ligase (New England Biolabs, Beverly, MA). Ligation reactions were incubated at 16°C for 16 hours. The ligase reactions were heat-inactivated, extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 20 µl of 10 mM Tris (pH 8.0), 1 mM EDTA. One microliter was used as template for PCR amplification with oligonucleotides ik213f (5’-GGGTGATTGCAATGCTTTCT) and ik214r (5’-TGTCTCTCATCCCAGGTACG) or ik224f (5’-AGCCTTGCTAAGGGAGACATCTAGTG) and ik225r (5’-GTTCTTCACCTCCTTCCCTGTA). DNA fragments were excised from agarose gels and cloned using the TA cloning vector pGEM T-easy (Promega, Madison, WI). DNA sequences were obtained from these cloned PCR products.
Statistical Analysis
Statistics were performed with JMP 9.0 statistical analysis software (SAS Institute Inc., Cary, NC). Data was analyzed using the Fisher's exact test and two-sample proportions analysis. P values <0.05 were considered statistically significant. For power analyses, statistical power was set at 0.8.
RT-PCR and gene expression analysis
RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol from iMSCs or iPSCs grown on matrigel. cDNA synthesis was performed from total RNA using the Superscript III First-Strand Synthesis System, as per the manufacturer’s instructions (Invitrogen). FV vector transgene expression was detected with primers Foamy-f and Foamy-r by PCR as previously described24. Quantitative RT–PCR was performed using a StepOnePlus Real-Time PCR System and TaqMan Gene Expression Assay (Applied Biosystems, Calsbad, CA) or the Bio-Rad MyiQ Single Color Real-Time PCR Detection System and Bio-Rad iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) using the manufacturer’s recommended conditions. Gene expression profiling by Illumina Bead Chip v3 was performed as described24.
Supplementary Material
Acknowledgements
This research was funded by NIH grants AR48328, AR48328-09S1, DK55759, GM86497, HL53750, and AR53917.
Footnotes
Conflict of Interest
The authors declare no conflict of interests.
Bibliography
- 1.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-Associated Clonal T Cell Proliferation in Two Patients after Gene Therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 2.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. The Journal of Clinical Investigation. 2008;118(9):3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 4.Krause D. Gene therapy for Wiskott–Aldrich syndrome: benefits and risks. The Hematologist. 2011;8(2):10. [Google Scholar]
- 5.Modlich U, Bohne J, Schmidt M, von Kalle C, Knoss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108(8):2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotech. 2006;24(6):687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 7.Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16(4):718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 8.Hargrove PW, Kepes S, Hanawa H, Obenauer JC, Pei D, Cheng C, et al. Globin Lentiviral Vector Insertions Can Perturb the Expression of Endogenous Genes in [beta]-thalassemic Hematopoietic Cells. Mol Ther. 2008;16(3):525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- 9.Winkler T, Cantilena A, Métais J-Y, Xu X, Nguyen A-D, Borate B, et al. No Evidence for Clonal Selection Due to Lentiviral Integration Sites in Human Induced Pluripotent Stem Cells. STEM CELLS. 2010;28(4):687–694. doi: 10.1002/stem.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronen K, Negre O, Roth S, Colomb C, Malani N, Denaro M, et al. Distribution of Lentiviral Vector Integration Sites in Mice Following Therapeutic Gene Transfer to Treat [beta]-thalassemia. Mol Ther. 2011;19(7):1273–1286. doi: 10.1038/mt.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biasco L, Ambrosi A, Pellin D, Bartholomae C, Brigida I, Roncarolo MG, et al. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Molecular Medicine. 2011;3(2):89–101. doi: 10.1002/emmm.201000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17(11):1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, Sartori D, et al. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol Ther. 2009;17(5):851–856. doi: 10.1038/mt.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane NM, Nowrouzi A, Mukherjee S, Blundell MP, Greig JA, Lee WK, et al. Lentivirus-mediated Reprogramming of Somatic Cells in the Absence of Transgenic Transcription Factors. Mol Ther. 2010;18(12):2139–2145. doi: 10.1038/mt.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trobridge G, Josephson N, Vassilopoulos G, Mac J, Russell DW. Improved foamy virus vectors with minimal viral sequences. Mol Ther. 2002;6(3):321–328. doi: 10.1006/mthe.2002.0672. [DOI] [PubMed] [Google Scholar]
