Abstract
Viral fusogens mediate the merger of the viral envelope and cellular membrane during viral entry. These proteins share little sequence similarity but all are thought to act by refolding through a series of conformational intermediates from the metastable prefusion form to the stable postfusion form. Crystal structures of both prefusion and postfusion forms have illuminated the conformational pathways of several viral fusogens. By contrast, only the structure of the postfusion form is available for glycoprotein B (gB), the conserved fusogen of herpesviruses. To gain insight into the nature of the fusogenic conformational changes in gB, we used several approaches aimed at engineering the prefusion form of the HSV-1 gB ectodomain, including modifications intended to stabilize the prefusion form and novel mutations aimed at destabilizing the postfusion form. We found that the postfusion conformation of gB is remarkably stable and resistant to perturbations. Several mutations successfully destabilized the gB trimer, identifying regions that are critical for the stability of the postfusion form. Yet, none of the constructs adopted the prefusion conformation. We propose that the soluble ectodomain of gB folds into the postfusion form without first adopting the prefusion intermediate. These results suggest that other regions of gB, including the transmembrane region and the cytoplasmic domain, may be necessary to establish and maintain the metastable prefusion conformation.
Keywords: membrane fusion, viral fusogen, structure, protein engineering
INTRODUCTION
Enveloped viruses enter host cells by fusing their envelopes with the cellular plasma membrane or the membrane of an endocytic vesicle. This process is initiated by binding of a virus to its cellular receptor and is catalyzed by a viral fusogen 1. In most enveloped viruses, the receptor binding and the fusogenic functions are carried out by two different subunits of a single glycoprotein. Conformational changes in the receptor-binding subunit upon receptor interaction are thought to trigger fusogenic conformational changes in the fusion subunit. In some viruses, such as paramyxoviruses, these two functions are distributed between two viral proteins 2
Herpesviruses are double-stranded-DNA, enveloped viruses that cause lifelong, latent infections and a variety of diseases, including skin lesions, encephalitis, cancers, and disseminated disease in the immunocompromised and neonates. Like all enveloped viruses, herpesviruses penetrate cells by fusing their envelopes with a host cell membrane: either the plasma membrane or an endosomal membrane 3. But, the entry machinery of herpesviruses is more complex than that of most other viruses and consists of three conserved viral proteins: gB, gH, and gL, plus additional non-conserved proteins. In herpesviruses, the receptor-binding and the fusogenic functions are distributed among multiple proteins 4.
The mechanism of herpesvirus cell entry is perhaps best understood for the prototypical Herpes Simplex viruses Type 1 and 2 (HSV-1 and HSV-2). Binding of the receptor-binding protein gD to its cellular receptors nectin-1, herpesvirus entry mediator (HVEM), or modified heparan sulfate 5; 6; 7 is thought to trigger the conserved membrane fusion machinery composed of gB and gH/gL. Receptor-bound gD probably interacts with and activates gH/gL 8; 9, which in turn is thought to interact with and activate gB 10; 11. gB is class III viral fusogen 12; its postfusion structure shares structural similarity with the postfusion forms of vesicular stomatitis virus (VSV) glycoprotein G 13 and baculovirus gp64 14 despite lack of any sequence similarity.
Viral fusogens mediate the merger of the viral envelope and the host membrane by refolding through a series of conformational intermediates from the initial prefusion form to the final postfusion form 15. This conformational pathway has been mapped out for several viral fusogens from all three known classes, including influenza hemagglutinin (class I) 16, Dengue E (class II) 17, and VSV G (class III) 18. Crystal structures of both prefusion and postfusion forms of these and other viral fusogens have been invaluable in illuminating their membrane fusion mechanisms. Despite different architectures and lack of any sequence similarity, viral fusogens undergo fundamentally similar conformational changes, refolding from the metastable prefusion conformation into the stable postfusion conformation that resembles a trimeric hairpin 15.
Understanding the structural basis for the prefusion-to-postfusion transition in gB and how it is triggered is one of the major challenges in the field of herpesvirus entry 3. The crystal structure of the recombinant HSV-1 gB ectodomain 12 probably represents the postfusion form because it more closely resembles the extended postfusion and not the more compact prefusion structure of VSV G 13; 18. But, very little is known about the structure of the prefusion form of gB, and, given that gB requires several additional proteins for function, it is unclear to what extent the fusogenic conformational changes in gB resemble those in other viral fusogens.
Attempts to characterize the prefusion form have been complicated by spontaneous refolding of the recombinantly expressed gB ectodomain into its postfusion conformation 12; 19; 20. Here, we have taken several approaches to engineer the prefusion form of HSV-1 gB. First, we have made modifications to stabilize the prefusion form of gB, similar to those successfully used to obtain the prefusion forms of several other viral fusogens 21; 22; 23; 24; 25; 26. In a novel approach, we also introduced a number of mutations designed to destabilize the postfusion form of gB. A large number of the resulting gB ectodomain constructs were characterized for antibody reactivity and the overall conformation. We found that the postfusion form of gB is very stable and resistant to structural perturbations. The remarkable stability of the postfusion form of gB probably provides the driving force for membrane fusion.
Mutations that successfully destabilized the gB trimer pinpoint domain dV as critical for the stability of the postfusion form. Yet even these mutants adopted the postfusion conformation instead of the prefusion conformation, supporting the idea that the soluble ectodomain of gB folds into the postfusion form directly without passing through a prefusion intermediate. We postulate that other regions of gB, including the transmembrane region and the cytodomain, may influence the folding energetics and are thus required for the stability of its prefusion conformation.
RESULTS
Mutant design
The recombinant gB ectodomain (residues 31–730) adopted the postfusion conformation when expressed in insect cells 12, implying that the initially formed prefusion form spontaneously refolded into the more stable postfusion form. To prevent or reduce spontaneous refolding of the gB ectodomain into the postfusion form, we used two orthogonal strategies: stabilization of the prefusion conformation, and destabilization of the postfusion conformation.
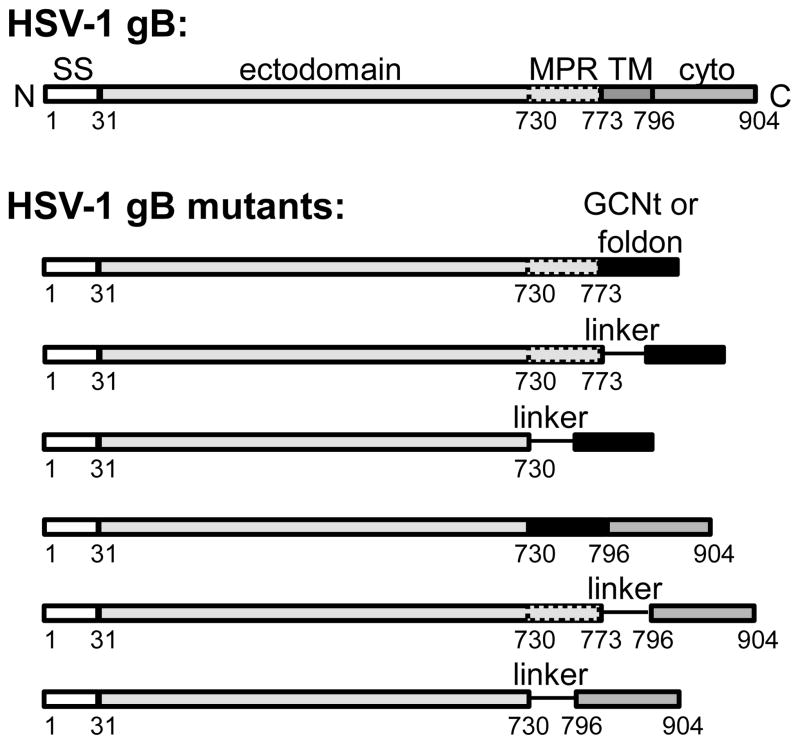
The prefusion forms of several viral fusogens have been successfully stabilized by the addition of C-terminal trimerization tags to the ectodomains 21; 22; 23; 24; 25; 26. To stabilize the prefusion form of HSV-1 gB, we used three trimerization tags: the GCN4-derived trimerization tag, GCNt 27; 28; the “foldon”, a C-terminal trimerization domain of T4 fibritin 29; 30; or the cytoplasmic domain (cytodomain) of gB itself 31. GCN4 is a transcription factor that normally forms dimers; however, the GCN4-pII mutant forms stable trimers 27; 28 and thus has been used as a trimerization tag (GCNt). The 27-amino-acid GCNt tag was successfully used to stabilize the soluble ectodomain of trimeric paramyxovirus F protein in its prefusion conformation 21. Foldon is a trimeric beta-propeller that can promote trimerization of heterologous proteins; it was previously used to stabilize the prefusion forms of HIV Env 24; 25; 26, influenza HA 22, and rabies viral fusion proteins 23. The gB cytodomain is trimeric 31 and is, in essence, a native trimerization tag. The GCNt tag, the foldon tag, or the cytodomain were placed following the last residue of the gB ectodomain, residue 730 (Fig. 1, Table 1). The length of the linker connecting gB ectodomain and tag sequences was varied. Some constructs also included the hydrophobic membrane-proximal region (MPR), residues 731–773 (Fig. 1, Table 1). In two constructs, both MPR and TM were replaced with GCNt but the cytodomain was preserved (Table 1).
