Abstract
Lysine methylation is one of the most prominent histone posttranslational modifications that regulate chromatin structure. Changes in histone lysine methylation status have been observed during cancer formation, which is thought to be a consequence of the dysregulation of histone lysine methyltransferases or the opposing demethylases. KDM4/JMJD2 proteins are demethylases that target histone H3 on lysines 9 and 36 and histone H1.4 on lysine 26. This protein family consists of three ~130 kDa proteins (KDM4A–C) and KDM4D/JMJD2D, which is half the size, lacks the double PHD and Tudor domains that are epigenome readers and present in the other KDM4 proteins, and has a different substrate specificity. Various studies have shown that KDM4A/JMJD2A, KDM4B/JMJD2B and/or KDM4C/JMJD2C are overexpressed in breast, colorectal, lung, prostate and other tumors and are required for efficient cancer cell growth. In part, this may be due to their ability to modulate transcription factors such as the androgen and estrogen receptor. Thus, KDM4 proteins present themselves as novel potential drug targets. Accordingly, multiple attempts are underway to develop KDM4 inhibitors, which could complement the existing arsenal of epigenetic drugs that are currently limited to DNA methyltransferases and histone deacetylases.
Keywords: Gene transcription, Histone demethylation, JMJD2, KDM4, Lysine methylation
Introduction
Negatively charged DNA wraps around a core of positively charged histones to allow for condensation of our genetic material. The state of compaction changes following specific alterations in histone posttranslational modifications. Acetylation and methylation are the two predominant covalent modifications, where acetylation of a positively charged lysine residue reduces the overall charge of a histone and generally leads to the relaxation of chromatin and thereby enhanced gene transcription. Methylation on arginine or lysine residues, in contrast, does not alter the charge of histones and can have repressive or activating consequences on gene expression, depending on which particular arginine or lysine residue becomes modified (1, 2).
Global as well as local changes in chromatin structure are characteristic for tumors, suggesting that such epigenetic changes are an underlying cause of cancer. Accordingly, enzymes involved in histone modification and also DNA methylation may be viable drug targets. And indeed, histone deacetylase and DNA methyltransferase inhibitors are already FDA-approved for the treatment of cutaneous T-cell lymphoma and myelodysplastic syndrome, respectively. However, targeting enzymes that methylate or demethylate histones has not yet progressed to standard clinical use (3).
JMJD Proteins
Not long ago, histone methylation was considered to be an irreversible mark. This dogma was finally laid to rest upon the discovery of the first lysine-specific demethylase (LSD1) in 2004 (4). Human LSD1 and its only paralog, LSD2, demethylate mono- and dimethylated histone H3 lysine 4 (H3K4) and H3K9 through a FAD-dependent amine oxidation reaction. The second known family of histone demethylases, the JMJD (Jumonji C domain-containing) proteins, is comprised of 30 members in humans based on the presence of the roughly 150 amino acid-long JmjC (Jumonji C) domain (5). However, while most of the JMJD proteins have been proven to demethylate H3K4, H3K9, H3K27, H3K36 or H4K20, the catalytic activity of several JMJD proteins remains to be uncovered. Notably, some JMJD proteins are predicted to have no catalytic activity at all. Furthermore, it remains controversial whether any JMJD protein can target methylated arginine residues (6).
JMJD proteins employ a different reaction mechanism compared to LSD1/2. They act through a dioxygenase reaction mechanism requiring Fe2+, O2 and 2-oxoglutarate to demethylate histones. The true catalytic step is the hydroxylation of a lysine methyl group, thereby converting it to a hydroxymethyl moiety that spontaneously disconnects from the nitrogen center resulting in the release of formaldehyde. This reaction mechanism allows JMJD proteins in principal to demethylate tri-, di- and monomethylated lysine residues, whereas LSD1/2 are prohibited from attacking trimethylated lysines due to the requirement of a free electron pair on the methylated nitrogen (5, 6). One of the largest JMJD subfamilies that has recently attracted much attention is comprised of the JMJD2A-D proteins (nowadays preferentially called KDM4A-D, for K demethylase 4 A-D), which are capable of recognizing di- and trimethylated H3K9 and H3K36 as well as trimethylated H1.4K26 as substrates (Fig. 1A and 1B).
