Abstract
The goal of our study is to investigate the contribution of promoter DNA methylation of α-adducin (ADD1) gene to the risk of essential hypertension (EH). Using the bisulphite pyrosequencing technology, DNA methylation levels of five CpG dinucleotides on ADD1 promoter were measured among 33 EH cases and 28 healthy controls. Significantly higher ADD1 DNA methylation levels were observed in the females than in the males (CpG1: P = 0.016; CpG2-5: P = 0.021). A breakdown analysis by gender showed that lower CpG1 methylation was associated with an increased risk of EH in females (adjusted P = 0.042). A much more significant association between lower CpG2-5 methylation levels and the increased risk of EH was found in males (adjusted P = 0.008). CpG1 methylation was inversely correlated with age in females (r = −0.407, P = 0.019) but not in males. ADD1 CpG1 and CpG2-5 methylation levels were significantly lower in post-menopausal (>50 years) women than pre-menopausal (≤50 years) women (CpG1: P = 0.006; CpG2-5: P = 0.034). A significant interaction between CpG1 methylation and age was found in females (CpG1*age: P = 0.029). CpG2-5 methylation was shown as a significant predictor of EH in males [area under curve (AUC) = 0.855, P = 0.001], in contrast that CpG1 methylation was a trend toward indicator in females (AUC = 0.699, P = 0.054). In addition, significant differences were observed between males and females for alanine aminotransferase (ALT, P = 0.001), aspartate aminotransferase (AST, P = 0.005) and uric acid (P<0.001). The concentration of AST was inversely correlated with ADD1 CpG2-5 methylation levels in female controls (r = −0.644, P = 0.024). These observations may bring new hints to elaborate the pathogenesis of EH.
Introduction
Essential hypertension (EH) is one of the most important causes of premature death worldwide. EH is a complex disorder resulting from both genetic and environmental factors [1], [2]. Approximately 20–60% of the blood pressure variability in general population is heritable [3]. Epidemiological studies have documented environmental factors such as physical inactivity, obesity, high sodium and low potassium diet, and alcohol consumption are associated with hypertension risk [4], [5]. Disorders in the metabolism of high-density lipoprotein cholesterol (HDL-C) and triglycerides (TG) play a key role in EH progression [6], [7].
A sexual dimorphism exists in the developmental origins of EH [8], [9]. Males are reported to be more susceptible to hypertension than females [10]. Gender difference in the risk of hypertension was observed to be associated with altered expression of hormone receptors such as renal alpha2-adrenergic receptors [11] and angiotensin receptors [12]. In addition, evidence has shown a gender dimorphism in the expression of renin [13], [14] and urinary angiotensinogen excretion [15] which are important risk factors for EH. The sexual dimorphism in mammalian gene expression have been observed to be linked to the gender differences in the amounts of sex hormones [16] (e.g., estrogens, androgens).
ADD1 gene encodes one of adducin subunits (α-adducin) [17]. Adducin modulates the surface expression of multiple transporters and ion pumps, and thus regulates cellular signal transduction and cytolemma ion transport [18]. Human and animal model studies have found that ADD1 gene is a candidate gene for EH [18], [19]. However, epidemiological studies have shown that the contribution of ADD1 Gly460Trp mutation (rs4961) to hypertension varies among different ethnic groups [20]–[24]. Meta-analyses were unable to reach a consensus for this mutation [25]–[28]. Moreover, gender dimorphism of the association between hypertension and ADD1 Gly460Trp was observed in Caucasians [29].
DNA methylation is a stable epigenetic mark and usually occurs at cytosine residues in the context of cytosine-phosphate-guanine dinucleotide (CpG) in mammalian cells [30]. Promoter DNA methylation is linked to transcriptional silencing of protein-coding genes [31] and thus regulates the function of protein. Aberrant methylation is shown to play important roles in the occurrence and development of diseases including colorectal cancer [32], [33], breast cancer [34], [35], coronary artery disease [36] and schizophrenia [37], [38]. The evidence of the association between DNA methylation and the risk of EH was scarce. A significant decline in global DNA methylation level is observed in EH patients and the trend continues along with the progression of hypertension [39]. Altered global DNA methylation in pre-eclampsia placentas was shown to be associated with maternal hypertension [40]. Aberrant DNA methylation of 11beta-HSD2 and Adrb1 genes were found to be associated with EH [41] and the outcome of medications [42], respectively.
We hypothesize that ADD1 promoter DNA methylation contributes to EH. Our goal is to study whether promoter DNA methylation of ADD1 gene is associated with EH, and to explore the interaction of promoter DNA methylation with gender and clinical indicators of lipid and amino acid metabolism.
Materials and Methods
Sample Collection
This study comprised 33 cases (14 males, 50.1±4.9 years; 19 females, 51.3±4.7 years) and 28 controls (14 males, 51.3±6.3 years; 14 females, 47.9±5.0 years) collected from the community residents in Zhenhai district of Ningbo city in Zhejiang province, China. All individuals are Han Chinese living in Ningbo city for at least three generations. Hypertensive patients were defined according to the golden standard [43]. All hypertensives have received antihypertensive medications for more than three months or have at least three consecutive records of systolic blood pressure (SBP) >140 mmHg and/or diastolic blood pressure (DBP) >90 mmHg (European Society of Hypertension-European Society of Cardiology Guidelines, 2003). Patients had SBP<120 mmHg and DBP<80 mmHg and had no family history of hypertension in the first degree relatives were recruited as controls. None of the controls has received antihypertensive therapy. The gender and age of controls were well matched with EH cases. All the individuals don’t have a history of diabetes mellitus, secondary hypertension, myocardial infarction, stroke, renal failure, drug abuse and other serious diseases. A calibrated mercury sphygmomanometer with appropriate adult cuff size was applied to measure blood pressures according to a standard protocol recommended by the American Heart Association [44]. Blood pressures were measured in supine position by two trained observers at an interval of at least 10 minutes. Blood samples were collected in 3.2% citrate sodium-treated tubes and then stored at −80°C for DNA extraction. The study protocol was approved by the ethical committee of Ningbo University. The informed written consent was obtained from all subjects.