Fig. 1.
Schematic view of the sequence of the full-length HSV-1 gB and constructs designed to stabilize the prefusion conformation. SS – signal sequence, MP – membrane-proximal region, TM – transmembrane region, Cyto – cytoplasmic domain are shown as rectangles of different shades of gray. GCNt (sequence: QIEDKIEEILSKIYHIENEIARIKKLIGE) and foldon (sequence: GYIPEAPRDGQAYVRKDGEWVLLSTF) are shown as black rectangles. Linkers GSGS, GSGTGS, or GGSGGTGGSG are shown as lines.
Table 1. Summary of mutants’ properties.
All gB constructs were expressed in Sf9 cells infected with recombinant baculoviruses. Expression was assessed by Western blots of infected supernatants using an anti-gB pAb R68. Trimerization was assessed by detection of trimeric form of protein by C226 or DL16 antibodies using P1 refeed samples prepared under mildly denaturing conditions (0.1% SDS, no reducing agent, no boiling). Xa - factor Xa protease cleavage site, His6 - hexahistidine tag, Strep - streptag.
| Construct | Expression | Trimer | C226/DL16 |
|---|---|---|---|
| Mutants to stabilize the prefusion conformation | |||
| gB773-foldon | + | + | +/+ |
| gB773-foldon-Xa-His6 | + | + | +/+ |
| gB773-GSGTGS-foldon-Xa-His6 | + | + | +/+ |
| gB773-GGSGGTGGSG-foldon | + | + | +/+ |
| gB773-GGSGGTGGSG-foldon-Xa-His6 | + | + | +/+ |
| gB773-foldon-strep | + | + | +/+ |
| gB773-GSGTGS-foldon-strep | + | + | +/+ |
| gB773-GGSGGTGGSG-foldon-strep | + | + | +/+ |
| gB773-GCNt | + | + | +/+ |
| gB773-GSGTGS-GCNt-Xa-His6 | + | + | +/+ |
| gB773-GGSGGTGGSG-GCNt-Xa-His6 | + | + | +/+ |
| gB773-GSGTGS-cyto(796-904)-His6 | + | + | +/+ |
| gB773-GGSGGTGGSG-cyto(796-904)-His6 | + | + | +/+ |
| gB730-GSGS-foldon | + | + | +/+ |
| gB730-GSGTGS-foldon-Xa-His6 | + | + | +/+ |
| gB730-GGSGGTGGSG-foldon-Xa-His6 | + | + | +/+ |
| gB730-GSGTGS-GCNt-Xa-His6 | + | + | +/+ |
| gB730-GGSGGTGGSG-GCNt-Xa-His6 | + | + | +/+ |
| gB730-GCNt-cyto(821-904) | + | + | +/+ |
| gB730-GCNt-cyto(796-904) | + | + | +/+ |
| gB730-GSGTGS-cyto(796-904) | + | + | +/+ |
| gB730-GSGTGS-cyto(796-904)-His6 | + | + | +/+ |
| gB730-GGSGGTGGSG-cyto(796-904)-His6 | + | + | +/+ |
| Mutants to destabilize the postfusion conformation | |||
| E502A | + | + | +/+ |
| R505A | + | + | +/+ |
| Q507A | + | + | +/+ |
| N511A | + | + | +/+ |
| I391P | + | + | +/+ |
| T394P | + | − | +/− |
| A504P/R505G/Q507G/N511G (Quad) | + | + | +/+ |
| A504P/R505G/L506F/Q507G/T509F/N511G (Quad-FF) | + | + | +/+ |
| A504P/R505G/L506W/Q507G/T509W/N511G (Quad-WW) | + | + | +/− |
| L506F | + | + | +/+ |
| T509F | + | + | +/+ |
| I513F | + | + | +/+ |
| L506K | + | + | +/+ |
| R515P | + | + | +/+ |
| L700G/L711G | + | + | +/− |
| gB670 | + | − | +/− |
| gB678 | + | + | +/− |
| gB683 | + | + | +/− |
| gB689 | + | + | +/− |
| gB700 | + | + | +/− |
| gB684-GGSGGTGGSG-GCNt-Xa-His6 | − | N/D | N/D |
| gB690-GGSGGTGGSG-GCNt-Xa-His6 | − | N/D | N/D |
| gB701-GGSGGTGGSG-GCNt-Xa-His6 | − | N/D | N/D |
The destabilizing mutations were aimed at disfavoring the formation of the postfusion conformation so that the ectodomain would retain the prefusion conformation. One set of destabilizing mutations targeted the N terminus of helix αC in domain III (dIII) of gB (Fig. 2A) and was based on the differences between the prefusion and the postfusion structures of VSV G 13; 18, another class III viral fusogen. Although VSV G undergoes large conformational changes between the prefusion and the postfusion forms, these primarily involve whole domain movements with very few changes in secondary structure. The most significant secondary structure changes occur within dIII of VSV G (HSV-1 gB domain numbering scheme) where residues 263–275, mostly unfolded in the prefusion structure, become the N terminus of the central helix in the postfusion structure. This helix is stabilized by interactions with its counterparts within the VSV G trimer as well as with residues in domain dII (Fig. 2A). The postfusion structure of the gB ectodomain also contains a central helix, αC, in dIII (Fig. 2A). We hypothesized that, by analogy with VSV G, the N terminus of helix αC is unfolded in the prefusion form. To destabilize the helical conformation of the N terminus of the helix αC, we designed mutations to destabilize the helix itself (R515P), to disrupt its hydrogen bonds with residues in dII and dV (E502A, R505A, Q507A, N511A, I391P, T394P), or both (A504P/R505G/Q507G/N511G, henceforth referred to as Quad) (Fig. 2B, Table 1). We also designed mutations to disrupt side chain packing at the trimeric coiled coil interface by introducing residues with bulky side chains such as phenylalanine or tryptophan, or both bulky and charged lysine (L506F, L506K, T509F, and I513F) (Fig. 2C). Finally, two mutants combined mutations disrupting hydrogen bonds with mutations disrupting helical packing, A504P/R505G/L506F/Q507G/T509F/N511G (Quad-FF) and A504P/R505G/L506W/Q507G/T509W/N511G (Quad-WW).