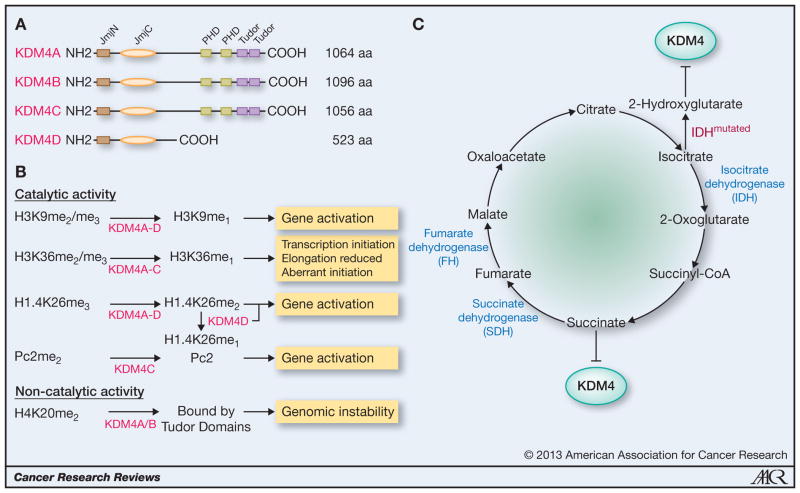
Figure 1.
(A) Schematic structure of the four KDM4 proteins. The JmjN domain is required for the activity of the JmjC catalytic center. (B) Modes of KDM4 function as demethylases or independent of enzymatic activity. (C) SDH, FH and IDH in the Krebs cycle. Succinate accumulates upon SDH or FH mutation, while neomorphic IDH mutations lead to 2-hydroxyglutarate production.
In general, H3K9 and H1.4K26 trimethylation are associated with transcription repression or heterochromatin formation, whereas H3K36 methylation has been perceived with activating gene expression (1, 3). However, this may be more nuanced, since crosstalking with other histone modifications influences the outcome of H3K9, H3K36 and H1.4K26 methylation (7). Also, H3K36 methylation shifts from mono- to trimethylation from the promoter to the end of transcribed genes. Thus, H3K36 trimethylation perhaps inhibits gene transcription at the start site, but facilitates transcription elongation and prevents unwanted transcription initiation within the body of the gene that can negatively interfere with transcription initiation from the regular start site (Fig. 1B). Moreover, the role of H3K36 methylation (and likely H3K9 and H1.4K26 methylation) is not limited to transcription control, but extends to alternative splicing, DNA replication, recombination and repair (8). Accordingly, modification of the histone methylation status due to changes in KDM4 activity may have profound effects on the transcriptome and other nuclear processes and therefore also on the initiation and progression of cancer. Here, we will focus on the potential oncogenic roles of the KDM4 proteins.
The Protagonist: KDM4A/JMJD2A
The most studied member of the KDM4 family is KDM4A/JMJD2A, also sometimes called JHDM3A (JmjC domain-containing histone demethylase 3A). Although KDM4A is capable of demethylating H3K9 and H3K36, qualitative and kinetic measurements indicated that it demethylates H3K9me3 ~5-fold more efficiently than H3K36me3. In addition, KDM4A is more efficient in demethylating tri- versus dimethylated H3K9/H3K36 (9–12) and, even more specifically, only demethylates trimethylated H1.4K26 (13). An explanation for its substrate specificity was provided by X-ray crystallographic structure analyses of KDM4A with and without peptide substrates. These studies revealed that amino acids surrounding lysines 9 and 36 on histone H3 primarily determine the binding specificity of KDM4A, while the space and the electrostatic environment in the methyl group-binding pocket allow tri-/di- but not monomethylated lysine residues to position a methyl group productively towards the iron-containing catalytic center (11, 14–16).
