Abstract
Urea destabilizes helical and folded conformations of nucleic acids and proteins, as well as protein-nucleic acid complexes. To understand these effects, extend previous characterizations of interactions of urea with protein functional groups, and thereby develop urea as a probe of conformational changes in protein and nucleic acid processes, we obtain chemical potential derivatives (μ23 = dμ2/dm3) quantifying interactions of urea (component 3) with nucleic acid bases, base analogs, nucleosides and nucleotide monophosphates (component 2) using osmometry and hexanol-water distribution assays. Dissection of these μ23 yields interaction potentials quantifying interactions of urea with unit surface areas of nucleic acid functional groups (heterocyclic aromatic ring, ring methyl, carbonyl and phosphate O, amino N, sugar (C,O)); urea interacts favorably with all these groups, relative to interactions with water. Interactions of urea with heterocyclic aromatic rings and attached methyl groups (as on thymine) are particularly favorable, as previously observed for urea-homocyclic aromatic ring interactions. Urea m-values determined for double helix formation by DNA dodecamers near 25°C are in the range 0.72 to 0.85 kcal mol−1 m−1 and exhibit little systematic dependence on nucleobase composition (17–42% GC). Interpretation of these results using the urea interaction potentials indicates that extensive (60–90%) stacking of nucleobases in the separated strands in the transition region is required to explain the m-value. Results for RNA and DNA dodecamers obtained at higher temperatures, and literature data, are consistent with this conclusion. This demonstrates the utility of urea as a quantitative probe of changes in surface area (ΔASA) in nucleic acid processes.
Introduction
Noncovalent self-assembly and function of proteins and nucleic acids involve large-scale changes in water accessible surface area (|ΔASA| >> 0). Solutes affect these processes because of their favorable or unfavorable interactions with the functional groups on the biopolymer surface that comprises the ΔASA. Once the interactions of solutes with protein and nucleic acid functional groups are quantified, these solutes can be used as probes to determine the amount and composition of the biopolymer surface exposed or buried in individual steps of a process.1 Myers et al. observed that urea protein unfolding m-values (derivatives of the standard free energy change ΔG°obs for unfolding with respect to urea concentration) are approximately proportional to the change in water accessible surface area (ΔASA) in unfolding, calculated using an extended chain model of the unfolded state. Analysis of effects of urea, other solutes and Hofmeister salts on unfolding and other processes using the solute partitioning model (SPM) predicts the proportionality of solute m-values to ΔASA, and demonstrates that the molecular basis of these thermodynamic effects is the accumulation (favorable interaction) or exclusion (unfavorable interaction, relative to water) of the solute or salt ion in the vicinity of the chemical functional groups exposed upon unfolding.1,3–6
Effects (m-values) of urea, glycine betaine and different Hofmeister salts on protein stability have been quantitatively interpreted in terms of the chemical interactions of these solutes and salt ions with the chemical functional groups exposed in unfolding (primarily aliphatic and aromatic hydrocarbon (~70%, with ~9:1 aliphatic: aromatic ratio) and amide oxygen and nitrogen (~20%, with ~2:1 O:N ratio ).1,3–9 We recently quantified the preferential interactions of urea with the seven most significant protein functional groups (amide O and N, aliphatic and aromatic C, hydroxyl O, carboxylate O, cationic N) relative to interactions with water and found that urea interacts most favorably with amide O and aromatic C.4 For amide O, this result indicates that hydrogen bonds formed with urea as the donor and amide O as the acceptor are more favorable than the corresponding hydrogen bonds with water; spectroscopic studies provide independent evidence for urea-amide hydrogen bonds.10,11 Urea m-values for protein unfolding and other protein processes are quantitatively predicted using these data.1,4 For globular protein unfolding, favorable interactions of urea with amide O, aromatic C and aliphatic C groups exposed in unfolding are predicted to make similar contributions to the urea m-value.4
In this work, we determine the chemical and thermodynamic basis of urea destabilization of DNA and RNA structures. Urea lowers the transition temperature of nucleic acid secondary and tertiary structures;12–16 for RNA duplexes, this destabilization has been correlated with the surface area exposed in the unfolding transition.17,18 Urea does not affect the cooperativity of melting or the structure of the folded form.17,19,20 While a perturbing effect of urea on stacking in single stranded DNA/RNA would be expected, evidence for this has not been obtained (see discussion).17
Carbonyl O and amino N groups on nucleic acid bases (nucleobases) are similar in hydrogen bonding potential to amide O and N of proteins, and the heterocyclic aromatic ring is analogous to the homocyclic aromatic ring of Phe or Tyr. Nucleic acid melting exposes ASA that is approximately 30% C and 70% O and N,1 the opposite composition to that exposed in unfolding globular proteins, so interactions of urea with polar (O, N) surface are expected to be more important determinants of the effect of urea on nucleic acid stability. By quantifying the interaction of urea with DNA and RNA backbone and nucleobase chemical functional groups, we gain insight into the molecular and thermodynamic origins of the effect of urea on DNA and RNA processes. Future comparative studies with primary and secondary amides will allow us to dissect these interactions into contributions from amide O and N, and provide insight into the thermodynamic contributions of interactions of amide groups on proteins with nucleobases and the sugar-phosphate backbone of nucleic acids.
