Summary
Steady-state levels of spontaneous DNA damage, the by-product of normal metabolism and environmental exposure, are controlled by DNA repair pathways. Incomplete repair or an age-related increase in damage production and/or decline in repair could lead to an accumulation of DNA damage, increasing mutation rate, affecting transcription and/or activating programmed cell death or senescence. These consequences of DNA damage metabolism are highly conserved and the accumulation of lesions in the DNA of the genome could, therefore, provide a universal cause of aging. An important corollary of this hypothesis is that defects in DNA repair cause both premature aging and accelerated DNA damage accumulation. While the former has been well-documented, the reliable quantification of the various lesions thought to accumulate in DNA during aging has been a challenge. Here, we quantified inhibition of long distance PCR as a measure of DNA damage in liver and brain of both normal and prematurely aging, DNA repair defective mice. The results indicate a marginal, but statistically significant, increase of spontaneous DNA damage with age in normal mouse liver but not in brain. Increased levels of DNA damage were not observed in the DNA repair defective mice. We also show that oxidative lesions do not increase with age. These results indicate that neither normal nor premature aging is accompanied by a dramatic increase in DNA damage. This suggests that factors other than DNA damage per se, e.g., cellular responses to DNA damage, are responsible for the aging phenotype in mice.
Keywords: DNA damage, aging, premature aging, long distance PCR, Ercc1, Ku80
Introduction
DNA damage has long been implicated as a causal factor in aging (Vijg 2007). Thousands of DNA lesions are introduced in the genome of a somatic cell each day, from such diverse sources as spontaneous hydrolysis, oxidation, spontaneous alkylation, ionizing and UV irradiation and a large variety of environmental chemicals (Lindahl 1993; De Bont & van Larebeke 2004). If not swiftly repaired, these lesions can interfere with transcription of genes that are critical for normal cellular function, activate cell death or senescence pathways or lead to mutations as a consequence of errors made during repair or replication. While very low under normal conditions (Collins et al. 1997), DNA damage levels may gradually increase during normal aging, as a consequence of increased genotoxic stress, or when genome maintenance is defective or suboptimal. Indeed, multiple symptoms of premature aging have been observed as a consequence of defects in DNA repair in both humans and mice (Hasty et al. 2003; Garinis et al. 2008), as well as in cancer patients after chemotherapy with DNA damage-inducing drugs (Maccormick 2006).
Because DNA damage levels are so low under normal conditions, the quantification of the many different types of DNA damage under physiological conditions remains a challenge. For example, estimates of what is likely the most frequent type of oxidative damage, i.e., 8-oxo-2'-deoxyguanosine (8-oxo-dG), differ by three orders of magnitude and it remains unclear what the true level of this lesion is (Collins et al. 2004). Accurate information on the amount of spontaneous DNA damage is important to estimate its potential functional consequence. Unless significantly increased it is unlikely that low, steady state levels of DNA lesions are able to significantly impact on cellular function. Indeed, while DNA repair defective mice display a broad range of premature aging phenotypes as well as a shorter life span, the type and level of DNA damage in these animals have not been determined.
A sensitive assay for quantifying DNA damage is based on long distance, quantitative polymerase chain reaction (QPCR). Based on the principle that many kinds of DNA lesions can slow down or block the progression of DNA polymerase, this assay has been demonstrated to detect lesions at concentrations of approximately 1 per 105 bp, permitting the study of DNA damage at biologically relevant doses (Santos et al. 2006; Furda et al. 2012a). Here, we present data obtained with this assay on spontaneous DNA damage in liver and brain of normal and DNA repair defective mice in relation to aging.
Results
Quantitative PCR (QPCR) method: sensitivity and reproducibility
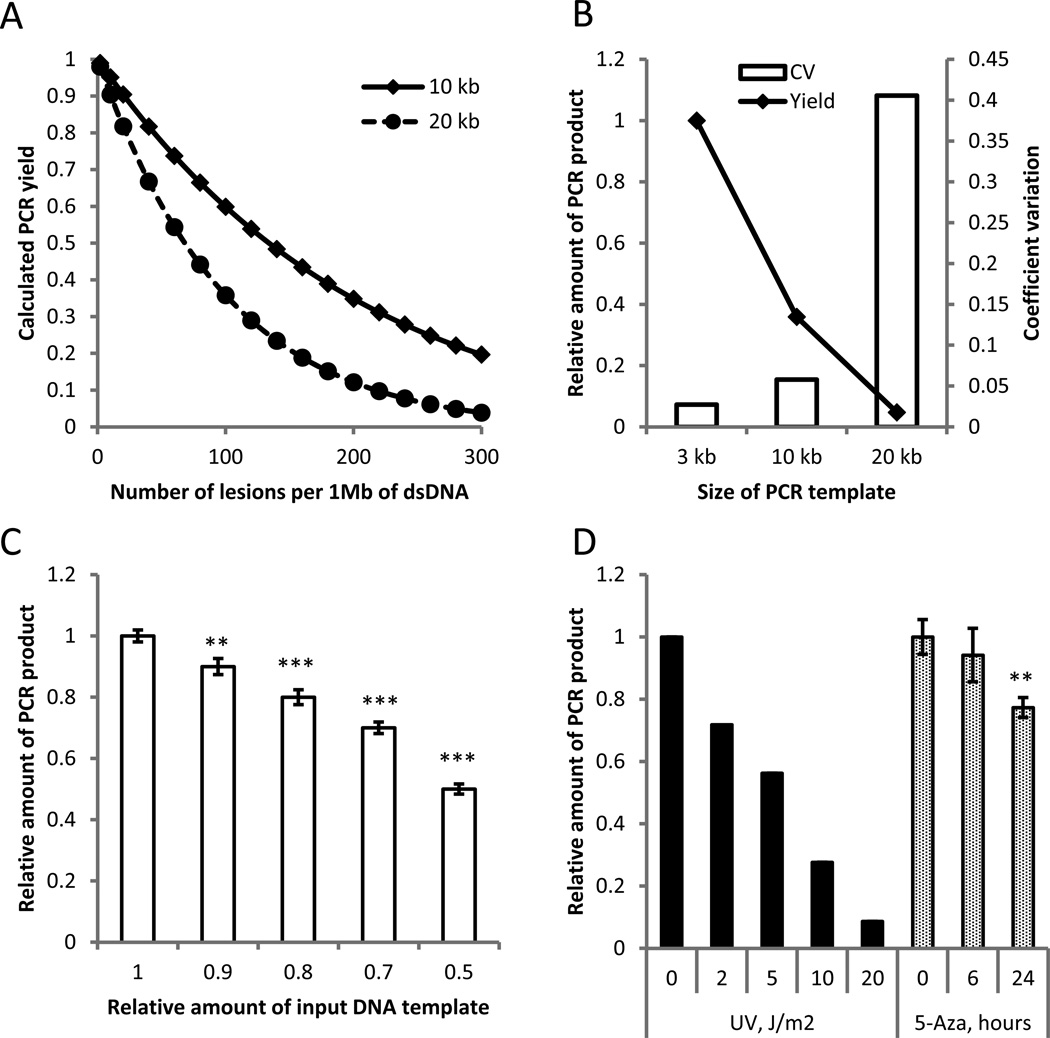
The QPCR method for detecting DNA damage is based on the assumption that if equal amounts of DNA from different samples are amplified under identical conditions, DNA with fewer lesions will amplify to a greater extent than more damaged DNA (Furda et al. 2012a). It is possible to estimate the density of DNA damage in the input DNA sample by quantitative assessment of the amount of amplified product. The predicted relationship between the amounts of DNA lesions and expected PCR product is shown in Fig. 1A for 10-kb and 20-kb templates (see also (Van Houten et al. 2000)). As it follows from the model, the sensitivity of the QPCR method entirely depends on the size of the targeted template, i.e., the longer the template, the more sensitive the assay. However, when the target fragments are too long, the efficiency of long-distance PCR becomes highly variable, which we experimentally determined for a 3-kb, 10-kb and 20-kb fragment (Fig. 1B). For our study we