A major study focus on KDM4A has been in transcription regulation, where it may either stimulate or repress gene transcription. The latter function may involve association with the nuclear receptor corepressor complex or histone deacetylases (17, 18) or direct binding to a transcription factor as observed for the p53 tumor suppressor (19), but it remains unresolved whether this repressing function requires KDM4A enzymatic activity. In contrast, KDM4A formed complexes with both the androgen and estrogen receptor (ER) and stimulated their activity, which was dependent on KDM4A catalytic activity (20, 21). Accordingly, depletion of KDM4A in ER-positive T47D breast cancer cells decreased the expression of ER targets such as the c-Jun and cyclin D1 oncogenes and led to reduced cell growth. Equally, JMJD2A knockdown inhibited proliferation of ER-negative MDA-MB-231 and ER-positive MCF7 breast tumor cells (22, 23), suggesting that KDM4A is critical for growth of both ER-positive and -negative breast tumors. Consistent with an oncogenic function of KDM4A, it is overexpressed at the protein level in ~60% of breast tumors (21) and further reports demonstrate KDM4A overexpression also at the mRNA level in this cancer (24, 25). Likewise, KDM4A is overexpressed in prostate (26) and lung cancer (27) (see also Supplementary Table 1).
Similar to breast cancer cells, downregulation of KDM4A in multiple colon cancer cell lines resulted in reduced cell proliferation and furthermore increased apoptosis and delayed the G2/M phase of the cell cycle (19). In U2OS human osteosarcoma cells, KDM4A depletion resulted in G0/G1 growth arrest, whereas senescence was induced in normal fibroblasts. On the other hand, overexpression of KDM4A cooperated with the H-Ras oncoprotein in transforming human IMR90 fibroblasts by suppressing Ras-induced senescence (27). However, no growth effect was observed upon KDM4A overexpression or downregulation in HeLa cervical carcinoma or 293T embryonal kidney cells, although S-phase progression was accelerated by wild-type, but not catalytically inactive KDM4A; seemingly, the S-phase acceleration was neutralized by delays in other cell cycle phases (28). Altogether, this implies that KDM4A overexpression will not generally stimulate tumor growth, but only in certain organs or cell types. It remains to be determined which circumstances allow KDM4A to exert a pro-oncogenic function, for instance the tissue-specific expression of cooperation partners.
Interestingly, overexpression of KDM4A, but not its H188A catalytic mutant, in mouse NIH3T3 cells reduced recruitment of heterochromatin protein 1 (HP1) to pericentric chromatin (10). Moreover, KDM4A overexpression reduced binding of HP1γ to chromatin during replication in 293T cells concomitant with a more open chromatin structure. On the other hand, depletion of HP1γ led to increased chromatin accessibility early in S phase, suggesting that KDM4A and HP1γ antagonize each other during DNA replication which may represent a possible mechanism how KDM4A promotes DNA replication. This antagonism appears to be conserved in the worm C. elegans, since loss of its sole KDM4 homolog led to slower DNA replication and this defect could be rescued by depletion of the C. elegans HP1 homolog (28).
In the fruitfly Drosophila, HP1a associates with KDM4A and stimulates its H3K36 demethlyation activity. Unlike mammalian KDM4A, the Drosophila homolog appears to be incapable of demethylating H3K9, although this is controversial (29, 30). Moreover, Drosophila KDM4A overexpression resulted in HP1a spreading from the chromocenter into the chromosome arms and HP1a was shown to recruit KDM4A to a subset of heterochromatic genes where it could demethylate H3K36me3 (29–31). Currently, it is unresolved if and how the interaction between HP1a and KDM4A affects DNA replication in Drosophila. However, it appears that these two proteins antagonize each other in the modulation of Drosophila gene transcription (32).
KDM4A is endowed with a double plant homeodomain (PHD) as well as a double Tudor domain (Fig. 1A). The latter binds to di- and trimethylated H3K4 and H4K20 (33–35), whereas the function of the KDM4A PHD domains has remained unknown. H3K4 trimethylation is a hallmark of active promoters and is normally mutually exclusive with H3K9 trimethylation, which is a mark of inactive chromatin (3). Thus, KDM4A may be recruited via the Tudor domains to active gene promoters and guarantee that H3K9 and also H1.4K26 become demethylated, which will amplify gene transcription by e.g. counteracting the binding of HP1 to these epigenetic marks. Although not yet studied for KDM4A, KDM4B is also part of the mixed-lineage leukemia 2 complex that has H3K4 methyltransferase activity, pointing at a potentially Tudor domain-independent way how KDM4B and possibly KDM4A concurrently demethylate H3K9 while H3K4 becomes trimethylated (36).