Fig. 2.
Regions targeted for mutagenesis. (A–C) Central coiled coil region as a target for mutagenesis. (A) The N terminus of the central alpha helix that undergoes refolding in VSV G is shown in magenta in the crystal structures of the prefusion and postfusion forms of the trimeric ectodomain 13; 18. The analogous region in the crystal structure of the postfusion form of HSV-1 gB ectodomain 12 is also shown in magenta. Domains in VSV G are labeled using gB domain numbering scheme, for clarity. Insets show close-up views of this region within single protomers. (B) A close-up view of the hydrogen bonds between the N-terminus of the central -helix αC in domain dIII (magenta) and domains dII and dV. Residues involved in hydrogen bonds are shown as sticks and labeled. Hydrogen bonds are shown as dashed lines. (C) Three symmetry-related C helices within the central coiled coil are shown in a top-to-bottom view. Residues L506, T509, and I513 at the coiled-coil interface are shown as sticks in all three protomers but labeled only in one protomer. (D–F) Domain dV as a target for mutagenesis. (D) A C-terminal helix in the ectodomain of paramyxovirus F protein, analogous to domain dV in HSV-1 gB, is shown in orange in the prefusion structure of PIV5 F and the postfusion structure of hPIV3 F 21; 32. Trimers are shown side by side with single protomers. (E) Domain dV in HSV-1 gB is shown in orange in the postfusion structure. The prefusion model of HSV-1 gB, including the hypothetical location of domain V, is shown schematically. Trimers are shown side by side with single protomers. dV truncations would occur within the orange stalk. (F) Truncation of dV (top) was expected to destabilize the postfusion form of gB with a minimal effect on the stability of the prefusion form of gB while the replacement of the dV with GCNt was expected to substitute the native helical stalk with an engineered one (bottom).
The other set of destabilizing mutations focused on domain dV of gB. This domain extends through almost the entire length of the molecule, making multiple interactions with dI, dII, and dIII, and is likely important for the stability of the postfusion conformation (Fig. 2E). In paramyxovirus fusion protein F, the postfusion structure is stabilized by a similarly extended domain 32 that in the prefusion structure forms an -helical stalk protruding away from the globular core 21 (Fig. 2D). We hypothesized that if dV forms α-helical protrusion in the prefusion form of gB (Fig. 2E), its modification or truncation might not affect the prefusion form but should destabilize the postfusion form. To test this, dV was either truncated (mutants gB670, gB678, gB683, gB689, gB700) or replaced with the GCNt sequence (mutants gB684- L10- GCNt-FactorXa-His6, gB690-L10-GCNt-FactorXa-His6, and gB701- L10-GCNt-FactorXa-His6, where L10 stands for linker GGSGGTGGSG) (Fig. 2F). We expected that trimeric GCNt would form a trimeric coiled-coil stalk in the prefusion form, replacing the deleted region of dV. Finally, we made a double mutation L700G/L711G to destabilize helix αF in dV in the postfusion structure, expecting a lesser destabilizing effect, if any, on the prefusion conformation.
The majority of gB mutant ectodomains are secreted
All gB constructs were expressed in Sf9 cells infected with recombinant baculoviruses. All proteins were secreted into culture media except gB684-L10-GCNt-Xa-His6, gB690-L10-GCNt-Xa-His6, and gB701-L10-GCNt-Xa-His6, suggesting that they were misfolded (Fig. 3, Table 1). These mutants were not pursued further.
Fig. 3.

Most constructs are secreted from insect cells and are not misfolded. Proteins secreted into the supernatant (A) or remaining inside the cells (B) were detected by Western blot with anti-gB polyclonal antibody R68. Under nonreducing denaturing conditions, gB migrates as a monomer (M). Positions of molecular weight standards are labeled. Lanes are labeled at the top. gB730-GCNt-cyto lane corresponds to construct gB730-GCNt-cyto(821–904).
Several mutations destabilize the trimer and abolish DL16 reactivity
To test if any of the mutants adopted the prefusion conformation, we performed using SDS-PAGE under mildly denaturing conditions and Western blot analysis with two anti-gB monoclonal antibodies, DL16 and C22633. Although a panel of neutralizing monoclonal antibodies with known epitopes, many of which are conformational, is available 33, all of these antibodies also bind the postfusion ectodomain 33, which is not surprising considering that their epitopes are exposed on the surface of the postfusion form 12. Thus none of these antibodies can be expected to reliably distinguish the prefusion form from the postfusion form. By contrast, the non-neutralizing mAb DL16 does not only recognize a conformational epitope but also requires the intact postfusion trimer: DL16 does not bind monomeric gB. Moreover, the DL16 epitope, which encompasses domains dI and dV, is very sensitive to disruptions 19; 33. The prefusion form of gB is expected to differ substantially from its postfusion form, considering that the known structures of the two forms in other viral fusion proteins are very different 15; 34. Given the high specificity of DL16 for the postfusion trimer, particularly, the high sensitivity of its epitope to even small conformational changes, we hypothesized that the DL16 antibody is highly unlikely to appreciably bind the prefusion form or any form other than the postfusion form. This was our rationale for using DL16 as a postfusion-form-specific reagent.
Under mildly denaturing SDS-PAGE conditions, the postfusion HSV-1 gB ectodomain, gB730, migrates as a high-molecular weight band that corresponds to a trimer (Fig. 4). This band reacts with the trimer-specific DL16. Lack of reactivity with DL16 was taken to mean that a gB mutant likely adopted a conformation different from the postfusion one (Table 1). All mutant proteins were also tested for their reactivity with the mAb C226 (Table 1). This antibody recognizes a conformational epitope within dII and reacts with both trimeric and monomeric species on SDS-PAGE. While C226 reacts with the postfusion conformation of the gB ectodomain, it is a neutralizing antibody and should react with the prefusion form as well. Reactivity with C226 was used to test for proper folding and trimerization of mutants not recognized by DL16.
Fig. 4.
Most constructs react with anti-gB monoclonal antibody DL16, specific for the postfusion form. (A) Mutants designed to stabilize the prefusion conformation. (B) Mutants designed to destabilize the postfusion conformation. Proteins secreted into the supernatant were detected by a Western blot with anti-gB monoclonal antibodies DL16 (top panels) or C226 (bottom panels). Under nonreducing mildly denaturing conditions, gB migrates as a trimer. T – trimer, M – monomer. Positions of molecular weight standards are labeled. In A, lanes are numbered and corresponding constructs are listed underneath the blots. “L6” stands for linker sequence GSGTGS; “L10” stands for linker sequence GGSGGTGGSG. “Cyto” corresponds to the entire gB cytoplasmic domain. In B, lanes are labeled at the top.
Under mildly denaturing SDS-PAGE conditions, all gB mutants designed to stabilize the prefusion form migrated as oligomers similar in size to the WT gB730 trimers (Fig. 4A). All mutants from this category reacted with both C226 and DL16 antibodies, indicating that the presence of GCNt or foldon sequences did not affect their respective epitopes (Fig. 4A). Moreover, DL16 reactivity suggested that these mutants retained the trimeric postfusion conformation of WT gB730. All 6 mutants containing the cytodomain were less stable trimers than gB730 (Fig. 4A).
Among mutants designed to destabilize the postfusion conformation, all but nine had WT gB730 electrophoretic mobility on mildly denaturing SDS-PAGE (Fig. 4B). The nine mutants that did not were T394P, gB670, gB678, gB683, gB689, gB700, L700G/L711G, Quad-FF and Quad-WW. Mutant T394P showed only high molecular weight species that entered the gel poorly and was not characterized further. Truncation mutants gB670, gB678, gB683, gB689 and gB700 migrated primarily as monomers (Fig. 4B). Truncations of dV thus destabilized the trimeric gB ectodomain. Weak trimer bands were seen for gB678, gB683, and gB689 but these did not react with DL16 probably because truncations of dV affected the DL16 epitope. Mutants L700G/L711G, Quad-FF, and Quad-WW also formed less stable trimers, and only Quad-FF reacted with DL16 (Fig. 4B). Because of the change in their electrophoretic and antigenic properties, mutants gB670, gB678, gB683, gB689, gB700, L700G/L711G, and Quad-WW were considered the most promising and were characterized further.