adopted target fragments of about 10-kb as most suitable in terms of sensitivity and reproducibility. Under these conditions the limit of detection is approximately 1 lesion per 105 bases of single-strand DNA. This is demonstrated in Fig. 1C, which shows the yield of a ~9-kb beta Hl globin (Hbb-bh1) gene, PCR-amplified from decreasing amounts of input DNA. A 10% decrease in input DNA, which corresponds to 20 lesions per 1 Mb of dsDNA (or 1 lesion per 105 bases of ssDNA), could still be detected at a significance level of 0.05. To avoid spurious DNA oxidation associated with phenol-based DNA extraction we used the column-based, solid phase extraction methods for DNA isolation from Zymo Research throughout this study, which were found to reproducibly give rise to a high yield of relatively high molecular weight genomic DNA. The quality of extracted DNA was routinely checked by spectroscopy and by running agarose gels under neutral and alkaline conditions (Supplementary Fig. S1). In all experiments both input DNA and the amounts of PCR product were measured by a PicoGreen assay, with the specificity of the PCR confirmed by agarose gel electrophoresis. Finally, to demonstrate the efficacy of the assay in detecting DNA damage we subjected mouse embryonic fibroblasts (MEFs) to different doses of UV radiation and, in a separate experiment, cultured MEFs in the presence of 5-aza-2'-deoxycytidine (5-Aza-dC), previously reported to induce DNA double-strand breaks (Maslov et al. 2012). As a read-out we measured the amount of ~9-kb long Hbb-bh1 PCR product. The results indicate a linear dose response relationship between UV treatment and the decline in QPCR efficiency (Fig. 1D), similar to what has been obtained previously (Van Houten et al. 2000), and a statistically significant decrease in PCR product yield after 24-h culture in the presence of 5-Aza-dC (Fig. 1D).
Figure 1.
The quantitative long-distance PCR (QPCR) is a reliable and sensitive method for DNA damage detection. A computational model, describing expected yield of PCR as a function of the DNA damage density, predicts increased sensitivity of QPCR assay with increased length of analyzed template (A). Experimental testing of PCR yield and reproducibility demonstrated a linear decline of PCR efficiency with larger templates, whereas the variability of PCR product quantity increased exponentially (B). Quantification of a 10-kb PCR product allows reliable detection of 10% difference in the amount of loaded template (C). The QPCR allows detection of effects of DNA damaging factors of different nature in a dose-dependent manner (D). All experiments were done in three replicates, except UV irradiation, where three independent experiments were performed and representative results shown. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01; *** - p<0.001).
Increased DNA damage in liver, but not brain of aged mice
Both brain and liver show a broad range of degenerative changes during normal aging, some of which could in principle be caused by transcriptional interference due to accumulating lesions (Vijg 2007). Having demonstrated the efficacy of the QPCR method in our laboratory for detecting different types of DNA lesions, we analyzed DNA damage levels in liver and brain collected from young (4 month-old) and aged (28 month-old) C57BL/6 mice. We designed a panel of primer pairs targeting genes with different levels of expression and located on different chromosomes. For each targeted region we tested at least 4 primer pairs and selected the one consistently giving a single distinct band of the predicted size, as assessed by agarose gel electrophoresis (not shown). Based on the PicoGreen assay, identical amounts of input DNA were used for all samples. All samples were randomized, beginning with DNA isolation. For each QPCR run we included a sample containing 50% of template DNA as an internal control.
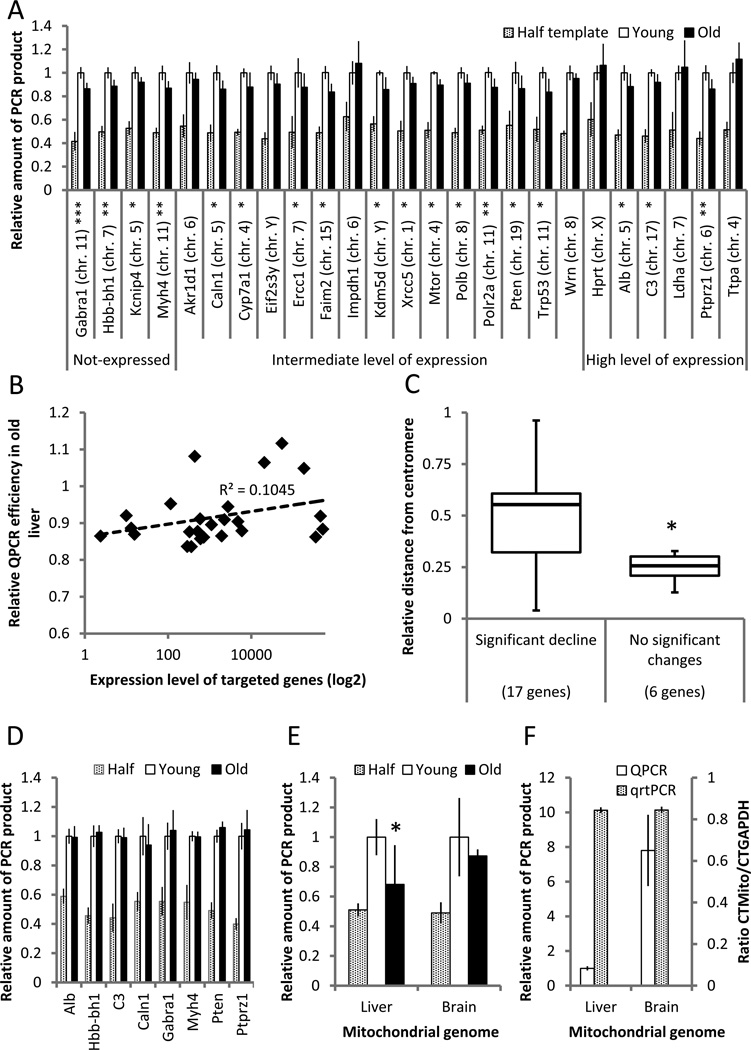
One of the advantages of the QPCR inhibition assay is that any gene (or region of DNA) that can be PCR-amplified can be studied. DNA repair rates are generally higher in actively transcribed genes as compared to silent genes (Bohr et al. 1985), which could lead to a relatively low rate of DNA damage accumulation. Alternatively, actively transcribed genes could be more susceptible to DNA damage accumulation because of their relatively open configuration. Thus, we decided to design QPCR assays for a range of genes differing in their level of expression from highly expressed to completely silent. The results for liver indicate a statistically significant reduction in QPCR efficacy in aged mice. A reduced QPCR efficacy was observed for the far majority of the tested nuclear genes (Fig. 2A). This average reduction in amplification efficiency in the old liver samples corresponds to about 33 lesions per million basepairs (Table 1). While there was a slight trend for highly expressed genes to accumulate less damage (Fig. 2B), this was not significant. We also tested for an effect of chromosomal position and while there was no obvious correlation between age-related decline in QPCR efficacy and chromosomal location of the tested gene, we did observe a statistically significant increase in age-related damage accumulation closer to the centromere of the mouse, acrocentric chromosomes (Fig. 2C).