Phenotypes Collection
Blood samples were obtained after a 12 h overnight fast from the antecubital vein using vacutainer tubes containing EDTA. Plasma levels of cholesterol, TG, ALT, AST, uric acid and glucose concentrations were enzymatically measured using CX7 biochemistry analyzer (Beckman, Fullerton, CA).
DNA Methylation Assay
Human genomic DNA was prepared from peripheral blood samples using the nucleic acid extraction automatic analyzer (Lab-Aid 820, Xiamen City, China). DNA was quantified using the PicoGreen® double strand DNA (dsDNA) Quantification Kit (Molecular Probes, Inc. Eugene, USA). Bisulphite pyrosequencing technology was used to determine the 5 CpG dinucleotides methylation levels on the fragment within ADD1 promoter (Figure 1). Pyrosequencing assays combine sodium bisulfite DNA conversion chemistry (EpiTech Bisulfite Kits; Qiagen; #59104), polymerase chain reaction (PCR) amplification (Pyromark PCR Kit; Qiagen; #978703) and sequencing by synthesis assay (Pyromark Gold Q24 Reagents; Qiagen; #978802) of the target sequence. Sodium bisulfite preferentially deaminates unmethylated cytosine residues to thymines (after PCR amplification), whereas methyl-cytosines remain unmodified. PCR primers were selected using PyroMark Assay Design software v2.0.1.15. The PCR and pyrosequencing primers for ADD1 gene promoter amplification were described in Table S1.
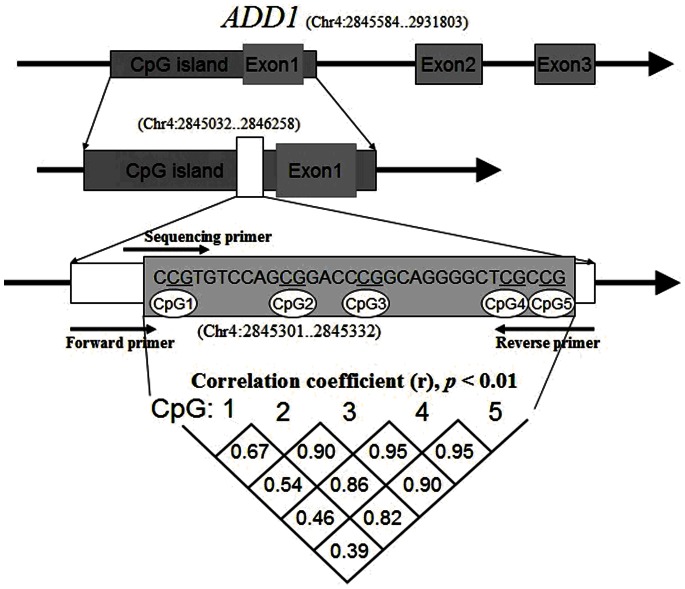
Figure 1. Correlation among five CpGs in ADD1 gene promoter.
Statistical Analysis
Statistical analyses were performed to investigate the association among ADD1 DNA methylation, metabolic profile and EH. Either Pearson chi-square or Fisher exact test was used for the association of EH with categorical variables including gender, smoking, and drinking. Two sample t-test was applied for the association of EH with continuous variables including age, body mass index (BMI), total cholesterol, total triglycerides, glucose, ALT, AST, and uric acid. Pearson correlation was used to determine the association between the ADD1 DNA methylation and metabolic characteristics. Receiver operating characteristic (ROC) curve was used to analyze the sensitivity of ADD1 DNA methylation in EH diagnosis. Logistic regression was implemented for the interaction of ADD1 methylation and age. A two-sided p-value <0.05 was considered statistically significant. All the above statistical analyses were performed with PASW Statistics 18.0 software (SPSS, Inc., Somers, NY, USA). Meanwhile, Power and Sample Size Calculation software (v3.0.43) was used to estimate the power of the study [45].
Results
A total of 33 cases and 28 age- and gender-matched controls were recruited in the current association study. As shown in Table 1, mean levels of body mass index (BMI) and all metabolic phenotypes were within normal ranges. The mean levels of age and BMI were well paired between males and females (Table 1).
Table 1. Characteristics of all subjects (n = 61).