An unexpected function of KDM4A (and apparently also KDM4B) independent of its enzymatic activity was revealed in the DNA damage pathway. Here, 53BP1 is recruited into DNA damage foci and required to orchestrate the DNA damage response. Both 53BP1 and the Tudor domains of KDM4A competed for binding to dimethylated H4K20. However, KDM4A became ubiquitylated by the E3 ligases RNF8/RNF168 upon DNA damage and thereby marked for degradation, allowing recruitment of 53BP1 to sites of DNA damage (37). Thus, KDM4A overexpression could impair DNA damage repair and induce genomic instability by suppressing 53BP1 recruitment to DNA damage foci, another mechanism by which KDM4A possibly promotes tumorigenesis. In addition, KDM4A may affect DNA repair by inhibiting the Tip60 acetyltransferase, which is involved in the activation of the ataxia telangiectasia mutated kinase, a key component of the DNA double-strand repair pathway. Interestingly, Tip60 recognizes trimethylated H3K9 at DNA double-strand breaks and is thereby activated, suggesting that in this case the catalytic activity of KDM4A is required to demethylate H3K9me3 to exert an inhibitory effect on Tip60-dependent DNA repair (38). The involvement of KDM4A/KDM4B in DNA damage repair appears to be evolutionary conserved, since loss of C. elegans KDM4 led to more DNA damage and altered progression of meiotic DNA double-strand break repair and heterozygous mutation of KDM4B in Drosophila resulted in more sensitivity to UV irradiation (9, 28, 39).
The Close Relatives: KDM4B/JMJD2B and KDM4C/JMJD2C
KDM4B and KDM4C are structurally very similar to KDM4A (Fig. 1A) and have the same target specificity and comparable enzymatic activities in vitro (12, 13). However, earlier reports indicated a much lower catalytic activity of KDM4B compared to other KDM4 proteins (9, 40); it is currently unclear what the cause of this discrepancy is, for instance the utilization of differently sized recombinant KDM4B proteins or dissimilar substrates to measure catalytic activity. Phylogenetically, KDM4B and KDM4C are present like KDM4A in all vertebrates studied, whereas Drosophila has only two KDM4 homologs and C. elegans just one (12).
Similar to KDM4A, KDM4B and KDM4C mRNA levels are upregulated in breast tumors. Interestingly, KDM4B expression is higher in ER-positive than -negative breast tumors, whereas the opposite holds true for KDM4C (41–43). In addition, KDM4B appears to be overexpressed in triple-negative breast tumors (25) (see also Supplementary Table 1). Like KDM4A, KDM4B formed complexes with ER and stimulated transcription of ER target genes (36, 42). Downregulation of KDM4B in ER-positive MCF7 or T47D cells reduced cell proliferation and tumor formation in nude mice, whereas no changes were reported for ER-negative MDA-MB-231 cells upon KDM4B knockdown (36, 42, 44). Further, KDM4C downregulation led to reduced proliferation of ER-negative HCC1954 and Colo24 breast cancer cells, while its overexpression in non-transformed MCF10A cells caused colony formation in soft agar and increased mammosphere formation, an indicator of cancer stem cells (41). Notably, KDM4C is a target gene of the pluripotency factor Oct4 and KDM4C downregulation induced the differentiation of embryonic stem cells (45), suggesting that KDM4C overexpression is particularly important in cancer stem cells. Collectively, KDM4C overexpression may contribute to tumor formation in ER-negative breast tumors, whereas KDM4B mediates neoplastic transformation of ER-positive cells. The fact that KDM4B is an ER target gene (36, 42, 44) lends further support to the view that KDM4B is involved in the genesis of ER-positive tumors.