Figure 2.
Analysis of DNA damage in liver and brain of old mice. (A) The majority of the nuclear genes tested demonstrated an age-related decrease in the amount of amplifiable template in liver of 28-month old animals as compared to 4-month old controls. (B) Relative QPCR efficiency, i.e., PCR yield in old vs. young mice did not correlate significantly with gene expression levels. Expression levels were obtained from the GEO database (www.ncbi.nlm.nih.gov/geo; dataset GDS3373). (C) QPCR efficiency is highest for genes closer to the centromere. On a scale of 0 to 1, 1 is the most distant position from the centromere. The loci are grouped based on observed age-related changes in QPCR efficiency of liver samples. (D) QPCR efficiency in brain nuclear DNA of 28-old mice versus 4-month old, young controls. (E) QPCR efficiency of a ~10kb mitochondrial fragment in liver and brain of old vs young, control mice. To compensate for copy number, the data shown are normalized to PCR yields of a short, 117 bp mtDNA fragment. (F) QPCR efficiency of the ~10 kb mtDNA fragment in liver and brain of young mice as compared to the 117-bp quantitative real-time PCR (qrtPCR) analysis yields. In all cases, the QPCR assay was performed at least 3 times for each analyzed target/sample; n=10 for young animals and n=8 for old animals. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01; *** - p<0.001).
Table 1.
Calculated number of DNA lesions in excess of those in control samples, i.e., from young mice. Data shown are averages ±SD.
| Studied group | Nuclear genome | Mitochondrial genome | ||||||
|---|---|---|---|---|---|---|---|---|
| PCR inhibiting lesions1 (Per MB of dsDNA) |
PCR inhibiting lesions after FPG (Per MB of dsDNA) |
PCR inhibiting lesions1 | PCR inhibiting lesions after FPG |
|||||
| Per MB of dsDNA2 |
Per 16.3 kb of dsDNA |
Per MB of dsDNA2 |
Per 16.3 dsDNA |
|||||
| C57Bl/6 | Y | Liver | 0 | 62.3±15.2 | 0 | 0 | 46.8±9.2 | 0.76±0.15 |
| Brain | 0 | 86.5±14.1 | 0 | 0 | 94.1±31.4 | 1.53±0.51 | ||
| O | Liver | 33.4±8.6 | 88.9±7.3 | 76.4±25.4 | 1.24±0.41 | 129.5±47.5 | 2.11±0.77 | |
| Brain | −8.2±2.9 | 89.2±19.4 | 26.0±8.6 | 0.44±0.14 | 97.4±13.8 | 1.59±0.22 | ||
| Wild type control |
Liver | 0 | 95.0±14.7 | 0 | 0 | 111.2±13.2 | 1.81±0.23 | |
| Brain | 0 | 79.1±11.9 | 0 | 0 | 61.7±7.2 | 1.01±0.12 | ||
| Ercc1−/Δ mutants |
Liver | −5.9±0.2 | 116.9±7.6 | −14.6±1.4 | −0.24±0.02 | 88.5±21.9 | 1.44±0.36 | |
| Brain | −0.9±0.0 | 83.9±13.6 | 6.3±0.2 | 0.10±0.00 | 72.1±5.3 | 1.18±0.09 | ||
| Ku80 mutants |
Liver | 0.7±0.0 | 120.1±16.8 | 111.6±65.8 | 1.82±1.07 | 209.0±46.1 | 3.42±0.75 | |
| Brain | −2.8±0.6 | 84.0±8.8 | −10.5±1.2 | −0.17±0.02 | 59.3±8.6 | 0.97±0.14 | ||
Control group is set to zero, all other samples are compared to this level of damage
To compare damage levels in nuclear and mtDNA
In the mouse brain we tested a much smaller number of genes, selected from those that demonstrated an age-related increase of DNA damage in liver. However, in contrast to the situation in liver, DNA samples obtained from aged mouse brain did not show a reduction in QPCR efficacy as compared to DNA from young brain (Fig. 2D).
Next, we analyzed mitochondrial DNA using the same approach, after correcting for copy number using an amplimer of 117 bp; amplification efficiency of such a small fragment is not dependent on DNA damage. For mitochondrial DNA very similar patterns of QPCR were observed as for nuclear DNA (Fig. 2E). However, the age-related increase in relative density of DNA lesions was higher in the mitochondrial genome – about 76 excess lesions per million basepairs (1.2 lesions per genome). Also, while not statistically significant, brain mtDNA showed an increase of DNA damage with age, i.e., with 26 excess lesions per million basepairs (0.4 lesions per genome) (Table 1). While QPCR of a mitochondrial target from DNA obtained from old liver was significantly less efficient than that from young liver, no significant differences were observed for brain. Of note, the QPCR assay also allowed us to compare the relative PCR yields from the same amount of template DNA from liver and brain. While PCR yields of nuclear genes were not different between brain and liver of young mice, the results indicated a substantially higher PCR yield from our mitochondrial DNA target in young brain (Fig. 2F, open bars). This is likely to indicate a smaller number of spontaneous DNA lesions in brain than in liver rather than fewer mitochondria in the latter, which was ruled out by quantitative real-time PCR of the 117-bp mtDNA fragment normalized to the 84-bp Gapdh fragment (Fig. 2F, grey bars).
No detectable increase of oxidative DNA lesions in old mice
A limitation of the QPCR assay is that DNA lesions that may not significantly stall progression of DNA polymerase may not be detected with high efficiency (Furda et al. 2012b). One such lesion is 8-oxo-2'-deoxyguanosine (8-oxo-dG), which is generally recognized as a marker for oxidative stress and likely to represent the most common oxidative DNA lesion (Cooke et al. 2003). To detect this lesion in the QPCR assay, we treated DNA with formamidopyrimidine glycosylase (FPG), a bacterial DNA repair enzyme that efficiently removes several types of oxidative damage, including 8-oxo-dG, from duplex DNA, leaving a single nucleotide gap. The phosphodiester bond at the abasic site is subsequently cleaved by the FPG-associated lyase activity (Tchou et al. 1994).