| Characteristics | Mean ± s.e. | Men (Mean ± s.e.) | Women (Mean ± s.e.) | pgender | EH (Mean ± s.e.) | Non-EH (Mean ± s.e.) | pEH |
| Age (years) | 50.2±5.3 | 50.7±5.5 | 49.8±5.1 | 0.527 | 50.8±4.7 | 49.6±5.8 | 0.361 |
| Gender (M/F) | 28/33 | NAc | NAc | 14/19 | 14/14 | 0.554 | |
| BMI (kg/m2)a | 22.50±2.55 | 22.63±1.77 | 22.41±3.01 | 0.781 | 22.95±2.72 | 21.84±2.19 | 0.159 |
| Smoking (Y/N) | NAc | 9/19 | 0/33 | 0.002 | 5/28 | 3/25 | 0.896 |
| Drinking (Y/N) | NAc | 6/22 | 0/33 | 0.018 | 4/29 | 2/26 | 0.826 |
| Total cholesterol (mmol/L) | 5.05±0.93 | 5.01±0.85 | 5.08±1.01 | 0.775 | 5.09±0.81 | 4.99±1.07 | 0.681 |
| Total triglycerides (mmol/L) | 1.50±0.91 | 1.44±0.84 | 1.56±0.97 | 0.615 | 1.69±1.08 | 1.28±0.58 | 0.066 |
| Glucose (mmol/L) | 5.18±0.58 | 5.15±0.59 | 5.20±0.58 | 0.735 | 5.31±0.59 | 5.02±0.53 | 0.055 |
| ALT (IU/L) | 21.4±15.5 | 28.6±17.9 | 15.3±9.9 | 0.001 | 23.7±13.5 | 18.8±17.5 | 0.228 |
| AST (IU/L)b | 23.9±7.0 | 26.6±7.7 | 21.5±5.4 | 0.005 | 25.3±5.7 | 22.2±8.3 | 0.089 |
| Uric Acid (µmol/L) | 298.8±88.8 | 351.5±91.0 | 254.0±57.3 | 0.000 | 312.0±96.4 | 283.2±77.7 | 0.210 |
| CpG1 methylation (%) | 9.97±2.24 | 9.25±1.40 | 10.58±2.63 | 0.016 | 9.52±1.46 | 10.50±2.85 | 0.091d |
| CpG2-5 methylation (%) | 29.33±6.81 | 27.17±7.56 | 31.16±5.58 | 0.021 | 27.54±7.48 | 31.44±5.30 | 0.026d |
n = 44 (18 men versus 26 women, 26 EH versus 18 Non-EH);
n = 59 (28 men versus 31 women, 33 EH versus 26 Non-EH).
NA denotes not applicable.
The p-values were adjusted for age, gender, smoking and drinking using a logistic regression analysis.
In this study, we selected a locus containing 5 CpG dinucleotides to explore the association of ADD1 gene promoter DNA methylation with EH (Figure 1). We found DNA methylation levels were closely correlated among CpG2-5 (Figure 1, r >0.80, P<0.001). As shown in Table 1 and Figure S1, our results showed that ADD1 CpG2-5 methylation levels were significantly associated with EH (cases versus controls (%): 27.54±7.48 versus 31.44±5.30, adjusted P = 0.026). Moreover, significantly higher ADD1 DNA methylation levels were observed in females than in males (CpG1: P = 0.016; CpG2-5: P = 0.021). CpG1 methylation was inversely correlated with age in females (Figure 2, r = −0.407, P = 0.019). ADD1 methylation levels were significantly higher in pre-menopausal (≤50 years) women than post-menopausal (>50 years) women (Figure 3, CpG1: P = 0.006; CpG2-5: P = 0.034).
Figure 2. Pearson correlation between ADD1 methylation and age in males and in females.
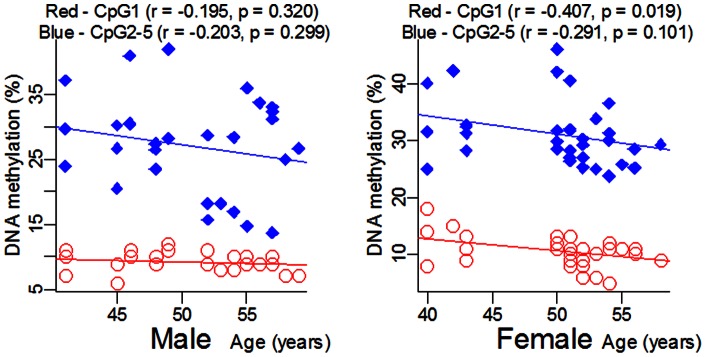
Figure 3. Significant ADD1 methylation difference between pre-menopausal and post-menopausal females.
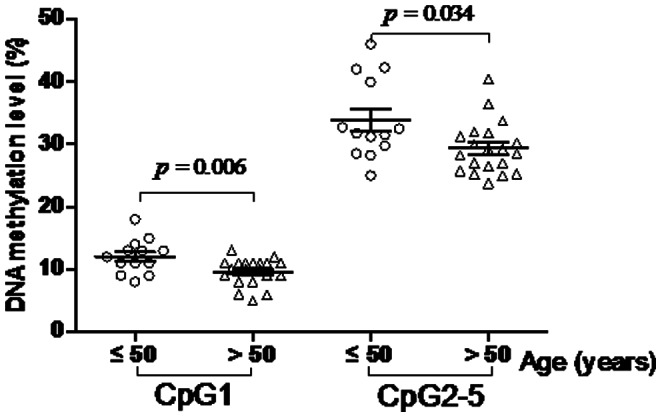
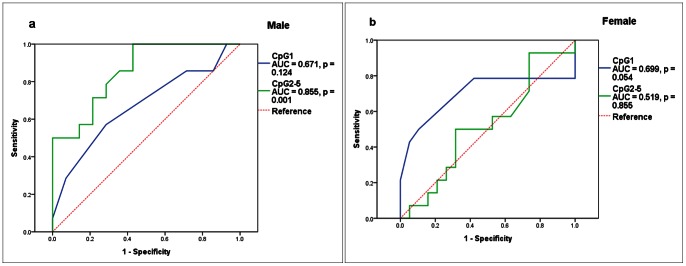
A breakdown association test by gender was also performed to explore the association between ADD1 methylation levels (including CpG1 and CpG2-5) and EH. As shown in Table 2 and Figure S1, ADD1 CpG1 methylation level was significantly associated with EH in females (cases versus controls (%): 10.00±1.41 versus 11.36±3.63, adjusted P = 0.042) but not in males (adjusted P = 0.133). As shown in Table 2 and Figure S1, lower levels of ADD1 CpG2-5 methylation were associated with increased risk of EH in males (cases versus controls: 22.48% versus 31.86%, adjusted P = 0.008). In contrast, no association of CpG2-5 methylation levels with EH was found in females (adjusted P = 0.557). Furthermore, significant interaction of CpG1 methylation and age was found to influence EH status in females (CpG1*age: P = 0.029). Prediction potential of EH for ADD1 CpG1 and CpG2-5 methylation levels was assessed by the ROC curves. CpG2-5 methylation was shown as a significant predictor of EH in males [Figure 4a, area under curve (AUC) = 0.855, P = 0.001]. CpG1 methylation was also shown with a trend toward indicator in females (Figure 4b, AUC = 0.699, P = 0.054).