Also similar to KDM4A, KDM4C formed complexes with and stimulated androgen receptor and promoted androgen-dependent growth of prostate cancer cells, thus implicating KDM4C in prostate tumorigenesis (46). Evidence emerged that KDM4B and KDM4C are overexpressed in prostate tumors and medulloblastomas (26, 47); additionally, KDM4B is overexpressed in gastric, bladder, lung and colorectal cancer and required for proliferation, colony formation ability, invasion or survival of respective cell lines (48–50) (see also Supplementary Table 1). Moreover, the KDM4C gene was found to be translocated in mucosa-associated lymphoid tissue lymphoma, resulting in its overexpression (51). While all this furthers the notion of KDM4B and KDM4C as oncoproteins, such evidence must be regarded with caution, since KDM4B/C overexpression in tumors may be a consequence rather than a cause of tumorigenesis and changes of in vitro physiology upon modulation of KDM4B or KDM4C levels in cancer cells are no proof that KDM4B/C overexpression initiates or supports tumor formation in the human body.
Maybe a more convincing case for KDM4C as an oncogene is present in primary mediastinal B cell lymphoma and Hodgkin lymphoma, where amplification of the 9p24 chromosomal region has been found. KDM4C and the tyrosine kinase JAK2 are encoded within this amplicon, and both proteins are capable of epigenetic modulation, including activation of the c-Myc oncogene. A potential mechanism is the eviction of HP1 from the c-Myc promoter, since binding of HP1 to trimethylated H3K9 or H1.4K26 would be inhibited by KDM4C-dependent demethylation as well as by JAK-mediated phosphorylation of H3Y41 that also suppresses HP1 binding to chromatin. Moreover, inhibition of JAK2 and KDM4C synergized to kill primary mediastinal B cell and Hodgkin lymphoma cells (52). These genetic and in vitro data combined strongly argue that, in addition to JAK2, KDM4C is pro-oncogenic and a potential drug target in these lymphomas. Notably, 9p24 amplification has also been found in esophageal squamous cell carcinomas, which coined the older name GASC1 (gene amplified in squamous cell carcinoma 1) for KDM4C (53), as well as in sarcomatoid carcinoma of the lung and desmoplastic medulloblastoma (54, 55), suggesting that KDM4C may cooperate with JAK2 in many different tissues to contribute to tumor formation.
Overexpression of KDM4A-C in the same cancer (e.g. prostate cancer) and similar physiological functions in cancer cells suggest that KDM4 proteins may perform overlapping functions. Moreover, the absence of obvious pathological phenotypes in KDM4B as well as KDM4D knockout mice hints at redundancy within the KDM4 family (42, 56). However, KDM4 proteins can also behave differently. For instance, hypoxia induced KDM4B and, to a lesser extent, KDM4C transcription, whereas the KDM4A gene appeared to be unaffected (57, 58). In fact, KDM4B was required for increased transcription of many hypoxia-inducible genes in colorectal cancer cell lines. Also, KDM4B was overexpressed in colorectal cancer specimens that concurrently were positive for carbonic anhydrase 9, a marker of hypoxia, and KDM4B overexpression correlated with larger tumor size and advanced clinical stage (50). In addition, KDM4C bound to and stimulated HIF-1α, the key transcription factor mediating the cellular response to hypoxia (59). Therefore, KDM4B and/or KDM4C may help tumors to thrive in a hypoxic environment. Another example of differences amongst KDM4 proteins comes from the study of the polycomb 2 protein (Pc2), which is associated with heterochromatin formation and epigenetic silencing. Pc2 was demonstrated to be dynamically methylated, and KDM4C was the only member of the KDM4 family capable of demethylating Pc2. This demethylation of Pc2 led to the activation of growth-control genes, providing a distinct mechanism how KDM4C could facilitate tumorigenesis independent of histone demethylation (60).