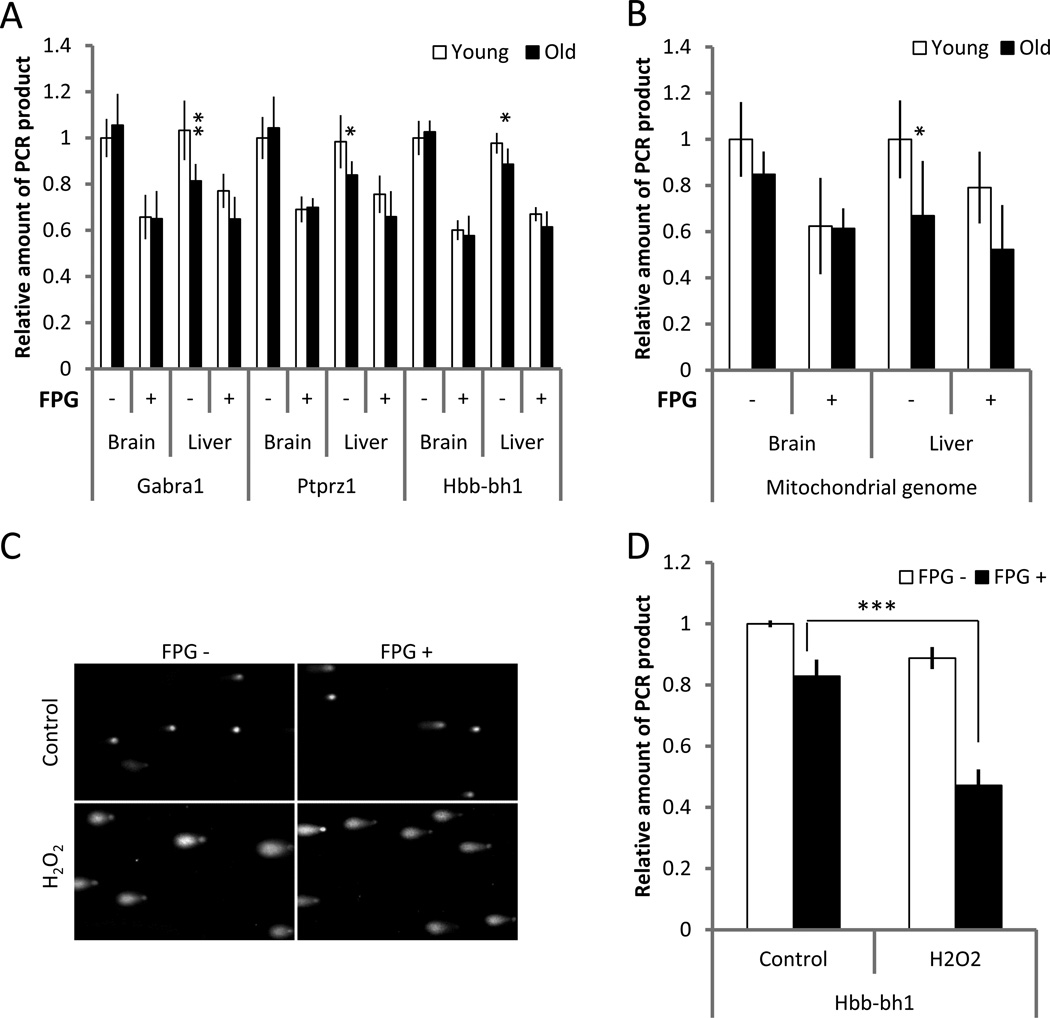
Fig. 3A shows the results of QPCR of brain and liver DNA from young and old mice treated with FPG. The reduced PCR efficacy in all cases indicates a substantial amount of oxidative lesions in both tissues, but without an effect of age. Of note, in the non-treated liver samples the reduced PCR efficacy on DNA from aged liver confirmed the significant increase in DNA damage with age in this organ (compare to Fig. 2A). Essentially the same picture emerged for mitochondrial DNA (Fig. 3B), which showed the same increase in DNA damage in aged liver as observed in the previous experiments (Fig. 2E), but no statistically significant increase in brain. While also for mitochondrial DNA FPG significantly reduced PCR efficacy in both organs, indicating increased oxidative DNA damage, there was no effect of age. To confirm that the FPG effect was specific for oxidative lesions and not simply a reflection of non-specific nuclease activity, we treated mouse embryonic fibroblasts with hydrogen peroxide (H2O2). For this purpose, the MEFs were cultured in serum-free medium supplemented with H2O2 (0.05 mM) for 15 min. Cells were trypsinized and DNA collected immediately after treatment. As an additional control we subjected some of these cells to a comet assay, a highly sensitive method to detect nuclear DNA breaks in the form of a comet tail (Liao et al. 2009). The comet assay was performed under alkaline conditions allowing detection of both single- and double-strand breaks. After embedding in agarose, cells were pretreated with FPG to create nicks at 8-oxo-dG sites. As expected the cells subjected to H2O2 demonstrated prominent comet tails (Fig. 3C). Treatment with FPG resulted in the appearance of longer tails, with comet heads substantially smaller (Fig. 3C), indicating a significant effect of FPG. Consistent with this result, the QPCR analysis showed a significant increase of FPG-sensitive sites in H2O2-treated MEFs, as demonstrated by a significant decline in yield of Hbb-bh1 gene amplification after treatment with FPG (Fig. 3D). Of note, while these results confirm the sensitivity of oxidative lesions to FPG, they do not rule out the possibility of non-specific cleavage of DNA by the FPG extract used. Hence, our measures of oxidative damage in liver and brain could be overestimates.
Figure 3.
Analysis of oxidative DNA damage in liver and brain of old mice. Although treatment with FPG led to decreased efficiency of QPCR in DNA samples obtained from both liver and brain, there is no evidence for higher amounts of oxidative damage in the old animals, neither in nuclear (A) nor mitochondrial (B) genome. To confirm specificity of this approach for detection of oxidative lesions, MEFs were subjected to H2O2 (0.05 mM, 15 min) and analyzed by alkaline comet assay (C) and QPCR targeted to the nuclear genome (D) with and without FPG pre-treatment. QPCR assay was performed at least 3 times for each analyzed target; n=10 for young animals and n=8 for old animals. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01).
No detectable increase of DNA damage in prematurely aged, DNA repair defective mice
Finally, we used QPCR to assess spontaneous DNA damage levels in liver and brain of mice harboring specific defects in DNA repair concurrent with multiple symptoms of premature aging. In such mice one would expect increased DNA damage underlying the premature aging symptoms. The two prematurely aging models included in this study were harboring defects in Ercc1 and Ku80, respectively. The Ercc1−/Δ mouse model carries a null mutation in one allele and a 7-amino acid truncation in the second allele. Its maximum life span is approximately 6 months and the animals show deficiencies in nucleotide excision repair, inter-strand crosslink repair and DNA double-strand break repair (Weeda et al. 1997). Ku80 mutant mice harbor two null alleles and are defective in the repair of DNA double-strand breaks through non-homologous end-joining (NHEJ). These animals have a maximum life span of 14 months (Vogel et al. 1999). Both mouse models were kept in a C57BL/6-FVB background. The 50% FVB background was necessary because C57BL/6 alone is embryonically lethal for the Ercc1−/Δ genotype and causes breeding problems with the Ku80 mice (not shown).