Table 2. Characteristics comparison between EH and Non-EH stratified by gender.
| Characteristics | EH Mean± s.e. | Non-EH Mean ± s.e. | p value |
| Men (28) | |||
| Age (years) | 50.1±4.9 | 51.3±6.3 | 0.595 |
| BMI (kg/m2)a | 22.70±2.09 | 22.53±1.41 | 0.846 |
| Smoking (Y/N) | 6/8 | 3/11 | 0.420 |
| Drinking (Y/N) | 4/10 | 2/12 | 0.648 |
| Total cholesterol (mmol/L) | 5.09±0.80 | 4.93±0.92 | 0.630 |
| Total triglycerides (mmol/L) | 1.54±0.98 | 1.34±0.71 | 0.546 |
| Glucose (mmol/L) | 5.31±0.60 | 4.99±0.55 | 0.146 |
| ALT(IU/L) | 30.4±13.9 | 26.9±21.6 | 0.607 |
| AST(IU/L) | 28.0±4.6 | 25.2±9.9 | 0.351 |
| Uric Acid (µmol/L) | 375.0±107.3 | 328.1±67.3 | 0.178 |
| CpG1 methylation (%) | 8.86±1.29 | 9.64±1.45 | 0.133d |
| CpG2-5 methylation (%) | 22.48±6.29 | 31.86±5.65 | 0.008d |
| Women (n = 33) | |||
| Age (years) | 51.3±4.7 | 47.9±5.0 | 0.051 |
| BMI (kg/m2)b | 23.11±3.11 | 21.29±2.60 | 0.137 |
| Smoking (Y/N) | 0/19 | 0/14 | |
| Drinking (Y/N) | 0/19 | 0/14 | |
| Total cholesterol (mmol/L) | 5.09±0.85 | 5.06±1.23 | 0.914 |
| Total triglycerides (mmol/L) | 1.81±1.17 | 1.22±0.44 | 0.058 |
| Glucose (mmol/L) | 5.31±0.60 | 5.06±0.54 | 0.235 |
| ALT (IU/L) | 18.7±11.2 | 10.8±5.2 | 0.012 |
| AST (IU/L)c | 23.4±5.6 | 18.7±3.6 | 0.008 |
| Uric Acid (µmol/L) | 265.6±53.4 | 238.2±60.6 | 0.180 |
| CpG1 methylation (%) | 10.00±1.41 | 11.36±3.63 | 0.042e |
| CpG2-5 methylation (%) | 31.26±6.04 | 31.02±5.11 | 0.557e |
n = 18 (10 EH versus 8 Non-EH);
n = 26 (16 EH versus 10 Non-EH);
n = 31 (19 EH versus 12 Non-EH);
The p-values were adjusted for age, smoking and drinking using logistic-regression analysis.
The p-values were adjusted for age, smoking, drinking, ALT and AST using logistic-regression analysis.
Figure 4. ROC Cure of ADD1 promoter DNA methylation in EH (a, b).
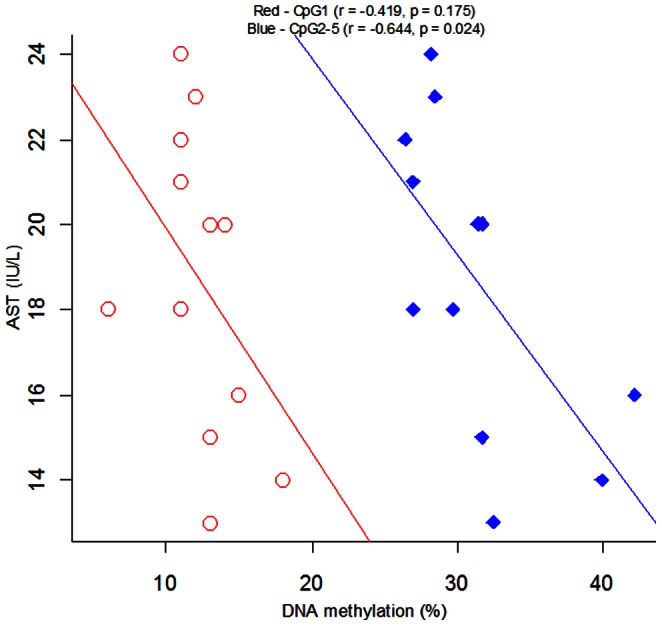
As shown in Table 1, significant differences were observed between males and females for ALT (P = 0.001), AST (P = 0.005) and uric acid (P<0.001). Subsequently, we performed correlation tests between ADD1 DNA methylation levels and metabolic phenotypes including uric acid, ALT, and AST in controls. However, we didn’t find any correlations in controls between ADD1 DNA methylation levels and these metabolic phenotypes (P>0.05, Figure S2a–c). A post hoc analysis was performed for the correlation between ADD1 CpG1 and CpG2-5 methylation levels and metabolic phenotypes (uric acid, ALT, and AST) in females and males. Other than a significant correlation between CpG2-5 and AST in females (r = −0.644, P = 0.024, Figure 5), no significant results observed in the rest of correlation tests (Figure S3a–e).