Purification of mutant protein constructs
We purified all mutant proteins with reduced DL16 reactivity and several mutant proteins with WT reactivity, namely, gB730-GSGS-foldon, gB730- L10-GCNt-Xa-H6, gB730-GCNt-cyto(821-904), gB730-L10-cyto(796-904)-His6, R505A, I391P, L506F, T509F, and Quad. Most mutants were purified using immunoaffinity chromatography with immobilized DL16 or C226 mAb followed by size-exclusion chromatography, except Quad-WW and mutants with truncated dV. As expected, all mutants with WT DL16 reactivity as well as mutant L700G/L711G eluted near the same elution volume as WT gB730 (Fig. 5).
Fig. 5.
(A). Overlay of size-exclusion chromatograms of WT gB ectodomain (gB730) and representative mutants. gB trimer elutes as a broad peak near the void volume. The elution volumes of the void and the molecular weight standards are shown (arrows).
gB mutants with truncated dV (gB670, gB678, gB683, gB689, and gB700) and Quad-WW were purified by gravity flow instead of peristaltic flow, to reduce aggregation. All truncated mutants were purified using Ni-NTA metal affinity chromatography, made possible by a histidine patch on the surface of the gB ectodomain, followed by immunoaffinity chromatography with immobilized C226 mAb and by hydroxyapatite chromatography. Quad- WW mutant was purified using Ni-NTA metal affinity chromatography followed by hydroxyapatite chromatography, in the presence of the detergent CHAPS during all purification stages.
All mutants adopt the WT postfusion conformation
The gB730 ectodomain represents the postfusion conformation 12; 35 and has a characteristic shape with distinct “crown” and “base” ends that can be easily discerned by negative-stain electron microscopy (EM) 19; 20. Detection of this shape in mutant proteins was used as a definitive test of the postfusion conformation. As expected, all DL16-reactive constructs adopted the WT postfusion conformation: R505A, I391P, L506F, gB730-L10-GCNt-Xa-H6, gB730-GCNt-cyto(821-904), gB730-GSGTGS-cyto(796-904), gB730-GSGS-foldon (Fig. 6A–G), T509F, and Quad (data not shown). Despite the lack of DL16 reactivity, both L700G/L711G and Quad-WW mutants adopted the WT postfusion conformation (Fig. 6H–I), indicating that in these two mutants, the DL16 epitope was disrupted by local rather than global conformational changes.
Fig. 6.
(A). All of the tested constructs adopted the postfusion conformation. gB molecules were visualized by negative-stain EM, and representative point mutants (A–C, H–I), “trimerization” constructs (D–G), and a truncation mutant (J) are shown. gB730-GCNt corresponds to construct gB730-GGSGGTGGSG-GCNt-Xa-His6, gB730-GCNt-cyto corresponds to construct gB730- GCNt-cyto(821-904), gB730-cyto corresponds to construct gB730-GSGTGS-cyto(796-904), and gB730-foldon corresponds to construct gB730-GSGS-foldon. Electron micrographs with multiple particles as well as close-up views of individual particles are shown. All large panels and all close-up panels are shown at the same magnification levels, respectively. (K) The crystal structure of gB730 is shown in surface representation, for comparison. Crown and base ends are labeled.
Mutant gB670 formed rosettes with 3 to 5 arms (Fig. 6J). The length of each arm was 17.2 ± 0.5 nm (n=9), which is similar to the length of HSV-1 gB ectodomain calculated from its atomic coordinates (16.7 nm) 12; 20. Moreover, the characteristic “crown” shape at the tip of each arm (Fig. 6J) resembled the “crown” end of the HSV-1 gB ectodomain in EM images 19; 20 and the crystal structure 12; 20 (Fig. 6K). This overall shape similarity suggested that gB670 adopted the postfusion gB conformation and that each rosette was composed of several gB trimers joined by their “base” ends. Unlike gB670, WT gB730 does not form rosettes. We hypothesize that truncation of domain dV in gB670 causes local conformational changes in dI, which interacts extensively with dV, causing normally buried hydrophobic residues to become exposed at the “base” end and to form rosettes. gB670 rosettes resemble rosettes formed by the ectodomains of gB from EBV 36; 37 and HCMV 38, which contain exposed hydrophobic residues in their fusion loops which are located at the tips of domains dI, indirectly supporting the above hypothesis. Although truncations of dV caused local conformational changes in the vicinity of the fusion loops and the DL16 epitope, none affected the global conformation of the gB ectodomain.
Mutations in dV reduce overall stability of the gB ectodomain
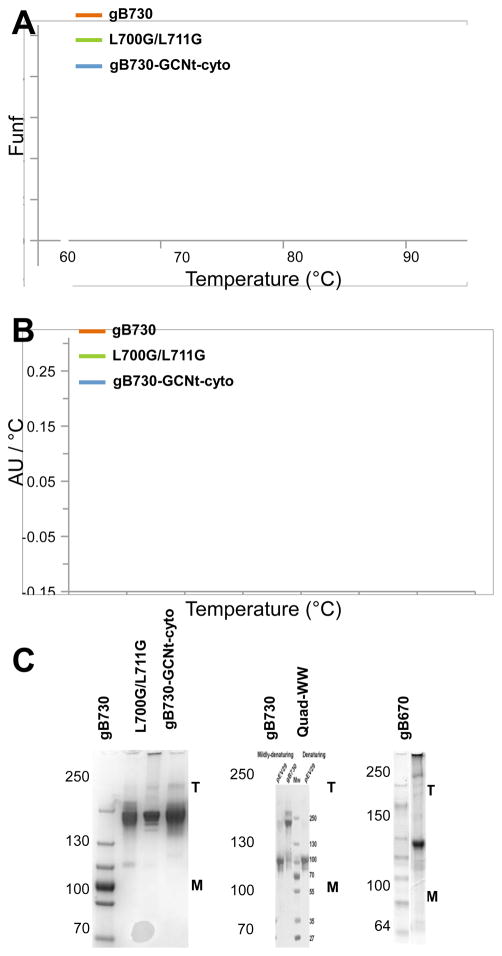
To assess the overall stability of the purified gB mutants, we carried out thermal denaturation studies using circular dichroism (CD) (Fig. 7A). CD signal provides an estimate of the secondary structure content. Due to a high β-strand content of the gB ectodomain, the CD signal at 218 nm (θ218) was used to monitor folding. The decrease of θ218 signal with increasing temperature reflects unfolding of the secondary structure. We found that gB730 was highly resistant to thermal denaturation, with a melting temperature (Tm) of 78.2±0.7°C. Since the Quad-WW mutant and mutants with truncated dV aggregated, we could not analyze their stability using CD. Instead, we chose to test mutants L700G/L711G and gB730-GCNt-cyto(821-904) because they appeared to form less stable trimers in Western blots of insect cell supernatants (Fig. 4). We found that gB730-GCNt- cyto(821-904) was slightly more resistant to denaturation than gB730 and had Tm = 79.5±0.7°C, which is consistent with the stabilizing effect of trimeric GCNt. By contrast, mutant L700G/L711G was noticeably less stable, with a Tm of 72.1±0.7°C. The stability of the ectodomain was thus sensitive to mutations in dV.
Fig. 7.