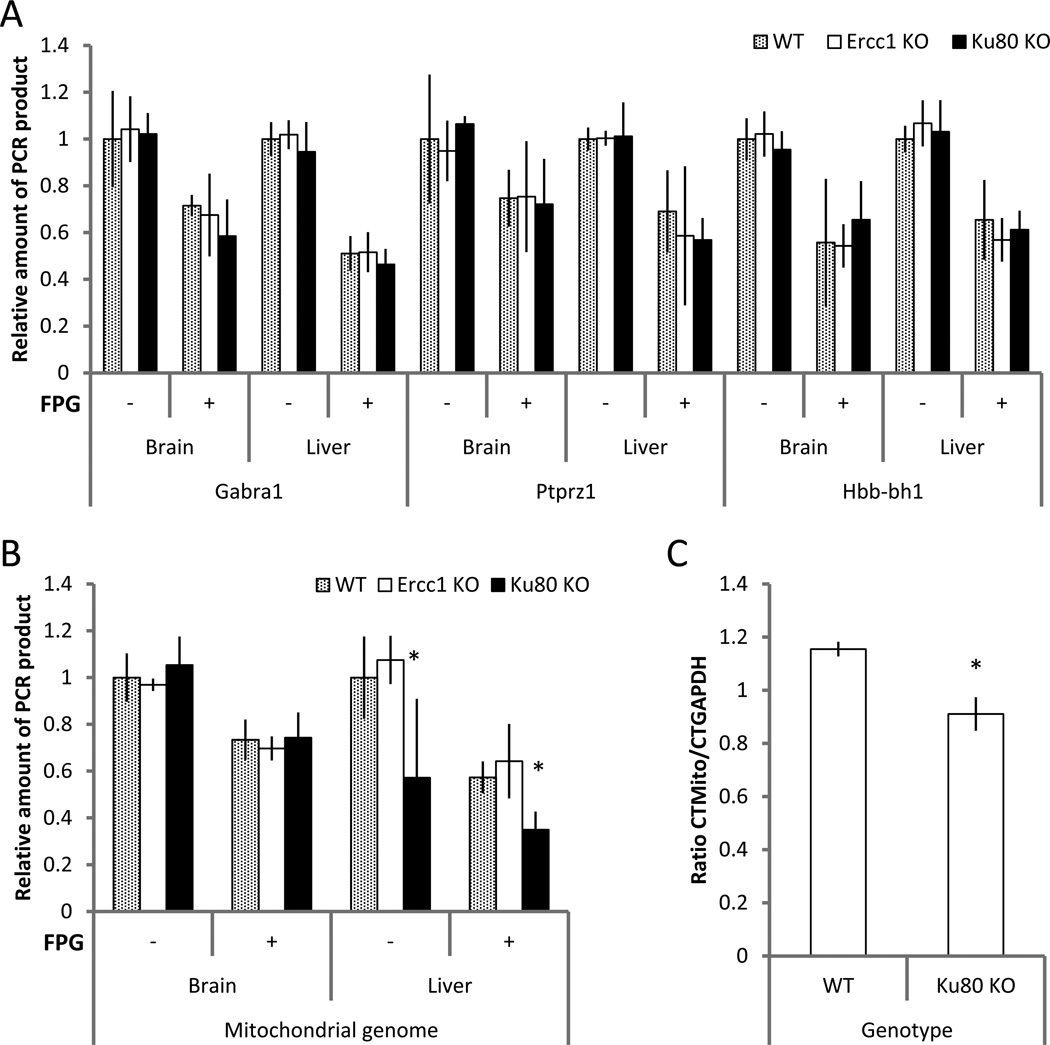
Fig. 4A compares QPCR results for the Ercc1 and Ku80 mutants at the end of their natural life span (i.e., about 5 and 12 months, respectively) as compared to ~3- and ~ 10-month old wild type control animals, respectively, in the same genetic background. The analyses include QPCR after FPG treatment. In the two DNA repair defective mouse models, QPCR efficacy, either with or without FPG treatment, was very similar for all 3 genotypes studied, indicating a lack of excess DNA damage within our limit of detection. The analysis of the long mitochondrial PCR target demonstrated decreased efficacy of QPCR for DNA obtained from the liver of Ku80 animals relative to wild-type and Ercc1−/Δ mutants (Fig. 4B). However, we found that the ratio of mitochondrial versus nuclear DNA in the liver of these mutant animals is shifted towards nuclear DNA (Fig. 4C). In this case this suggests that the observed decline in the efficacy of QPCR is not due to accumulation of DNA damage in the mitochondrial genome, but rather due to a decrease of mtDNA copy number in the liver of Ku80 mutants.
Figure 4.
Analysis of non-oxidative and oxidative DNA damage in liver and brain of mouse models of premature aging. QPCR efficacy of nuclear genome targets, with and without FPG treatment, was similar for DNA obtained from DNA-repair deficient mouse models of premature aging at terminal ages and DNA from young wild type animals (A). Although QPCR for the long mitochondrial genome target from the liver of Ku80 mutants was significantly less efficient than for DNA from either wild type or Ercc1−/Δ mutant animals (B), this difference is due to relative underrepresentation of mitochondrial DNA in the liver of Ku80 mutants, as demonstrated by real-time qPCR (C). QPCR assay was performed at least 3 times for each analyzed target; n=5 for each genotype. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (p<0.05).
Discussion
Our present results indicate that spontaneous DNA damage levels in the nuclear genome of mouse liver, but not brain are significantly higher in old animals, with an excess number of lesions of about 33 per Mb. The mitochondrial genome was found to accumulate age-related damage in liver at a higher extent, i.e., about 76 excess lesions per Mb (equivalent to about 1.2 lesions per mitochondrial genome). Although the level of DNA damage in brain mitochondria also had a tendency to increase with age, i.e., about 26 excess lesions per Mb (or about 0.4 lesions per mitochondrial genome), this difference was not statistically significant. These results are in reasonable agreement with previous work, both from our laboratory and others. Using a different method, i.e., alkaline elution, we previously reported a similar increase of spontaneous lesions in nuclear DNA of liver (Mullaart et al. 1988), but not brain (Mullaart et al. 1990) of aging rats. Furthermore, as we also showed, DNA mutations accumulate with age in liver but not in brain (Dollé et al. 1997). Of note, using the same QPCR method, Acevedo-Torres et al. (Acevedo-Torres et al. 2009) observed an age-related increase in PCR blocking lesions in both nuclear and mitochondrial DNA in the striatum of mouse brain, with more damage in the mitochondrial than in the nuclear DNA. While we also observed a trend towards an increase in mtDNA damage in the brain, there was no age effect on nuclear DNA. It is possible that this discrepancy is due to intra-organ specificity of damage accumulation. Indeed, we previously reported an age-related mutation accumulation in mouse hippocampus and hypothalamus, which was absent in whole brain analysis (as we did in this present study). In this present work we also show that DNA damage levels in two DNA repair-deficient mouse models are not elevated as compared to littermate controls or such an increase is below the detection level of the QPCR method.
In liver we analyzed a substantial number of genes spread across the genome. Somewhat surprisingly, we did not observe a correlation between DNA damage accumulation and the level of gene expression. One could expect that actively transcribed genes are less susceptible to the accumulation of DNA damage with age because of transcription-coupled repair. However, while there was a weak trend towards such a correlation (Fig. 2C) a reduction in damage accumulation as a function of the level of transcription was not significant. We did observe a correlation between damage accumulation and chromosomal position. That is, genes closer to the centromere tended to be less vulnerable to DNA damage accumulation (Fig. 2D). This was not due to a generally higher expression level of the genes close to the centromere (not shown). We have no explanation for this result, but it is possible that a generally more compact chromatin organization closer to the centromere, independent of gene expression level, has a protective effect on the induction of DNA damage.