Figure 5. Pearson correlation between ADD1 methylation and AST in females.
Discussion
The goal of the current study is to evaluate the contribution of ADD1 gene promoter DNA methylation to EH. We found that DNA methylation of ADD1 gene was significantly higher in females than in males. In addition, DNA methylation was shown to be a risk factor of EH in males (CpG2-5) and females (CpG1). To our knowledge, this is the first study showing the association of ADD1 gene promoter DNA methylation with EH. Our comprehensive analysis on the role of ADD1 methylation in the risk of EH may provide new hints to clarify the pathogenesis of EH in future.
Adducin was implicated in the pathogenesis of EH by modulating Na+-K+-ATPase activity [46]–[48]. Evidence indicated that adducin might be a candidate protein to explain genetic alterations in ion transport associated with EH [47]. A previous study reported that hypertensive rat had an increased activity and expression of Na+-K+-pump [48]. In this study, we hypothesized that the aberrant ADD1 methylation may cause hypertension. We speculated that lower ADD1 methylation led to higher expression of α-adducin which resulted in an increased activity and expression of Na+-K+-pump and eventually caused high Na+-reabsorption and hypertension. In accordance with the speculation, we did observe a significant lower level of ADD1 gene promoter methylation in EH cases than in controls.
Interestingly, CpG1 methylation was associated with EH in females, while the CpG2-5 methylation was significantly associated with EH in males. We also found ADD1 methylation was significantly higher in females than in males. In the present study, all subjects were recruited from Han Chinese residents in Ningbo city for at least three generations and diagnosed by generally recognized protocol. The gender and age were well matched between EH cases and Non-EH controls. The power of our association test of EH was 86.5% for ADD1 CpG2-5 in all samples, 66.7% for ADD1 CpG1 in females, and 84.4% for ADD1 CpG2-5 in males. Additionally the power of the analysis stratified by menopausal status was 71% for ADD1 CpG1 and 61.7% for ADD1 CpG2-5. However, our study was only involved with 61 samples and we could not exclude a possibility of spurious association due to the hidden structures in the tested samples. Future replication study with larger size of samples is warranted to confirm our findings.
Sexual dimorphism was found in the whole genome analysis for the risk loci of hypertension in rats [49], [50] and humans [51]. Gender dimorphism was also observed in the association studies of hypertension. These studies addressed ADD1 Gly460Trp polymorphism in female Caucasians [29], CYP19A1 polymorphisms (rs700518, rs10046 and rs4646) in male Japanese [52], SELE T1559C polymorphism [53] and PNMT G390A polymorphism [54] in male Chinese, and other P2RY2 polymorphism (rs4944831) [55]. In the current study, we found that ALT and AST were associated with EH only in females, and that ADD1 CpG1 and CpG2-5 methylation levels were associated with EH in females and males, respectively. Moreover, CpG2-5 was correlated with AST in females, but not in males. These results may be associated with the difference of sex hormones. The role for gender (and/or sex hormones) has been described in mediating differential epigenetic effects on cardiomyocytes [56], effects of radiation [57], [58] and the endocrine system [59]. Significant higher ADD1 methylation levels were found in pre-menopausal women than in post-menopausal ones. Our results provide new clues to explain the sexual dimorphism of EH.
DNA methylation level in humans is reported to alter along with changes of environmental factors such as nutrients [60] and drugs [61], [62]. Therefore, major risk factors for hypertension including physical inactivity, high sodium diet, alcohol consumption and obesity [4], [5] may alter the DNA methylation levels of EH risk genes and cause EH over time. Since lifestyle factors such as smoking, drinking, and physical activity are different between males and females, our findings of gender-dependent ADD1 methylation may reflect the difference in these non-heritable risk factors of EH.
Uric acid was reported positively correlated to the incidence of EH [63]. Previous study also showed uric acid was the risk factor associated with the mortality in hypertensive stroke patients [64]. In addition, uric acid has been reported associated with the prevalence of chronic kidney disease [65]. Raised plasma ALT level was associated with hypertension in Hong Kong Chinese [66]. Another study also reported that elevated ALT and (or) AST were associated with increased risk of pulmonary arterial hypertension [67]. However, ALT and AST have mostly been reported to be associated with hepatic disease [68]–[70], and they were seldom reported to related with EH. In the current study, we didn’t find the associations of uric acid and ALT with ADD1 DNA methylation levels in controls, whereas CpG2-5 was associated with AST in females. Future study is needed to investigate the mechanism underlining this association.
In summary, this study presents a lower ADD1 promoter methylation increases EH risk. ADD1 CpG2-5 methylation is able to predict EH risk in males with higher fidelity than CpG1 methylation does in females. The loss of power for CpG1 may be due to a significant interaction between CpG1 and age in females. Our findings are likely to bring new hints to elaborate the pathogenesis of this complex disease.
Supporting Information
Subgroup analysis in ADD1 promoter DNA methylationa. a: Triangles and circles stand for males and females respectively; blue and red stand for cases and controls, respectively.
(TIF)
Pearson correlation between ADD1 methylation and metabolic phenotypes in controls (A–C).
(TIF)
Pearson correlation between ADD1 methylation and metabolic phenotypes in males (A–C) and in females (D–F).
(TIF)
Primers for ADD1 gene CpG island loci analysis.