Several mutations affect the stability of the postfusion trimer. (A) Thermal denaturation stability of gB730, L700G/L711G, and gB730-GCNt-cyto(821-904) as monitored by measuring change in the CD signal at 218 nm with increasing temperature. Data are shown as fraction unfolded vs. temperature. (B) Thermal denaturation stability of gB730, L700G/L711G, and gB730-GCNt-cyto(821-904) as determined by Thermofluor assay. Data are shown as the first derivative of absorbance at 610 nm, in absorbance units per C, vs. temperature. (C) Oligomeric state of gB730 and several purified mutant proteins as analyzed by SDS-PAGE under mildly denaturing conditions (0.1% SDS, no boiling). Similar protein amounts were loaded into each lane. T – trimer, M – monomer. Positions of molecular weight standards are labeled.
To obtain an additional assessment of the overall protein stability, we used the Thermofluor assay, which determines protein stability by monitoring the exposure of hydrophobic regions during unfolding by measuring binding of a hydrophobic dye 39(Fig. 7B). The Tm values estimated by Thermofluor were slightly higher for all proteins than those estimated from CD but remained consistent: gB730 had a Tm = 80.2±0.1°C, while gB730-GCNt-cyto(821-904) was more stable than the WT, with Tm = 81.7±0.2°C, and mutant L700G/L711G was less stable than the WT, with Tm = 75.5±0.1°C.
While assaying protein resistance to thermal denaturation by measuring either the CD signal or the binding of a hydrophobic dye provides an estimate of the overall protein stability, it does not directly test stability of the trimer. To correlate the overall stability of the ectodomain with the trimer stability, we analyzed the purified proteins on mildly denaturing gels (Fig. 7C). Similarly to gB730, both gB730-GCNt-cyto(821-904) and L700G/L711G migrated as trimers on SDS-PAGE under mildly denaturing conditions. This result contrasts with our observations of protein stability in supernatants where gB730-GCNt-cyto(821-904) and L700G/L711G migrated as both trimers and monomers (Fig. 4). One possible explanation could be the difference between slightly acidic pH of the insect cell media (pH 6.1–6.4) as opposed to pH 8.0 of the purified protein storage buffer. Low pH affects the electrophoretic mobility of gB730 under mildly denaturing conditions: while at pH 8.0, only the trimer band can be detected; at pH 6.0 both the monomer and the trimer can be observed; and at pH 5.0 only the monomer and not the trimer can be detected 20. This effect has been attributed to the destabilizing effect of the low pH on the trimeric gB ectodomain, which in combination with SDS and electric current causes trimer dissociation 20. Regardless of the cause of trimer dissociation of protein in viral supernatants, both mutants form stable trimers when they are purified, which is in agreement with thermal denaturation studies (Fig. 7A, B).
Importantly, both truncation mutant gB670 and the Quad-WW mutant primarily migrated as monomers under mildly denaturing conditions (Fig. 7C). Reduced trimer stability of these mutants as purified proteins demonstrates that the trimeric gB ectodomain is sensitive to deletions of dV and large perturbations of the coiled-coil interface.
A504P/R505G/Q507G/N511G mutations do not perturb the central helix αC
To understand how the gB ectodomain can maintain a postfusion conformation despite the presence of multiple destabilizing mutations within helix αC of dIII, we determined the crystal structure of the gB Quad mutant (Fig. 8A). Although proline and glycine residues are known to introduce kinks or breaks into helices 40; 41; 42, the mutation of three amino-acid residues to glycines and one to a proline within the N terminus of helix αC did not change its overall conformation (Fig. 8A), probably because it is restricted by the neighboring domains dII and dV. The conformational changes within helix αC were limited to two side chains. The side chain of F503 in the Quad mutant switched its rotamer, relocating into the empty space introduced by elimination of the side chain of R505 (Fig. 8C). The side chain of E502 also relocated (Fig. 8C) but probably not as a consequence of any of the mutations in the gB Quad mutant because this alternative conformation of the E502 side chain has been observed in other gB structures. Thus, the introduction of multiple glycine and proline residues was not sufficient to destabilize the N-terminus of helix αC. Initial analysis of the WT gB730 structure suggested that the hydrogen bonds between the side chains of R505, Q507, and N511 and domains dII and dV could stabilize helix αC. Although mutations R505G, Q507G, and N511G disrupted the majority of hydrogen bonds between the N terminus of helix αC and domains dII or dV (Fig. 8D), they did not destabilize the helix, probably because the backbone hydrogen bonds are sufficient for its stability.
Fig. 8.
Mutations in the gB Quad mutant do not affect the overall conformation of the gB ectodomain and cause only small local changes. (A) Crystal structure of the gB Quad mutant ectodomain (yellow) overlaid onto the structure of the WT gB (cyan). Only a single protomer (chain A) is shown. (B) A close-up view of the mutated region. Mutated residues are shown as sticks and labeled. (C) In gB Quad mutant, the side chain of F503 relocates to fill the void formed by R505G mutation. (D) A close-up view of the hydrogen bonds between the N-terminus of the central -helix αC in domain dIII (magenta) and domains dII and dV in the WT gB and gB Quad. Mutations eliminated all but one hydrogen bond. Residues involved in hydrogen bonds are shown as sticks and labeled. Hydrogen bonds are shown as dashed lines.
DISCUSSION
The postfusion conformation of gB is very stable
Overall, it was very difficult to destabilize the trimeric postfusion form of gB ectodomain as it was resistant to perturbations by a number of approaches used here. Introducing glycines and prolines into the N terminus of central helix αC, which forms the coiled-coil core of the postfusion trimer, did not destabilize even helix αC itself, let alone the trimer. Likewise, mutations disrupting hydrogen bonds between the N terminus of helix αC and dII or dV did not have any obvious effect on the formation of the postfusion gB trimer. The T394P mutant aggregated under SDS-PAGE conditions, so the T394P mutation probably destabilized the protein (T394 is located within a β-strand), but because this mutant was not amenable to purification and biochemical characterization we could not assess the effect of the mutation on stability directly.
Initially, we expected that introducing bulky residues at the coiled coil interface within the N terminus of helix αC would successfully destabilize the trimer given the tight side chain packing at the coiled coil interface. We found, however, that none of the single phenylalanine or lysine mutations, each of which introduced three bulky side chains at the trimeric coiled coil interface, destabilized the postfusion trimer. Only a double phenylalanine and especially double tryptophan mutations (Quad-FF and Quad-WW), which introduced six bulky side chains at the coiled coil interface, destabilized the trimer to some extent. Nevertheless, even these mutations failed to prevent the formation of the postfusion form. These observations point to remarkable elasticity of the gB ectodomain, which can accommodate a number of mutations without a deleterious effect on its overall structure. This elasticity manifested itself in the relocation of the F303 side chain in the Quad mutant to compensate for the removal of the R505 side chain.
Truncations of dV and, to a lesser extent, the L700G/L711G double mutation destabilized the trimeric postfusion form of the gB ectodomain. dV is not an independently folded domain but rather a long extension spanning nearly the entire length of the ectodomain (Fig. 2E). Residues within dV have no contact with the rest of the polypeptide chain of the same protomer and instead make multiple contacts with domains dI, dII, and dIII of the other two protomers within the trimer, which strengthens trimer interactions. dV plays an important role in the stability of the postfusion trimer, and it is not surprising that truncations of dV destabilize the trimer. Nevertheless, even the complete removal of dV in mutant gB670 did not abrogate the formation of the postfusion trimer. This suggests that multiple regions within the molecule contribute to the stability of the postfusion conformation such that perturbation of any single region is not sufficient to destroy the postfusion conformation completely. The remarkable stability of the postfusion form of gB probably provides the driving force for membrane fusion.