QPCR to detect spontaneous DNA damage has two limitations. First, DNA lesions, such as 8-oxo-dG, that may not significantly stall progression of DNA polymerase, will not be detected at high sensitivity. While this is unlikely to explain the lack of an increase in DNA damage in the brain during normal aging, since there are many oxidative lesions that do stall DNA polymerase, it could readily explain the negative results for the Ercc1−/Δ and Ku80 mice. Indeed, the highly toxic inter-strand crosslinks and double-strand breaks may only need to be increased slightly in causing the premature aging effects. Since the limit of detection of the QPCR assay is approximately 1 lesion per 105 bases, it is conceivable that slight increases in these lesions would go undetected. Alternatively, it is possible that there is a significant DNA damage increase, but limited to specific genomic regions not covered by the genes analyzed. While some regional and sequence specificity for DNA damage is known, there is no evidence that most DNA damage is not introduced randomly along the genome.
Second, the QPCR assay does not detect specific lesions but rather infers their presence from PCR inhibition as compared to a reference sample, in this case DNA from the same organ taken from the young animal. Thus, whenever using the QPCR assay for mitochondrial DNA damage, care must be taken to normalize the long mitochondrial amplimer with a short non-damaged amplimer, which corrects for copy number differences. Our results indicate that Ku80 liver DNA loses copies of mtDNA over time and that the reduction in QPCR “efficiency” was simply due to a reduced ratio of mtDNA to nuclear DNA. While artifacts with assays measuring spontaneous DNA damage can never be completely ruled out, we believe we carefully calibrated the QPCR assay and cannot readily identify factors other than DNA damage to explain reduced QPCR efficiency in these experiments. At any rate, while PCR inhibiting factors in the aged genome other than DNA lesions could theoretically explain the reduced QPCR efficacy in aged liver, they cannot explain the negative results obtained for the premature aging mice or normal brain.
Since the QPCR method is based on relative efficiency of PCR amplification in different samples, it is impossible to measure absolute numbers of DNA lesions using this approach. However, by arbitrarily setting the number of lesions in the control sample at zero, it is possible to calculate the excess number of lesions in the sample analyzed (Table 1). In this way, while the QPCR assay was able to detect a significant age-related increase of DNA damage in liver (33 lesions/Mb of nuclear DNA and about 1.2 excess lesions per mitochondrial genome), there was no evidence that oxidative damage as monitored by FPG sensitive sites contributed to increased damage levels at old age in this organ. While the FPG data clearly indicate a high steady-state level of 8-oxo-dG in both liver and brain, which can be expected since both organs exhibit a high oxygen metabolism, there was no change with age. This is surprising since oxidative damage is often considered a major cause of aging (Beckman & Ames 1998). It is also in contrast with most published studies in which young and old tissues were compared (Moller et al. 2010). However, not all studies reported an age-related increase in oxidative DNA damage (Anson et al. 1999).
Our results also indicate the lack of an age-related increase in DNA damage in mice harboring defects in DNA crosslink and/or double-strand break repair. This was also true for FPG-susceptible lesions, i.e., 8-oxo-dG. Ercc1−/Δ mutant mice are also defective in nucleotide excision repair, which may play some role in the repair of 8-oxo-dG and other oxidative lesions (Reardon et al. 1997). Indeed, our data are in contrast to data indicating increased levels of 8-oxo-dG in liver of Ercc1 null mice (Selfridge et al. 2001). However, the about 2.5 8-oxo-dG lesions per 105 dG’s as compared to ~0.3 of such lesions per 105 dG’s in controls could very well be just below our detection limit. Alternatively, it is possible that this discrepancy is due to a difference between actual Ercc1 null mice, as used by Selfridge et al., and our Ercc1 model, which still has some remaining activity left in the form of one, truncated allele. Increased levels of 8,5’-cyclopurine-2’-deoxynucleosides, which comprise another category of endogenous oxidative lesions, were reported for the same Ercc1−/Δ mutant mice in liver, kidney and brain (Wang et al. 2012). The levels of these lesions, as detected by a sensitive combination of HPLC and mass spectrometry, were about two orders of magnitude lower than normal levels of 8-oxo-dG, which clearly brings them well below our detection limit. While one may wonder how likely it is that at such low, steady state levels DNA damage exerts adverse effects, the possibility of transcription interference and apoptosis signaling even by very rare lesions should not be ignored.
There is no doubt that both liver and brain are undergoing degenerative changes during normal aging, and similar changes occur prematurely and in a more severe fashion in the two DNA repair defective mice. Accelerated age-related cognitive decline and neurodegeneration has been observed in the Ercc1−/Δ mutant mice (Borgesius et al. 2011), while in Ku-deficient mice neuronal apoptosis has been demonstrated (Gu et al. 2000). The latter can provide some clue to the immediate causes of premature aging in the two DNA repair-deficient animals. Indeed, apart from a direct effect of accumulating DNA damage, which may simply be too low to detect with our current assay, cellular responses to these highly toxic types of DNA damage could lead to increased apoptosis and/or cellular senescence. Hepatocytes of Ercc1−/Δ mice show profound cellular senescence (Gregg et al. 2012) and so do embryonic fibroblasts isolated from these mice (Weeda et al. 1997). Moreover, in conditionally induced Ercc1 deficiency in murine hepatocytes in vitro increased apoptosis has been reported (Kirschner et al. 2007). For Ku80 mutant mice we have shown previously increased senescence in liver, based on the detection of an increase in persistent 53BP1 and gamma-H2AX DNA damage foci as compared to liver from littermate control animals (Busuttil et al. 2008). Also increased mutation frequency, both in the Ercc1 and Ku80 mutant mice, could contribute to the progeroid phenotype in these animals (Dollé et al. 2006; Busuttil et al. 2008).
Our present data underscore the high preservation during organismal life span of DNA repair efficacy in maintaining genome structural integrity up until old age. Even with major pathways affected, as is the case in the Ercc1 and Ku80 mutant mice, the consequences in terms of DNA damage accumulation are minor. Indeed, any adverse effects are more likely to be a consequence of the cellular responses to DNA damage rather than the damage per se. This is not surprising in view of the very large number of lesions induced in living cells continuously. Indeed, under physiological conditions DNA is highly unstable and without the emergence of DNA repair early in the evolution of cellular life it could never have become the universal carrier of genetic information. In view of the important role of highly proficient DNA repair pathways and their extensive overlap and excessive capacity, a build-up of DNA damage over time would be highly unlikely.
Experimental procedure
Cell culture and treatment
MEFs were obtained from D13.5 embryos of C57Bl/6 transgenic mice and maintained in 10% CO2 and 3% O2 atmosphere at 37°C in DMEM (GIBCO, CA, USA) supplemented with 10% FBS (GIBCO). The UV and hydrogen peroxide (H2O2) treatments were performed as described previously (Busuttil et al. 2007). In short for UV treatment the cells covered with a thin layer of PBS were exposed to different dosages of irradiation and collected immediately after exposure; for H2O2 treatment, cells were subjected to 0.05 mM H2O2 in serum-free medium and harvested after 15 min of exposure. For 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma, MO, USA) treatment, the medium with 5-Aza-dC was prepared at the time of application and applied for 24 hours.
Tissue samples
Samples of young (4-month old) and old (28-month old) brain and liver were obtained from the NIA Rodent and Tissue Bank (National Institute on Aging, Bethesda, MD, USA). Samples of brain and liver were collected from Ercc1−/Δ (5-month old) and Ku80 mutant mice (12-month old), both in a C57BL/6-FVB background and kept in the animal facilities of the RIVM in Bilthoven, The Netherlands. Thin coronal sections in the frontal area of the whole brain and random samples of liver tissue weighing 30–40 mg were collected for DNA extraction.