(DOC)
Funding Statement
The research was supported by the grants from National Natural Science Foundation of China (31100919), Zhejiang Provincial Natural Science Foundation of China under Grant No.R13H020004, K.C. Wong Magna Fund in Ningbo University, Natural Science Foundation of Ningbo City (2011A610037), Ningbo social development research projects (2012C50032), and Key Program of Education Commission of Zhejiang Province (Z201017918). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. He J, Bazzano LA, Chen CS, Chen J, Rao DC, et al. (2007) GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 21: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Svetkey LP, Harris EL, Martin E, Vollmer WM, Meltesen GT, et al. (2011) Modulation of the BP response to diet by genes in the renin-angiotensin system and the adrenergic nervous system. Am J Hypertens 24: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kurtz TW, Spence MA (1993) Genetics of essential hypertension. Am J Med 94: 77–84. [DOI] [PubMed] [Google Scholar]
- 4. Binder A (2007) A review of the genetics of essential hypertension. Curr Opin Cardiol 22: 176–184. [DOI] [PubMed] [Google Scholar]
- 5. Whelton PK, He J, Appel LJ, Cutler JA, Havas S, et al. (2002) Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. JAMA 288: 1882–1888. [DOI] [PubMed] [Google Scholar]
- 6. Zhang X, Giovannucci EL, Wu K, Smith-Warner SA, Fuchs CS, et al. (2012) Magnesium intake, plasma C-peptide, and colorectal cancer incidence in US women: a 28-year follow-up study. Br J Cancer 106: 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tohidi M, Hatami M, Hadaegh F, Azizi F (2012) Triglycerides and triglycerides to high-density lipoprotein cholesterol ratio are strong predictors of incident hypertension in Middle Eastern women. J Hum Hypertens 26: 525–532. [DOI] [PubMed] [Google Scholar]
- 8. Gilbert JS, Nijland MJ (2008) Sex differences in the developmental origins of hypertension and cardiorenal disease. Am J Physiol Regul Integr Comp Physiol 295: R1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fisher ND, Ferri C, Bellini C, Santucci A, Gleason R, et al. (1997) Age, gender, and non-modulation. A sexual dimorphism in essential hypertension. Hypertension 29: 980–985. [DOI] [PubMed] [Google Scholar]
- 10.Dzudie A, Kengne AP, Muna WF, Ba H, Menanga A, et al.. (2012) Prevalence, awareness, treatment and control of hypertension in a self-selected sub-Saharan African urban population: a cross-sectional study. BMJ Open 2. [DOI] [PMC free article] [PubMed]
- 11. Coatmellec-Taglioni G, Dausse JP, Giudicelli Y, Ribiere C (2002) Gender difference in diet-induced obesity hypertension: implication of renal alpha2-adrenergic receptors. Am J Hypertens 15: 143–149. [DOI] [PubMed] [Google Scholar]
- 12. Silva-Antonialli MM, Tostes RC, Fernandes L, Fior-Chadi DR, Akamine EH, et al. (2004) A lower ratio of AT1/AT2 receptors of angiotensin II is found in female than in male spontaneously hypertensive rats. Cardiovasc Res 62: 587–593. [DOI] [PubMed] [Google Scholar]
- 13. Danser AH, Derkx FH, Schalekamp MA, Hense HW, Riegger GA, et al. (1998) Determinants of interindividual variation of renin and prorenin concentrations: evidence for a sexual dimorphism of (pro)renin levels in humans. J Hypertens 16: 853–862. [DOI] [PubMed] [Google Scholar]
- 14. Bachmann J, Feldmer M, Ganten U, Stock G, Ganten D (1991) Sexual dimorphism of blood pressure: possible role of the renin-angiotensin system. J Steroid Biochem Mol Biol 40: 511–515. [DOI] [PubMed] [Google Scholar]
- 15. Rands VF, Seth DM, Kobori H, Prieto MC (2012) Sexual dimorphism in urinary angiotensinogen excretion during chronic angiotensin II-salt hypertension. Gend Med 9: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rinn JL, Snyder M (2005) Sexual dimorphism in mammalian gene expression. Trends Genet 21: 298–305. [DOI] [PubMed] [Google Scholar]
- 17. Matsuoka Y, Li X, Bennett V (2000) Adducin: structure, function and regulation. Cell Mol Life Sci 57: 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tripodi G, Valtorta F, Torielli L, Chieregatti E, Salardi S, et al. (1996) Hypertension-associated point mutations in the adducin alpha and beta subunits affect actin cytoskeleton and ion transport. J Clin Invest 97: 2815–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casari G, Barlassina C, Cusi D, Zagato L, Muirhead R, et al. (1995) Association of the alpha-adducin locus with essential hypertension. Hypertension 25: 320–326. [DOI] [PubMed] [Google Scholar]
- 20. Manunta P, Cusi D, Barlassina C, Righetti M, Lanzani C, et al. (1998) Alpha-adducin polymorphisms and renal sodium handling in essential hypertensive patients. Kidney Int 53: 1471–1478. [DOI] [PubMed] [Google Scholar]
- 21. Iwai N, Tamaki S, Nakamura Y, Kinoshita M (1997) Polymorphism of alpha-adducin and hypertension. Lancet 350: 369. [DOI] [PubMed] [Google Scholar]
- 22. Kamitani A, Wong ZY, Fraser R, Davies DL, Connor JM, et al. (1998) Human alpha-adducin gene, blood pressure, and sodium metabolism. Hypertension 32: 138–143. [DOI] [PubMed] [Google Scholar]
- 23. Busch CP, Harris SB, Hanley AJ, Zinman B, Hegele RA (1999) The ADD1 G460W polymorphism is not associated with variation in blood pressure in Canadian Oji-Cree. J Hum Genet 44: 225–229. [DOI] [PubMed] [Google Scholar]