Lack of DL16 reactivity does not reliably indicate the absence of the postfusion conformation
DL16 is a conformational antibody that specifically recognizes the postfusion gB trimer and can serve as an indicator of the postfusion conformation. The DL16 epitope must span at least two protomers and probably includes residues within domains dI and dV 33 because it is sensitive to mutations within these domains. For example, a 5-amino-acid residue insertion after a buried residue E187 in dI abrogated DL16 binding 19. Here, we found that the L700G/L711G double mutation, all partial and full truncations of dV, and the L506W/T509W double mutation in the Quad-WW mutant affected the DL16 epitope. Although the Quad-WW mutant also contains mutations A504P/R505G/Q507G/N511G, these four mutations did not affect the DL16 epitope in the Quad mutant. Residues L700 and L711 lie within domain dV, buried underneath the putative DL16 epitope, and probably buttress domain dI by interacting with several residues in this domain. Substitution of L700 and L711 with glycines likely caused local conformational changes in dI that disrupted the DL16 epitope. Truncations of domain V had a more direct effect on the DL16 epitope, probably by removing a part of it. It is harder to explain why mutations L506W/T509W disrupted the DL16 epitope given that residues L506 and T509 are located far from its putative location within domains dI and dV. We hypothesize that the L506W/T509W double mutation probably splayed apart the trimeric coiled coil and that these conformational changes propagated to dI and dV, disrupting DL16 binding. While DL16 reactivity is a reliable indicator of the postfusion conformation, the high sensitivity of the DL16 epitope to proximal as well as distal structural perturbations leads us to conclude that the absence of DL16 reactivity cannot be taken as the absence of the postfusion form.
The soluble ectodomain folds into the postfusion conformation directly
While it was possible to destabilize the postfusion conformation, none of the mutants adopted the prefusion conformation or any conformation other than postfusion. Surprisingly, addition of the trimerization tags GCNt and foldon, which successfully stabilized the prefusion forms of other viral fusion proteins 21; 22; 23; 24; 25; 26, did not yield the prefusion form of gB. We have some indirect evidence that the TM helices of gB probably do not interact within the plane of the membrane 43. Replacing the native, non-trimerizing, TM sequence in the context of full-length gB constructs with trimerizing GCNt or foldon may interfere with proper folding of the prefusion form, which may potentially explain why trimeric GCNt and foldon are poor substitutes for the native TM sequence. More puzzling is the inability of GCNt and foldon sequences to lock the isolated gB ectodomain into its prefusion form even in the presence of long linkers connecting the gB and the GCNt (or foldon) sequences. We hypothesize that the folding pathway for the soluble gB ectodomain produces the postfusion form directly, without a prefusion intermediate. That is, when expressed on its own, the gB ectodomain folds into the stable postfusion form without first adopting the metastable prefusion conformation.
In support of this, three mutants in which various regions of dV were replaced with GCNt were not secreted, probably due to misfolding. This substitution cannot be accommodated in the postfusion form and would lead to misfolded protein; yet, we expected that it would be tolerated in the prefusion form, possibly stabilizing it. Although we cannot exclude the possibility that the GCNt substitution of dV cannot be tolerated in the prefusion form, our preferred explanation for misfolding of these constructs is that they fold into the postfusion conformation directly.
The observed inability of the soluble gB ectodomain to adopt the prefusion conformation suggests that that other regions of gB, including the transmembrane region and the cytoplasmic domain, may be necessary to establish and maintain the metastable prefusion conformation. The presence of the transmembrane region is particularly important considering that all constructs containing the cytoplasmic domain in addition to the ectodomain adopted the postfusion conformation. Future efforts to obtain the prefusion conformation of gB should focus on full-length gB. Alternatively, interactions with other viral proteins, such as gH/gL, may be critical to maintaining the gB prefusion form at the virion surface.
MATERIALS AND METHODS
Cloning
HSV-1 (strain KOS) gB was used in all cloning. Point, truncation and insertion mutants were generated by “splicing by overlap extension” PCR (SOE-PCR) method 44. Resulting PCR products were subcloned into the gB gene (residues A31–A730) in vector pFastBac1. In all signal sequence was replaced with the signal sequence from honeybee constructs, the endogenous melittin, to improve secretion. The resulting plasmids are pKH1 (gB773-foldon), pKH2 (gB773- GCNt), pKH6 (gB730-GSGS-foldon), pKH8 (gB730-GSGTGS-foldon-Xa-His6), pKH10 (gB730-GGSGGTGGSG-foldon-Xa-His6), pKH11 (gB773-GGSGGTGGSG-cyto(796-904)-His6), pKH12 (gB730-GSGTGS-cyto(796-904)-His6), pKH13 (gB730-GGSGGTGGSG- cyto(796-904)-His6), pKH14 (gB773-GSGTGS-GCNt-Xa-His6), pKH15 (gB730-GSGTGS- GCNt-Xa-His6), pKH17 (gB730-GGSGGTGGSG-GCNt-Xa-His6), pKH19 (gB773-GSGTGS- foldon), pKH20 (gB773-GGSGGTGGSG-foldon), pKH21 (gB773-foldon-strep), pKH22 (gB773-GSGTGS-foldon-strep), pKH23 (gB773-GGSGGTGGSG-foldon-strep), pKH30 (L506F), pKH31 (I513F), pKH34 (gB730-GSGTGS-cyto(796-904)), pKH35 (T509F), pKH36 (R515P), pKH78 (gB684-GGSGGTGGSG-GCNt-Xa-His6), pKH79 (gB690-GGSGGTGGSG- GCNt-Xa-His6), pKH80 (gB701-GGSGGTGGSG-GCNt-Xa-His6), pDM1 (gB773-GSGTGS- cyto(796-904)-His6), pEV1 (R505A), pEV2 (E502A), pEV3 (Q507A), pEV4 (I391P), pEV5 (L506K), pEV6 (N511A), pEV8 (L700G/L711G), pEV9 (gB670), pEV10 (gB689), pEV11 (gB678), pEV12 (gB683), pEV13 (gB700), pEV18 (A504P/R505G/Q507G), pEV19 (A504P/R505G/Q507G/N511G), pEV21 (T394P), pEV29 (A504P/R505G/L506W/Q507G/T509W/N511G), pEV30 (A504P/R505G/L506F/Q507G/T509F/N511G), pEV35 (gB730-GCNt-cyto(821-904), pEV36 (gB730-GCNt-cyto(796-904).
Antibodies
Hybridoma cell lines expressing anti-HSV-1 gB monoclonal antibodies (mAbs) DL16 or C226 were a gift from Drs. Roselyn J. Eisenberg and Gary H. Cohen (University of Pennsylvania). The mAbs were purified at the GRASP facility at Tufts Medical Center. DL16 is a conformational mAb that recognizes the trimeric form of the HSV-1 gB ectodomain 33. C226 is a neutralizing antibody that recognizes a conformational epitope located in domain II of HSV-1 gB ectodomain 33. Polyclonal antibody R68 was a gift from Drs. Roselyn J. Eisenberg and Gary H. Cohen (University of Pennsylvania).
Viruses and cells
Spodoptera frugiperda (Sf9) cells were grown in SF-900 II SFM (Invitrogen) in suspension at 27°C. All recombinant baculoviruses for gB expression were generated using Bac-to-Bac technology (Invitrogen). Sf9 cells were transfected with each recombinant bacmid DNA and passage 1 (P1) viral stock was harvested at 72 hours post transfection. After two rounds of amplification, passage 3 (P3) stocks of baculoviruses were harvested and stored at 4°C in the dark in the presence of 2% FBS.