DNA isolation and normalization
Isolation of total (including mitochondrial) DNA from cultured cells and samples of tissue was performed with the Quick-gDNA™ MiniPrep (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer’s protocol. Tissue samples were first homogenized in 0.2 ml of PBS using a FastPrep-24 instrument and lysing matrix D tubes (MP Biomedicals, Solon, OH, USA). After DNA isolation, the quantity of DNA was measured with Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies, Grand Island, NY, USA) and adjusted with TE solution to have the concentration 50 ng/µl. The adjustment was repeated until the difference between most and less concentrated samples did not exceed 3%.
DNA template preparation
For detection of oxidative damage 100 ng of sample DNA (2 µl of stock solution) were incubated in 50 µl of reaction mixture containing 4 units of FPG enzyme (NEB, Ipswich, MA, USA) and 5 µl of NEBuffer 4 (NEB) at 37°C for 1 hour followed by 20 min incubation at 65°C. For non-oxidative damage detection sample DNA was prepared in similar way, but in the absence of FPG. Additionally, in parallel with FPG treatment, samples intended for analysis of the mitochondrial genome were cleaved with XhoI restriction endonuclease (NEB), which has a unique recognition side outside of the amplified region, which helps to relax supercoiled mtDNA, making it more suitable for PCR amplification.
Agarose gel electrophoresis under neutral and alkaline conditions
Electrophoresis was performed in 0.8% agarose (Bio-Rad, Hercules, CA, USA) gels prepared and run with standard 1x TBE buffer (Fisher Scientific, Pittsburgh, PA, USA). For alkaline agarose gel electrophoresis, samples were denaturated in 1x running buffer (50 mM NaOH, 1 mM EDTA) and loaded into the gel after mixing with 5x loading buffer (300 mM NaOH, 6 mM EDTA, 18% Ficoll 400, 0.125% bromophenol blue). Electrophoresis was performed in 0.8% agarose (Bio-Rad) gels in 1x running buffer at 3V/cm until bromophenol blue migrated approximately two thirds of the length of the gel. Gels were soaked with neutralization solution (1 M Tris-Cl, pH 7.6; 1.5 M NaCl) for 45 minutes and stained with ethidium bromide before analysis.
Quantitative real-time PCR
Quantitative real-time PCR was performed with Fast SYBR® Green Master Mix (Life Technologies) using the StepOnePlus Real-Time PCR System (Life Technologies). Primers used were: 5’-GGCTCCCTAGGCCCCTCCTG-3’, 5’-TCCCAACTCGGCCCCCAACA-3’ for GAPDH as a reference target and 5’-CCCAGCTACTACCATCATTCAAGT-3’, 5’-GATGGTTTGGGAGATTGGTTGATGT-3’ for mitochondrial DNA. The relative amount of mitochondrial DNA in DNA samples was determined as a ratio CTMito/CTGAPDH.
Quantitative long-distance PCR and PCR yield quantification
Long-distance PCR was performed with the Takara LA PCR Kit (Takara Bio Inc., Otsu, Shiga, Japan) according to the manufacturer's protocol. Reaction conditions were optimized to provide a linear increase of the product quantity in response to increase of template amount and were as follows: 3 min pre-run denaturation at 94°C, followed by 30 (for nuclear targets) or 20 (for mitochondrial targets) cycles including 30 sec denaturation at 94°C and 9 min extension at 68°C. The primers used were:
| Targeted gene |
Primer 1 | Primer 2 | Product size (bp) |
|---|---|---|---|
| Akr1d1 | GCATTTCCAACGGCTGCCTG | GCAGGAACCTCTAGCCCTGG | 9934 |
| Alb | GCTGGTTGGGAGAGCCACTT | AGAGCAGAGAAGCATGGCCG | 9931 |
| C3 | GGCGATGAGGTTGGCAGCTA | TCCCACCTCTTAGTGCCCCT | 9762 |
| Caln1 | CATCCCCCTCTTCCCAGCAG | GGGAAATGCCTGCCCCCATA | 9798 |
| Cyp7a1 | CCCTGCCTGGGGACTCATTC | GCCAGAGGTCAGGGTCACAG | 9958 |
| Eif2s3y | TGGGGCAGCAGTGATGGATG | AGTGCAGGAGGCAGAAGAGG | 10263 |
| Ercc1 | AACTCAAAGCCCCCGAGTGG | GCTGGGGAGAGAGACAGCAC | 9984 |
| Faim2 | ACTGAGAACCCCGGGAGGAT | TCCTGGGGGACCAGCTAAGT | 10385 |
| Gabra1 | CACGCTTTTGCCATCCCACG | CTCTGCCCTCAGCTTTGCCT | 10350 |
| Hbb-bh1 | CAACCTGTGTCAGAAGCAGATGCA | ACTTCAAGGTCAGTTCTGAGCATGCT | 8943 |
| Hprt | GTGAGCCAAGGGGACTCCAG | ATGCCCAGAGCCCAGCATAC | 9550 |
| Impdh1 | ACACAATTGTGACCCCAAAGTGTGC | GGATTCCAGCCTTTCTGCTCCTCTA | 8903 |
| Kcnip4 | AGAGCTGCATGGAGCGCATT | CCACCTCCCAATGGGTTGCT | 10099 |
| Kdm5d | CAGGGCATCACGGAAGCTGA | TCCACACGAAGCTGCTGACC | 10225 |
| Xrcc5 | TGCCTAAGGAGGCAGGGAGA | CCCAGAGCCACTCAGTCACC | 9806 |
| Ldha | TAGGGCTCTGGGTGATGGGA | CTGGTTGGCTCCGCTCTCTC | 9675 |
| Mtor | ATGAGTGTAGGGGTTGCCGC | CTGTCACAGCAGTGCGGAGT | 10244 |
| Myh4 | CTACAGGGCATGCAGCCACT | TTCATGAAGGCGCGGACGTT | 8926 |
| Trp53 | CTTAGGGGCCCGTGTTGGTT | CCAGTGGAGGAGCACCTGTC | 10195 |
| Polb | CACTCACCAGTGACCCCCAG | CCTGCTGACCCACCCAAACT | 9641 |
| Polr2a | AGGGGAGTAACCTGGGCTGA | GTGTGTGGCAGGGGAGAGAG | 9553 |
| Pten | AGGGGAGGGAAGAACGGAGT | TGGGTAGCACTCACACACGC | 10246 |
| Ptprz1 | ACCGAAGTGACACCACAGGC | GCACACCTCCCTACCTGCTC | 9882 |
| Ttpa | ACAAGGAGGCCAGCAGGTTC | CTCCACACAGCCTGACCCTC | 9791 |
| Wrn | AAGGCCACCCTGAGACATGC | AGAAGGAAGGGGTGGGGACT | 10001 |
| Mito Long | GCCAGCCTGACCCATAGCCATAATAT | GAGAGATTTTATGGGTGTAATGCGG | 10090 |
| Mito Short | CCCAGCTACTACCATCATTCAAGT | GATGGTTTGGGAGATTGGTTGATGT | 117 |
The quantification of PCR product was performed with the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies) according to the manufacturer's protocol. In case of mitochondrial targets the data were normalized before analysis by calculating a ratio between the amounts of long and short product to compensate for possible differences in the ratio of mitochondrial versus nuclear DNA in the samples.