- 24. Ramu P, Umamaheswaran G, Shewade DG, Swaminathan RP, Balachander J, et al. (2010) Gly460Trp polymorphism of the ADD1 gene and essential hypertension in an Indian population: A meta-analysis on hypertension risk. Indian J Hum Genet 16: 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu K, Liu J, Huang Y, Liu Y, Lou Y, et al.. (2010) Alpha-adducin Gly460Trp polymorphism and hypertension risk: a meta-analysis of 22 studies including 14303 cases and 15961 controls. PLoS One 5. [DOI] [PMC free article] [PubMed]
- 26. Niu W, Qi Y (2011) Association of alpha-adducin and G-protein beta3 genetic polymorphisms with hypertension: a meta-analysis of Chinese populations. PLoS One 6: e17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu K, Liu Y, Liu J, Wang Z, Lou Y, et al. (2011) alpha-adducin Gly460Trp polymorphism and essential hypertension risk in Chinese: a meta-analysis. Hypertens Res 34: 389–399. [DOI] [PubMed] [Google Scholar]
- 28. Li YY (2012) alpha-Adducin Gly460Trp gene mutation and essential hypertension in a Chinese population: a meta-analysis including 10,960 subjects. PLoS One 7: e30214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang JG, Staessen JA, Barlassina C, Fagard R, Kuznetsova T, et al. (2002) Association between hypertension and variation in the alpha- and beta-adducin genes in a white population. Kidney Int 62: 2152–2159. [DOI] [PubMed] [Google Scholar]
- 30. Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, et al. (1984) Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci U S A 81: 2275–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morita S, Takahashi RU, Yamashita R, Toyoda A, Horii T, et al. (2012) Genome-Wide Analysis of DNA Methylation and Expression of MicroRNAs in Breast Cancer Cells. Int J Mol Sci 13: 8259–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grutzmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, et al. (2008) Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One 3: e3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeVos T, Tetzner R, Model F, Weiss G, Schuster M, et al. (2009) Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 55: 1337–1346. [DOI] [PubMed] [Google Scholar]
- 34. Muller HM, Fiegl H, Widschwendter A, Widschwendter M (2004) Prognostic DNA methylation marker in serum of cancer patients. Ann N Y Acad Sci 1022: 44–49. [DOI] [PubMed] [Google Scholar]
- 35. Muller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, et al. (2003) DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res 63: 7641–7645. [PubMed] [Google Scholar]
- 36. Guay SP, Brisson D, Munger J, Lamarche B, Gaudet D, et al. (2012) ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics 7: 464–472. [DOI] [PubMed] [Google Scholar]
- 37.Xu H, Wang B, Su D, Yu Q, Li Q, et al.. (2012) The DNA methylation profile of PLA2G4C gene promoter in schizophrenia. Psychiatry Res. [DOI] [PubMed]
- 38.Kinoshita M, Numata S, Tajima A, Shimodera S, Ono S, et al.. (2012) DNA Methylation Signatures of Peripheral Leukocytes in Schizophrenia. Neuromolecular Med. [DOI] [PubMed]
- 39. Smolarek I, Wyszko E, Barciszewska AM, Nowak S, Gawronska I, et al. (2010) Global DNA methylation changes in blood of patients with essential hypertension. Med Sci Monit 16: CR149–155. [PubMed] [Google Scholar]
- 40. Kulkarni A, Chavan-Gautam P, Mehendale S, Yadav H, Joshi S (2011) Global DNA methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell Biol 30: 79–84. [DOI] [PubMed] [Google Scholar]
- 41. Friso S, Pizzolo F, Choi SW, Guarini P, Castagna A, et al. (2008) Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis 199: 323–327. [DOI] [PubMed] [Google Scholar]
- 42. Jiang Q, Yuan H, Xing X, Liu J, Huang Z, et al. (2011) Methylation of adrenergic beta1 receptor is a potential epigenetic mechanism controlling antihypertensive response to metoprolol. Indian J Biochem Biophys 48: 301–307. [PubMed] [Google Scholar]
- 43. 2003 European Society of Hypertension-European Society of Cardiology guidelines for the management of arterial hypertension. J Hypertens 21: 1011–1053. [DOI] [PubMed] [Google Scholar]
- 44. Perloff D, Grim C, Flack J, Frohlich ED, Hill M, et al. (1993) Human blood pressure determination by sphygmomanometry. Circulation 88: 2460–2470. [DOI] [PubMed] [Google Scholar]
- 45. Dupont WD, Plummer WD Jr (1990) Power and sample size calculations. A review and computer program. Control Clin Trials 11: 116–128. [DOI] [PubMed] [Google Scholar]
- 46. Ferrandi M, Salardi S, Tripodi G, Barassi P, Rivera R, et al. (1999) Evidence for an interaction between adducin and Na(+)-K(+)-ATPase: relation to genetic hypertension. Am J Physiol 277: H1338–1349. [DOI] [PubMed] [Google Scholar]
- 47. Bianchi G, Tripodi G, Casari G, Salardi S, Barber BR, et al. (1994) Two point mutations within the adducin genes are involved in blood pressure variation. Proc Natl Acad Sci U S A 91: 3999–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferrandi M, Tripodi G, Salardi S, Florio M, Modica R, et al. (1996) Renal Na,K-ATPase in genetic hypertension. Hypertension 28: 1018–1025. [DOI] [PubMed] [Google Scholar]