Protein expression and purification
Initially, expression was tested in P1 “refeed” supernatants made by incubating infected cells in fresh media for 72 hours after harvesting P1. For large-scale protein expression, P3 stocks of baculoviruses were used to infect Sf9 cells. The amount of viral inoculum was optimized to yield the highest protein expression. Typically, 10 mL of viral stock was added to 1 L of SF9 cells at 1.5×106 cells/mL, and cell supernatants were harvested 72 hours post-infection. Cells were pelleted using centrifugation at 3750 rpm at 4°C for 20 minutes. The supernatant was collected, filtered through a 0.22 μm filter, and concentrated using a Tangential Flow Filtration system (Millipore) using a 30-kDa PLTK cartridge (Millipore). Buffer exchange was done three times into phosphate-buffered saline pH 7.4 to remove media components, and 0.1 mM PMSF was added to inhibit proteolytic activity.
With few exceptions, all gB mutants were purified by immunoaffinity chromatography using mAb DL16 immobilized on CNBr-activated Sepharose 4B (GE Healthcare) according to the protocol developed earlier for the purification of WT gB73012. Mutant L700G/L711G was purified by immunoaffinity chromatography using immobilized mAb C226 instead of DL16. After extensive washing with 10 mM Tris-HCl, pH 8.0, and 500 mM NaCl, the protein was eluted with 3M KSCN and concentrated on an Amicon Ultra-15 concentrator (Millipore) with a 50-kDa MW cutoff. Proteins were further purified by size-exclusion chromatography using Superdex S200 column (GE Healthcare) equilibrated with 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM EDTA. The column was calibrated using blue dextran (~2000 kDa), thyroglobulin (667 kDa), ferritin (440 kDa), catalase (232 kDa), and aldolase (158 kDa).
gB mutants with truncations in domain dV were purified by Ni-NTA metal affinity chromatography (GE Healthcare) in 50 mM Tris-HCl, pH 8.0, 300 mM NaCl. Proteins were eluted from the Ni-NTA resin with 250 mM Imidazole in 50 mM Tris-HCl, pH 8.0, 300 mM NaCl. After buffer exchange into 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, protein was further purified by immunoaffinity chromatography using immobilized mAb C226. Protein eluted from the immunoaffinity column was exchanged into 10 mM sodium phosphate, pH 7.5, 50 mM NaCl, loaded onto a hydroxyapatite column equilibrated in the same buffer, and eluted using a gradient of sodium phosphate, pH 7.5. All chromatography steps were done using gravity flow instead of the peristaltic pump, to reduce aggregation.
The A504P/R505G/L506W/Q507G/T509W/N511G (Quad-WW) mutant was purified by Ni-NTA metal affinity chromatography followed by hydroxyapatite column. 0.1% detergent CHAPS was added to all solutions during purification to reduce protein aggregation. All chromatography steps were done using gravity flow.
SDS-PAGE and Western blot analysis
Three conditions were used in preparation of samples for SDS-PAGE: mildly denaturing (0.1% SDS, no reducing agent, no boiling), non-reducing denaturing (1% SDS, no reducing agent, boiling), and reducing denaturing (1% SDS, reducing agent, boiling). To test expression by Western blot analysis using pAb R68, samples were prepared under reducing denaturing conditions. To test reactivity of gB mutants with conformational mAbs, samples were prepared under mildly denaturing conditions to preserve gB oligomers.
For Western blot analysis, protein samples were resolved by SDS-PAGE (4–15% acrylamide gel, Bio-Rad) and transferred to a nitrocellulose membrane. Membranes were blocked with 5% skim milk in Tris-buffered saline containing 0.05% Tween-20. To test the expression of gB constructs, membranes were probed with a pAb R68 at a 1:10,000 dilution, followed by incubation with a secondary anti-rabbit IgG conjugated to horseradish peroxidase at a 1:10,000 dilution. To test for reactivity with conformational antibodies, membranes were probed with a mAb DL16 at a 1:1,000 dilution or a mAb C226 at a 1:5,000 dilution, followed by incubation with a secondary anti-mouse IgG conjugated to horseradish peroxidase at a 1:10,000 dilution. Blots were developed using a Western Pico chemiluminescence kit (Pierce).
Electron microscopy
Purified protein samples (5 μM) were incubated on glow-discharged formvar/carbon-coated 200 mesh copper grids (SPI) at room temperature and stained with 0.75% solution of uranyl formate. EM images were obtained at room temperature using a Tecnai G2 Spirit BioTWIN equipped with an AMT 2k charge-coupled device camera microscope at the Harvard Medical School EM core facility.
Thermal Denaturation using Circular Dichroism (CD) Spectroscopy
Measurements were acquired using a JASCO-810 spectropolarimeter equipped with Peltier cell (Elfin model ELDC5D4) and a 1 cm quartz cuvette. Thermal denaturation was monitored at 218 nm (θ218). All measurements were performed in 20 mM phosphate pH 8.0 and 100 mM NaF using protein at a concentration of 0.2 mg/ml, as determined by 280 nm absorbance (Nanodrop ND-1000). 218 nm measurements were taken every 0.1°C with an 8 s response time and 1 nm width. Rapid heating of wild-type gB and mutants at 2°C/min was used to identify temperatures where each began to denature. Final melting curves were obtained by slow heating (0.1°C/min) starting at least 2°C below where protein started to denature. Melting curves were processed for noise elimination by averaging CD signals from all points in a 6°C range for each measurement. Each θ218 measurement was converted to fraction unfolded (Funf) using the equation: (θ218 − θU)/(θF − θU), where θ218 is the observed ellipticity at 218 nm for a given temperature, θU is the θ218 value when the protein is unfolded and θF is the θ218 value when the protein is folded. The θU and θF values were determined in each experiment by averaging unsmoothed θ218 from all readings spanning at least 5°C at temperatures either just prior to (θF) or just after (for θU) protein melting. The Tm was obtained from the point of the melting curve corresponding to Funf = 0.5. A 95% confidence interval for θ218 at the Tm was calculated by combining standard error of the baselines, converting this error to a Tm range using smoothed θ218 values, and then adding the error range of the Peltier unit (accurate to 0.6°C) to that range.
Thermal Denaturation with Thermofluor
The Thermofluor assay 45 uses a fluorescent dye which gives an enhanced signal upon exposure to hydrophobic environments. As the temperature is raised, the protein is denatured, exposing hydrophobic regions to which the dye binds. This causes an increase in the dye fluorescence. Protein was diluted into 20 mM sodium phosphate pH 8.0 and 100 mM NaF to a final concentration of 0.15 mg/ml, approximately 10-fold. SYPRO Orange (Invitrogen), the fluorescent dye, was added at a 1:2000 dilution. Each experiment was done in triplicate by aliquoting 20 μl samples into a 96-well PCR microplate, which was sealed and then centrifuged for 1 minute at 500 g at 23°C. Samples were analyzed in a Roche Light Cycler 480 qPCR instrument using an excitation wavelength of 465 nm with emission detected at 610 nm. The emission signal was analyzed from 23°C to 95°C at a continuous acquisition rate of 3 measurements per C. The raw spectra of fluorescence vs temperature were converted to the first derivative spectra, and Tm was determined as the temperature corresponding to the maximum value in the first derivative spectrum. The reported Tm represents the average of 3 experiments. Reported errors represent the standard deviation.