Comet assay under alkaline conditions
Direct visualization of DNA damage in MEFs was performed with the CometAssay Kit (R & D Systems, Minneapolis, MN, USA) according to the manufacturer's protocol. Briefly, cells were embedded in low–melting point agarose and lysed and treated with alkaline unwinding solution (pH>13). After electrophoresis in alkaline electrophoresis solution, comets were visualized by staining with SYBR Green.
Statistical analysis and in silico modeling
The p-values were calculated using a two-sample t-test (Microsoft Excel software package). Each experiment was repeated a minimum of 3 times; representative results are shown. For in silico modeling we assumed that there could not be more DNA lesions than 1 per 1 kb and used the formula Y=(1-0.002*L)S, where Y is the expected amount of PCR product, L the number of lesions per 1 Mb of dsDNA, and S the size of the amplified fragment in kb. The frequency of DNA lesions was calculated using the formula λ=-ln(AD/AO), where λ is the lesion frequency per amplified fragment size, AO is the amount of amplified product from the control sample, AD the amount of amplified product from analyzed sample (Van Houten et al. 2000).
Supplementary Material
Supplementary Figure S1. Quality analysis of DNA samples obtained from different organs of animals of different age. Representative examples of electrophoresis in agarose gels performed under neutral (A) and alkaline (B) conditions are shown.
Acknowledgments
This research was supported by NIH grant AG 17242 (JV), PA CURE (BVH).
Footnotes
Author Contribution
Conceived and designed the experiments: AYM, BVH and JV. Performed the experiments: AYM, SG, MW, RRW and WQ. Provided tissues: METD, ER, HvS, PH and JHJH. Analyzed the data: AYM, WQ, BVH, METD, HvS, PH, JHJH and JV. Wrote the paper: JV, AYM and BVH.
References
- Acevedo-Torres K, Berrios L, Rosario N, Dufault V, Skatchkov S, Eaton MJ, Torres-Ramos CA, Ayala-Torres S. Mitochondrial DNA damage is a hallmark of chemically induced and the R6/2 transgenic model of Huntington's disease. DNA Repair (Amst) 2009;8:126–136. doi: 10.1016/j.dnarep.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson RM, Senturker S, Dizdaroglu M, Bohr VA. Measurement of oxidatively induced base lesions in liver from Wistar rats of different ages. Free Radic Biol Med. 1999;27:456–462. doi: 10.1016/s0891-5849(99)00091-x. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Borgesius NZ, de Waard MC, van der Pluijm I, Omrani A, Zondag GC, van der Horst GT, Melton DW, Hoeijmakers JH, Jaarsma D, Elgersma Y. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J Neurosci. 2011;31:12543–12553. doi: 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Garcia AM, Reddick RL, Dolle ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS ONE. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Munoz DP, Garcia AM, Rodier F, Kim WH, Suh Y, Hasty P, Campisi J, Vijg J. Effect of Ku80 deficiency on mutation frequencies and spectra at a LacZ reporter locus in mouse tissues and cells. PLoS ONE. 2008;3:e3458. doi: 10.1371/journal.pone.0003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Cadet J, Epe B, Gedik C. Problems in the measurement of 8-oxoguanine in human DNA. Report of a workshop, DNA oxidation, held in Aberdeen, UK, 19–21 January, 1997. Carcinogenesis. 1997;18:1833–1836. doi: 10.1093/carcin/18.9.1833. [DOI] [PubMed] [Google Scholar]
- Collins AR, Cadet J, Moller L, Poulsen HE, Vina J. Are we sure we know how to measure 8-oxo-7,8-dihydroguanine in DNA from human cells? Arch Biochem Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, Niedernhofer LJ, van der Horst G, Hoeijmakers JH, van Steeg H, Vijg J. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Dollé ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- Furda AM, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol Biol. 2012a;920:111–132. doi: 10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furda AM, Marrangoni AM, Lokshin A, Van Houten B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair (Amst) 2012b;11:684–692. doi: 10.1016/j.dnarep.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg SQ, Gutierrez V, Robinson AR, Woodell T, Nakao A, Ross MA, Michalopoulos GK, Rigatti L, Rothermel CE, Kamileri I, Garinis GA, Stolz DB, Niedernhofer LJ. A mouse model of accelerated liver aging caused by a defect in DNA repair. Hepatology. 2012;55:609–621. doi: 10.1002/hep.24713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt FW. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci U S A. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- Kirschner K, Singh R, Prost S, Melton DW. Characterisation of Ercc1 deficiency in the liver and in conditional Ercc1-deficient primary hepatocytes in vitro. DNA Repair (Amst) 2007;6:304–316. doi: 10.1016/j.dnarep.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Liao W, McNutt MA, Zhu WG. The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods. 2009;48:46–53. doi: 10.1016/j.ymeth.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Maccormick RE. Possible acceleration of aging by adjuvant chemotherapy: a cause of early onset frailty? Medical hypotheses. 2006;67:212–215. doi: 10.1016/j.mehy.2006.01.045. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Lee M, Gundry M, Gravina S, Strogonova N, Tazearslan C, Bendebury A, Suh Y, Vijg J. 5-Aza-2'-deoxycytidine-induced genome rearrangements are mediated by DNMT1. Oncogene. 2012 doi: 10.1038/onc.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller P, Lohr M, Folkmann JK, Mikkelsen L, Loft S. Aging and oxidatively damaged nuclear DNA in animal organs. Free Radic Biol Med. 2010;48:1275–1285. doi: 10.1016/j.freeradbiomed.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Mullaart E, Boerrigter ME, Boer GJ, Vijg J. Spontaneous DNA breaks in the rat brain during development and aging. Mutat Res. 1990;237:9–15. doi: 10.1016/0921-8734(90)90027-o. [DOI] [PubMed] [Google Scholar]
- Mullaart E, Boerrigter ME, Brouwer A, Berends F, Vijg J. Age-dependent accumulation of alkali-labile sites in DNA of post-mitotic but not in that of mitotic rat liver cells. Mech Ageing Dev. 1988;45:41–49. doi: 10.1016/0047-6374(88)90017-6. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Selfridge J, Hsia KT, Redhead NJ, Melton DW. Correction of liver dysfunction in DNA repair-deficient mice with an ERCC1 transgene. Nucleic Acids Res. 2001;29:4541–4550. doi: 10.1093/nar/29.22.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- Van Houten B, Cheng S, Chen Y. Measuring gene-specific nucleotide excision repair in human cells using quantitative amplification of long targets from nanogram quantities of DNA. Mutat Res. 2000;460:81–94. doi: 10.1016/s0921-8777(00)00018-5. [DOI] [PubMed] [Google Scholar]
- Vijg J. Aging of the Genome. Oxford: Oxford University Press; 2007. [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci U S A. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Clauson CL, Robbins PD, Niedernhofer LJ, Wang Y. The oxidative DNA lesions 8,5'-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell. 2012;11:714–716. doi: 10.1111/j.1474-9726.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D, Hoeijmakers JH. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Quality analysis of DNA samples obtained from different organs of animals of different age. Representative examples of electrophoresis in agarose gels performed under neutral (A) and alkaline (B) conditions are shown.