- 49. Deng AY, Menard A, Xiao C, Roy J (2008) Sexual dimorphism on hypertension of quantitative trait loci entrapped in Dahl congenic rats. Clin Exp Hypertens 30: 511–519. [DOI] [PubMed] [Google Scholar]
- 50. Yagil C, Sapojnikov M, Kreutz R, Katni G, Lindpaintner K, et al. (1998) Salt susceptibility maps to chromosomes 1 and 17 with sex specificity in the Sabra rat model of hypertension. Hypertension 31: 119–124. [DOI] [PubMed] [Google Scholar]
- 51. Seda O, Tremblay J, Gaudet D, Brunelle PL, Gurau A, et al. (2008) Systematic, genome-wide, sex-specific linkage of cardiovascular traits in French Canadians. Hypertension 51: 1156–1162. [DOI] [PubMed] [Google Scholar]
- 52. Shimodaira M, Nakayama T, Sato N, Saito K, Morita A, et al. (2008) Association study of aromatase gene (CYP19A1) in essential hypertension. Int J Med Sci 5: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang ZG, Niu QL, Gu W, Liu K, Lou YQ, et al. (2011) [Association of C602A and T1559C polymorphisms of E-selectin gene and essential hypertension]. Zhonghua Yi Xue Za Zhi 91: 1238–1241. [PubMed] [Google Scholar]
- 54. Chen A, Chen X, Shi R, Guo Y, Chen L, et al. (2009) [Association of genetic polymorphism in phenylethanolamine-N-methyl transferase with essential hypertension in Changsha Han people]. Zhong Nan Da Xue Xue Bao Yi Xue Ban 34: 1120–1125. [PubMed] [Google Scholar]
- 55. Wang Z, Nakayama T, Sato N, Izumi Y, Kasamaki Y, et al. (2010) The purinergic receptor P2Y, G-protein coupled, 2 (P2RY2) gene associated with essential hypertension in Japanese men. J Hum Hypertens 24: 327–335. [DOI] [PubMed] [Google Scholar]
- 56. Sebag IA, Gillis MA, Calderone A, Kasneci A, Meilleur M, et al. (2011) Sex hormone control of left ventricular structure/function: mechanistic insights using echocardiography, expression, and DNA methylation analyses in adult mice. Am J Physiol Heart Circ Physiol 301: H1706–1715. [DOI] [PubMed] [Google Scholar]
- 57. Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O (2004) Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem Biophys Res Commun 325: 39–47. [DOI] [PubMed] [Google Scholar]
- 58. Koturbash I, Kutanzi K, Hendrickson K, Rodriguez-Juarez R, Kogosov D, et al. (2008) Radiation-induced bystander effects in vivo are sex specific. Mutat Res 642: 28–36. [DOI] [PubMed] [Google Scholar]
- 59. Zhang X, Ho SM (2011) Epigenetics meets endocrinology. J Mol Endocrinol 46: R11–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reynolds E (2006) Vitamin B12, folic acid, and the nervous system. Lancet Neurol 5: 949–960. [DOI] [PubMed] [Google Scholar]
- 61. Dong E, Nelson M, Grayson DR, Costa E, Guidotti A (2008) Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A 105: 13614–13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Melas PA, Rogdaki M, Osby U, Schalling M, Lavebratt C, et al. (2012) Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J 26: 2712–2718. [DOI] [PubMed] [Google Scholar]
- 63.Chen DL, Zhang CJ, Fu YH, Mo YJ, Chen FR (2010) [Correlation of angiotensin-converting enzyme 2 gene polymorphisms to essential hypertension and ischemic stroke]. Nan Fang Yi Ke Da Xue Xue Bao 30: 1890–1892, 1895. [PubMed]
- 64. Mo YJ, Huang WH, Chen DL, Chen FR (2010) [Relationship of angiotensin-converting enzyme 2 gene polymorphism with the prognosis of hypertensive stroke patients]. Nan Fang Yi Ke Da Xue Xue Bao 30: 84–87. [PubMed] [Google Scholar]
- 65.Jalal DI, Chonchol M, Chen W, Targher G (2012) Uric Acid as a Target of Therapy in CKD. Am J Kidney Dis. [DOI] [PMC free article] [PubMed]
- 66. Cheung BM, Ong KL, Tso AW, Cherny SS, Sham PC, et al. (2011) Gamma-glutamyl transferase level predicts the development of hypertension in Hong Kong Chinese. Clin Chim Acta 412: 1326–1331. [DOI] [PubMed] [Google Scholar]
- 67. Benza RL, Barst RJ, Galie N, Frost A, Girgis RE, et al. (2008) Sitaxsentan for the treatment of pulmonary arterial hypertension: a 1-year, prospective, open-label observation of outcome and survival. Chest 134: 775–782. [DOI] [PubMed] [Google Scholar]
- 68. Razavizade M, Jamali R, Arj A, Talari H (2012) Serum parameters predict the severity of ultrasonographic findings in non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int 11: 513–520. [DOI] [PubMed] [Google Scholar]
- 69.Ma L, Xie C, Ran Y, Liang X, Huang L, et al.. (2012) Synthesis, and Biological Evaluation of 5-Benzylidenepyrimidine-2,4,6(1H,3H,5H)-trione Derivatives for the Treatment of Obesity-related Nonalcoholic Fatty Liver Disease. J Med Chem. [DOI] [PubMed]
- 70. Ikai E, Ishizaki M, Suzuki Y, Ishida M, Noborizaka Y, et al. (1995) Association between hepatic steatosis, insulin resistance and hyperinsulinaemia as related to hypertension in alcohol consumers and obese people. J Hum Hypertens 9: 101–105. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Subgroup analysis in ADD1 promoter DNA methylationa. a: Triangles and circles stand for males and females respectively; blue and red stand for cases and controls, respectively.
(TIF)
Pearson correlation between ADD1 methylation and metabolic phenotypes in controls (A–C).
(TIF)
Pearson correlation between ADD1 methylation and metabolic phenotypes in males (A–C) and in females (D–F).
(TIF)
Primers for ADD1 gene CpG island loci analysis.
(DOC)