- 17.Russell DW, Miller AD. Foamy virus vectors. J Virol. 1996;70(1):217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, et al. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997;71(10):7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu SF, Sullivan MD, Linial ML. Evidence that the human foamy virus genome is DNA. J Virol. 1999;73(2):1565–1572. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, et al. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci U S A. 2006;103(5):1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrie PC, Huo Y, Stolitenko RB, Russell DW. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther. 2008;16(3):534–540. doi: 10.1038/sj.mt.6300398. [DOI] [PubMed] [Google Scholar]
- 22.Bauer TR, Allen JM, Hai M, Tuschong LM, Khan IF, Olson EM, et al. Successful treatment of canine leukocyte adhesion deficiency by foamy virus vectors. Nat Med. 2008;14(1):93–97. doi: 10.1038/nm1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohmine K, Li Y, Bauer TR, Jr, Hickstein DD, Russell DW. Tracking of specific integrant clones in dogs treated with foamy virus vectors. Hum Gene Ther. 2011;22(2):217–224. doi: 10.1089/hum.2010.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deyle DR, Khan IF, Ren G, Wang PR, Kho J, Schwarze U, et al. Normal collagen and bone production by gene-targeted human osteogenesis imperfecta iPSCs. Mol Ther. 2012;20(1):204–213. doi: 10.1038/mt.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukiyama T, Niwa O, Yokoro K. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol Cell Biol. 1989;9(11):4670–4676. doi: 10.1128/mcb.9.11.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang NS, Varghese S, Lee HJ, Zhang Z, Ye Z, Bae J, et al. In vivo commitment and functional tissue regeneration using human embryonic stem cell-derived mesenchymal cells. Proc Natl Acad Sci U S A. 2008;105(52):20641–20646. doi: 10.1073/pnas.0809680106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41(10):1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soung YH, Lee JW, Kim SY, Wang YP, Jo KH, Moon SW, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. International Journal of Cancer. 2006;118(6):1426–1429. doi: 10.1002/ijc.21507. [DOI] [PubMed] [Google Scholar]
- 31.Recchia A, Bonini C, Magnani Z, Urbinati F, Sartori D, Muraro S, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci U S A. 2006;103(5):1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li CL, Xiong D, Stamatoyannopoulos G, Emery DW. Genomic and Functional Assays Demonstrate Reduced Gammaretroviral Vector Genotoxicity Associated With Use of the cHS4 Chromatin Insulator. Mol Ther. 2009;17(4):716–724. doi: 10.1038/mt.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arumugam PI, Higashimoto T, Urbinati F, Modlich U, Nestheide S, Xia P, et al. Genotoxic potential of lineage-specific lentivirus vectors carrying the beta-globin locus control region. Mol Ther. 2009;17(11):1929–1937. doi: 10.1038/mt.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almarza D, Bussadori G, Navarro M, Mavilio F, Larcher F, Murillas R. Risk assessment in skin gene therapy: viral-cellular fusion transcripts generated by proviral transcriptional read-through in keratinocytes transduced with self-inactivating lentiviral vectors. Gene Ther. 2011;18(7):674–681. doi: 10.1038/gt.2011.12. [DOI] [PubMed] [Google Scholar]
- 35.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106(12):3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moiani A, Paleari Y, Sartori D, Mezzadra R, Miccio A, Cattoglio C, et al. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. The Journal of Clinical Investigation. 2012;122(5):1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight S, Zhang F, Mueller-Kuller U, Bokhoven M, Gupta A, Broughton T, et al. Safer, Silencing-Resistant Lentiviral Vectors: Optimization of the Ubiquitous Chromatin-Opening Element through Elimination of Aberrant Splicing. J Virol. 2012;86(17):9088–9095. doi: 10.1128/JVI.00485-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamberlain JR, Deyle DR, Schwarze U, Wang P, Hirata RK, Li Y, et al. Gene targeting of mutant COL1A2 alleles in mesenchymal stem cells from individuals with osteogenesis imperfecta. Mol Ther. 2008;16(1):187–193. doi: 10.1038/sj.mt.6300339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker KL, Wang Y, Dausman J, Jaenisch R. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25(18):3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.