The Outlier: KDM4D/JMJD2D
KDM4D is unique within the KDM4 family in that it lacks both the PHD and Tudor domains and thus is only half the size of KDM4A-C (Fig. 1A). Phylogenetically, KDM4D evolved recently, since it has only been found in placental mammals (12). Bioinformatical analyses revealed the presence of two further human genes, KDM4E and KDM4F, whose gene products are very similar to KDM4D. However, KDM4E and KDM4F are most likely pseudogenes (9, 61). Furthermore, in contrast to KDM4A-C, KDM4D has a different substrate specificity: it does not demethylate H3K36 due to several differences in its substrate binding cleft, yet has gained the ability to attack dimethylated in addition to trimethylated H1.4K26 (9, 13, 62). Also, KDM4D attacks H3K9me2 with similar efficiency as H3K9me3 and may, albeit inefficiently, even demethylate H3K9me1 (11, 12, 63). Another difference of KDM4D is manifest in its association behavior: while KDM4A and KDM4C homo- and heteromerize amongst each other, KDM4D is only capable of forming homomers (63).
Similar to KDM4A and KDM4C, KDM4D functioned as a coactivator of the androgen receptor (20). Also, like KDM4A, KDM4D was required for colon cancer cell proliferation and survival. However, in contrast to KDM4A being a repressor of p53 transcriptional activity, KDM4D stimulated p53-dependent gene expression (64). Whereas the stimulation of the androgen receptor and colon cancer cell growth points to a pro-oncogenic function, KDM4D’s role in activating p53-dependent gene transcription would suggest the opposite, highlighting that further research is needed to resolve the net effect, if any, of KDM4D in tumorigenesis.
Interestingly, the pro-inflammatory tumor necrosis factor α (TNFα) induced KDM4D expression in dendritic cells and macrophages. Moreover, the ability of KDM4D to demethylate H3K9 was shown to be involved in the TNFα response (65). Therefore, KDM4D may influence tumorigenesis not only in cancer cells, but potentially also in the tumor microenvironment and immune cells by mediating inflammatory responses elicited by cytokines such as TNFα.
Therapeutic Implications and Perspective
Several lines of evidence suggest that enhanced catalytic activity of KDM4 proteins is associated with cancer. If KDM4 proteins are indeed drivers of tumorigenesis, they represent viable novel targets for cancer therapy. Many efforts are underway or were already undertaken to design specific KDM4 inhibitors, but their specificity and utility as anti-cancer drugs requires rigorous preclinical testing in the future (66–68). As a cautionary note, however, genetically engineered mouse models are needed to definitively prove that overexpression of KDM4 proteins is an underlying cause of tumor formation. Also, such mouse models would represent an invaluable tool for eventually testing KDM4 inhibitors in vivo.
Apart from being histone demethylases, KDM4 proteins are predicted to demethylate non-histone proteins, and Pc2 was the first such example (60). Many more non-histone proteins that are targeted by KDM4 proteins will likely be discovered and thereby provide a deeper understanding of how KDM4 proteins can contribute to cancer formation. Not in the least, KDM4 proteins are dioxygenases and could therefore hydroxylate lysine or asparagine residues, as already shown for other JMJD proteins (6). Thereby, KDM4 proteins may regulate the function of a much broader spectrum of proteins by affecting different kinds of posttranslational modifications. Also, as shown for the role of KDM4A/B in the DNA damage response pathway (37) and for many KDM4A target genes in Drosophila (32), there are instances when KDM4 proteins act independent of their enzymatic activity and thus inhibitors targeting their catalytic center would not completely shut down KDM4 biological activities. Another open question is the function of the PHD domains in KDM4A-C: do they, like the Tudor domains, bind to specific histone modifications as observed for other proteins (69) and how does this relate to the function of KDM4 proteins?
Tumors often develop in a hypoxic environment, which is a major limiting factor to their survival and proliferation. Especially KDM4B is induced by hypoxia and involved in the upregulation of hypoxia-inducible genes (44, 50, 57, 58). Would inhibition of KDM4B therefore aggravate the hypoxic tumor microenvironment, which could lead to tumor necrosis? Also, since KDM4 proteins are O2-dependent enzymes, is their catalytic activity suppressed by hypoxia in tumors and what physiological consequences would that have?