Crystallization, data collection, and structure determination of the A504P/R505G/Q507G/N511G (Quad) mutant
For crystallization, gB mutant protein in 10 mM Tris·HCl, pH 7.6, 100 mM NaCl, and 1 mM EDTA was concentrated to 4.0 mg/mL using a Millipore Ultra-4 concentrator (MW cutoff 50 kDa). Crystals were grown by vapor diffusion at room temperature in hanging drops with 2 μl protein sample and 2 μl reservoir solution containing 15% PEG 4000, 0.2 M NaCl, and 0.1 M Na·citrate, pH 5.5. Hexagonal rods appeared after several days and grew to their final size over 2–3 weeks. For data collection, crystals were transferred stepwise to a solution identical to the well solution plus 15% mesoerythritol, 0.1 M NaCl, and 1 mM EDTA and plunged into liquid N2. Data for the mutant crystals were collected at 100 K at X25 beamline at the National Synchrotron Light Source. The dataset was processed using HKL200046 and indexed in space group P3 (Table 2). Prior to refinement, 5% of the data were set aside for cross-validation by transferring test set flags from the 3NWA dataset 20 to the A504P/R505G/Q507G/N511G mutant dataset. The 3NWA coordinates were used as a starting model for refinement of the A504P/R505G/Q507G/N511G mutant structure except that residues 500–515 were omitted. Model was refined using phenix. refine 47 with iterative rounds of rebuilding in Coot 48. The omit density was used to rebuild the conformation of residues 500–515. Molprobity 49 was used to assess the stereochemical quality of all models. Final statistics are listed in Table 2. The final model is missing residues 28-104, 474-491, and 724-730 in chain A; residues 28-102, 259-261, 478-490, and 725-730 in chain B; residues 28-104, 474-491, and 725-730 in chain C; and residues 28-102, 259-260, 479-493, and 725-730 in chain D. The final model contains carbohydrate moieties (N-acetylglucosamine) modifying residues N398 and N430 in chains A and C, and residues N398 and N674 in chains B and D, as well as 2 molecules of mesoerythritol, a cryoprotectant.
Table 2.
Data collection and refinement statistics for the gB Quad mutant Data collection
| Space group | P3 |
| Cell dimensions, (Å, °) | 117.295, 117.295, 321.872, 90, 90, 120 |
| Wavelength, (Å) | 1.1000 |
| Resolution (Å) | 45.91-3.10 (3.21-3.10)a |
| Total reflections | 149,982 |
| Unique reflections | 74,374 (7834)a |
| Completeness (%) | 82.8 (87.5)a |
| I/σI | 10.25 (3.00)a |
| Redundancy | 2.0 (1.9)a |
| Rmerge | 0.067 (0.236)a |
| Refinement | |
| Resolution (Å) | 45.91-3.10 |
| Rwork/Rfree | 0.2052/0.2549 |
| No. atoms | |
| Protein | 19615 |
| Ligand/ion | 20 |
| Solvent | 18 |
| Average B-factors (Å2) | |
| Protein | 45.4 |
| Ligand/ion | 41.4 |
| Solvent | 19.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.205 |
| Ramachandran plot (%)b | |
| Most favored region | 90.8 |
| Additionally allowed region | 10.0 |
| Disallowed region | 0.2 |
Values in parentheses are for the highest-resolution shell
Determined using Molprobity 43
Highlights.
The ectodomain of herpesvirus fusogen gB spontaneously folds into the postfusion form
Postfusion conformation of gB is remarkably stable and resistant to perturbations
Domain dV is critical for the stability of the postfusion form
The isolated gB ectodomain folds into postfusion form without prefusion intermediate
The TM and the cytodomain may be necessary to maintain the metastable prefusion form
Acknowledgments
We thank Drs. Roselyn J. Eisenberg and Gary H. Cohen (University of Pennsylvania) for providing antibodies and hybridomas used in this work and Maria Eriksson at the HMS EM Core Facility for help with sample preparation and data acquisition. We also thank Jared Pitts for expressing, purifying, and performing negative-stain EM on gB730-GSGTGS-cyto(796-904). We thank A. Héroux of the National Synchrotron Radiation Source for collecting and processing x-ray diffraction data on crystals of gB Quad mutant. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. This work was funded by the NIH grant 1DP20D001996 and by the Pew Scholar Program in Biomedical Sciences (E.E.H.).
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors for the gB mutant A504P/R505G/Q507G/N511G have been deposited to the RCSB Protein Data Bank under accession number 4HSI.
AUTHOR CONTRIBUTIONS
E.E.H. and E.V. designed the experiments and cloned the constructs, E.V., S.S., and S.D.S. expressed, purified, and characterized proteins, E.V. and S.S. carried out electron microscopy, S.D.S. performed and analyzed thermal denaturation experiments, E.V. crystallized the Quad mutant, E.E.H. determined the crystal structure of the Quad mutant, E.E.H. and E.V. analyzed the data, and E.E.H. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Plemper RK. Cell entry of enveloped viruses. Current opinion in virology. 2011;1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4:613–36. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. Herpes virus fusion and entry: a story with many characters. Viruses. 2012;4:800–32. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stampfer SD, Heldwein EE. Stuck in the middle: structural insights into the role of the gH/gL heterodimer in herpesvirus entry. Current opinion in virology. 2012 doi: 10.1016/j.coviro.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 2004;6:401–10. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 6.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 7.Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS pathogens. 2011;7:e1002277. doi: 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A. 2007;104:18718–23. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avitabile E, Forghieri C, Campadelli-Fiume G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17:882–8. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. Journal of virology. 2010;84:12292–9. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 13.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 14.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–30. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 15.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 17.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 18.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 19.Silverman JL, Sharma S, Cairns TM, Heldwein EE. Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation. J Virol. 2010;84:2001–12. doi: 10.1128/JVI.01791-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. Journal of virology. 2010;84:12924–33. doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–70. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 23.Sissoeff L, Mousli M, England P, Tuffereau C. Stable trimerization of recombinant rabies virus glycoprotein ectodomain is required for interaction with the p75NTR receptor. J Gen Virol. 2005;86:2543–52. doi: 10.1099/vir.0.81063-0. [DOI] [PubMed] [Google Scholar]
- 24.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3739–44. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. Journal of virology. 2000;74:4746–54. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbury PB, Kim PS, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–3. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 28.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–7. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 29.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–98. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 30.Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bachinger HP, Engel J. Stabilization of short collagen-like triple helices by protein engineering. J Mol Biol. 2001;308:1081–9. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- 31.Chowdary TK, Heldwein EE. Syncytial phenotype of C-terminally truncated herpes simplex virus type 1 gB is associated with diminished membrane interactions. Journal of virology. 2010;84:4923–35. doi: 10.1128/JVI.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol. 2007;81:3827–41. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Critical reviews in biochemistry and molecular biology. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65:1653–68. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Backovic M, Jardetzky TS, Longnecker R. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol. 2007;81:9596–9600. doi: 10.1128/JVI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106:2880–5. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma S, Wisner TW, Johnson DC, Heldwein EE. HCMV gB shares structural and functional properties with gB proteins from other herpesviruses. Virology. 2013;435:239–49. doi: 10.1016/j.virol.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips K, de la Pena AH. The combined use of the Thermofluor assay and ThermoQ analytical software for the determination of protein stability and buffer optimization as an aid in protein crystallization. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit10. Chapter 10. 2011. p. 28. [DOI] [PubMed] [Google Scholar]
- 40.Piela L, Nemethy G, Scheraga HA. Proline-induced constraints in alpha-helices. Biopolymers. 1987;26:1587–600. doi: 10.1002/bip.360260910. [DOI] [PubMed] [Google Scholar]
- 41.O’Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–51. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 42.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophysical journal. 1998;75:422–7. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverman JL, Greene NG, King DS, Heldwein EE. Membrane Requirement for Folding of the Herpes Simplex Virus 1 gB Cytodomain Suggests a Unique Mechanism of Fusion Regulation. Journal of virology. 2012;86:8171–84. doi: 10.1128/JVI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–32. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 45.Phillips K, de la Pena AH. The combined use of the Thermofluor assay and ThermoQ analytical software for the determination of protein stability and buffer optimization as an aid in protein crystallization. Curr Protoc Mol Biol. 2011;Chapter 10(Unit10):28. doi: 10.1002/0471142727.mb1028s94. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: CCW Jr, MSR, editors. Methods in Enzymology. Vol. 276. Academic Press; New York, NY: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]