Lastly, KDM4 proteins are 2-oxoglutarate dependent enzymes, so any perturbation of the endogenous 2-oxoglutarate pool will affect KDM4 activity. Or does overexpression of KDM4 deplete 2-oxoglutarate and thereby inhibit other 2-oxoglutarate-dependent enzymes? It is known that mutations of succinate dehydrogenase (SDH) and fumarate hydratase (FH) found in various cancers impair the Krebs cycle (Fig. 1C), leading to accumulation of succinate that is an end-product inhibitor of JMJD proteins, including KDM4A and KDM4D (70, 71). Moreover, cancer-associated mutations in isocitrate dehydrogenase (IDH) not only reduce the formation of 2-oxoglutarate, but have also gained a new catalytic activity of converting isocitrate to 2-hydroxyglutarate (Fig. 1C), which is an inhibitor of KDM4 proteins (72). Thus, could KDM4 overexpression be required in tumors to balance the inhibition of their catalytic activity upon SDH, FH or IDH mutation? Regardless, KDM4 activity appears to be intricately controlled by the cancer metabolome, but how this will affect KDM4 function and cancer cells requires more study.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by a grant to R. J. from the National Cancer Institute (R01 CA154745). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 3.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 7.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–5. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–6. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 11.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–95. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 12.Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, et al. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J Biol Chem. 2011;286:41616–25. doi: 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, et al. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284:8395–405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci USA. 2007;104:10818–23. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Yoon HG, Wong J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2) Mol Cell Biol. 2005;25:6404–14. doi: 10.1128/MCB.25.15.6404-6414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray SG, Iglesias AH, Lizcano F, Villanueva R, Camelo S, Jingu H, et al. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem. 2005;280:28507–18. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 19.Kim TD, Shin S, Berry WL, Oh S, Janknecht R. The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J Cell Biochem. 2012;113:1368–76. doi: 10.1002/jcb.24009. [DOI] [PubMed] [Google Scholar]
- 20.Shin S, Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun. 2007;359:742–6. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 21.Berry WL, Shin S, Lightfoot SA, Janknecht R. Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int J Oncol. 2012;41:1701–6. doi: 10.3892/ijo.2012.1618. [DOI] [PubMed] [Google Scholar]
- 22.Li BX, Zhang MC, Luo CL, Yang P, Li H, Xu HM, et al. Effects of RNA interference-mediated gene silencing of JMJD2A on human breast cancer cell line MDA-MB-231 in vitro. J Exp Clin Cancer Res. 2011;30:90. doi: 10.1186/1756-9966-30-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li BX, Luo CL, Li H, Yang P, Zhang MC, Xu HM, et al. Effects of siRNA-mediated knockdown of jumonji domain containing 2A on proliferation, migration and invasion of the human breast cancer cell line MCF-7. Exp Ther Med. 2012;4:755–61. doi: 10.3892/etm.2012.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patani N, Jiang WG, Newbold RF, Mokbel K. Histone-modifier gene expression profiles are associated with pathological and clinical outcomes in human breast cancer. Anticancer Res. 2011;31:4115–25. [PubMed] [Google Scholar]
- 25.Slee RB, Steiner CM, Herbert BS, Vance GH, Hickey RJ, Schwarz T, et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;31:3244–53. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 26.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 27.Mallette FA, Richard S. JMJD2A Promotes Cellular Transformation by Blocking Cellular Senescence through Transcriptional Repression of the Tumor Suppressor CHD5. Cell Rep. 2012;2:1233–43. doi: 10.1016/j.celrep.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol Cell. 2010;40:736–48. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res. 2008;36:2852–63. doi: 10.1093/nar/gkn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, et al. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin CH, Paulson A, Abmayr SM, Workman JL. HP1a targets the Drosophila KDM4A demethylase to a subset of heterochromatic genes to regulate H3K36me3 levels. PLoS One. 2012;7:e39758. doi: 10.1371/journal.pone.0039758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crona F, Dahlberg O, Lundberg LE, Larsson J, Mannervik M. Gene regulation by the lysine demethylase KDM4A in Drosophila. Dev Biol. 2013;373:453–63. doi: 10.1016/j.ydbio.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–51. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15:109–11. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci USA. 2011;108:7541–6. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–78. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–82. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palomera-Sanchez Z, Bucio-Mendez A, Valadez-Graham V, Reynaud E, Zurita M. Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. J Biol Chem. 2010;285:31370–9. doi: 10.1074/jbc.M110.128462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–62. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, et al. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene. 2009;28:4491–500. doi: 10.1038/onc.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6:e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berdel B, Nieminen K, Soini Y, Tengstrom M, Malinen M, Kosma VM, et al. Histone demethylase GASC1 - a potential prognostic and predictive marker in invasive breast cancer. BMC Cancer. 2012;12:516. doi: 10.1186/1471-2407-12-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Jubb AM, Pike L, Buffa FM, Turley H, Baban D, et al. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70:6456–66. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–53. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 47.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–72. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Zhao L, Zang W, Liu Z, Chen L, Liu T, et al. Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is overexpressed in gastric cancer. Biochem Biophys Res Commun. 2011;416:372–8. doi: 10.1016/j.bbrc.2011.11.045. [DOI] [PubMed] [Google Scholar]
- 49.Toyokawa G, Cho HS, Iwai Y, Yoshimatsu M, Takawa M, Hayami S, et al. The histone demethylase JMJD2B plays an essential role in human carcinogenesis through positive regulation of cyclin-dependent kinase 6. Cancer Prev Res. 2011;4:2051–61. doi: 10.1158/1940-6207.CAPR-11-0290. [DOI] [PubMed] [Google Scholar]
- 50.Fu L, Chen L, Yang J, Ye T, Chen Y, Fang J. HIF-1alpha-induced histone demethylase JMJD2B contributes to the malignant phenotype of colorectal cancer cells via an epigenetic mechanism. Carcinogenesis. 2012;33:1664–73. doi: 10.1093/carcin/bgs217. [DOI] [PubMed] [Google Scholar]
- 51.Vinatzer U, Gollinger M, Mullauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 2008;14:6426–31. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- 52.Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, et al. Identification of a novel gene, GASC1, within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60:4735–9. [PubMed] [Google Scholar]
- 54.Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, et al. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23 approximately p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–30. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Ehrbrecht A, Müller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–63. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- 56.Iwamori N, Zhao M, Meistrich ML, Matzuk MM. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod. 2011;84:1225–34. doi: 10.1095/biolreprod.110.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pollard PJ, Loenarz C, Mole DR, McDonough MA, Gleadle JM, Schofield CJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416:387–94. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 58.Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–52. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci USA. 2012;109:E3367–E76. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh M, Katoh M. Identification and characterization of JMJD2 family genes in silico. Int J Oncol. 2004;24:1623–8. [PubMed] [Google Scholar]
- 62.Krishnan S, Trievel RC. Structural and Functional Analysis of JMJD2D Reveals Molecular Basis for Site-Specific Demethylation among JMJD2 Demethylases. Structure. 2013:21. doi: 10.1016/j.str.2012.10.018. Epub, in press. [DOI] [PubMed] [Google Scholar]
- 63.Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353:973–7. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 64.Kim TD, Oh S, Shin S, Janknecht R. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One. 2012;7:e34618. doi: 10.1371/journal.pone.0034618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Y, van Essen D, Saccani S. Cell-type-specific control of enhancer activity by H3K9 trimethylation. Mol Cell. 2012;46:408–23. doi: 10.1016/j.molcel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 66.Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, et al. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett. 2009;19:2852–5. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki T, Miyata N. Lysine demethylases inhibitors. J Med Chem. 2011;54:8236–50. doi: 10.1021/jm201048w. [DOI] [PubMed] [Google Scholar]
- 68.Hoffmann I, Roatsch M, Schmitt ML, Carlino L, Pippel M, Sippl W, et al. The role of histone demethylases in cancer therapy. Mol Oncol. 2012;6:683–703. doi: 10.1016/j.molonc.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith EH, Janknecht R, Maher LJ., 3rd Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum Mol Genet. 2007;16:3136–48. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- 71.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.