At sub-denaturing concentrations, urea exhibits very large effects on protein-nucleic acid processes including binding of lac repressor protein to operator DNA21 and the late steps in the mechanism of formation of a transcription complex between RNA polymerase and promotor DNA.22 Knowledge of how urea interacts with both protein and nucleic acid functional groups is allowing us to predict and interpret the effect of urea on these processes in terms of interface formation and coupled folding or other conformational changes. In the longer term, urea will be used as one of a set of solute probes (including, glycine betaine, proline, trifluoroethanol, glycerol, and tetraethyleneglycol, once characterization of their interactions with protein and nucleic acid functional groups is complete) to quantify the amount and chemical composition of the ΔASA in individual steps of mechanisms of action of protein molecular machines.
Here we determine the interaction of urea with nucleic acid functional groups by quantifying the interactions of urea with model compounds representing nucleic acid surface types (nucleic acid bases/base analogs, nucleosides and nucleotide monophosphates) using vapor pressure osmometry (VPO) and measurements of the distribution of nucleobase between water and hexanol. We dissect interactions into interactions with individual functional groups using an ASA based analysis, which are interpreted in terms of noncovalent interactions.4 We determine the effect of urea on RNA and DNA duplex formation and interpret the m-values using individual functional group interactions to gain insight into the conformational changes involved in this process.
Thermodynamic Background and Analysis
The effect of solutes on biopolymer processes like nucleic acid helix formation in aqueous solution are quantified using m-values, derivatives of the standard free energy change (AG°obs=−RTlnKobs) for the process with respect to solute concentration:
| (1) |
where Kobs is the equilibrium concentration quotient of products and reactants in the process, Kγ is the corresponding quotient of activity coefficients (γ) describing the interaction of products and reactants with solutes and μ23 is the chemical potential derivative (the subscripts 1, 2 and 3 refer to water, biopolymer/model compound, and urea, respectively, throughout this article). The value of μ23 is a measure of the preferential interaction of a solute with a biopolymer or model compound where a negative value indicates a favorable preferential interaction and a positive value indicates an unfavorable preferential interaction relative to interactions with water.
Values of μ23/RT for interactions of urea with model compounds containing protein surface types have been obtained by solubility, micelle formation or VPO assays4; none of these assays are useful to quantify interactions of urea with nucleobases and base analogs. Here, a two-phase assay is developed to measure KDWH = (m2hex/m2aq)eq, the distribution of a nucleic acid base or base analog between hexanol- and water-rich solutions. Paralleling the thermodynamic analysis in Eq. 1:
| (2) |
where m3aq is the urea concentration in the aqueous phase. The approximation in Eq. 2 is discussed in supplemental.
Values of μ23/RT for protein model compounds were dissected in an ASA based analysis (motivated by refs 3,6 ) to determine interactions of urea with individual protein functional groups and inorganic ions.4 Here, we use this same analysis to determine interactions of urea with individual nucleic acid functional groups by dissecting experimentally determined μ23/RT values for the interaction of urea with Na2NMPs, nucleosides, nucleobases and base analogs determined by VPO and distribution assays into additive contributions from the interaction of urea with each nucleic acid functional group, as well as with the two Na+ ions for the 5’-NMPs:
| (3) |
where αi represents the contribution to μ23/RT from the interaction of urea with a unit surface area of surface type i. The term νNa+βNa+ represents the contribution of ν Na+ ions per formula unit (2 for the 5’-NMP salts, 0 for nonelectrolytes), where βNa+ is the contribution of the interaction of urea with a Na+ ion to μ23/RT, determined by analysis of VPO data for the interaction of urea with Na carboxylates, amino acids and NaCl4. The assumption of additivity of contributions to μ23 is supported by recent studies of interactions of Hofmeister salts,5 urea,4 and glycine betaine6 with model compounds displaying the functional groups of proteins, as well as interactions of oligomers of ethylene glycol with the surface exposed in melting nucleic acid oligomers.23 Deviations from additivity are of interest as examples of context-dependence arising from simultaneous interactions of a solute with two or more functional groups on the model compound or biopolymer.
Experimental Section
5'-NMP Vapor Pressure Osmometry
Details of sample preparation are described in supplemental. A Wescor Vapro 5520 vapor pressure osmometer operating at ambient temperature was calibrated using standard osmolality solutions from Wescor. All osmolalities were corrected for discrepancies between accepted literature values and the Wescor standard solutions of sodium chloride.24 Triplicate osmolality measurements were made on each sample and the standard errors determined as in Hong et al.24
Values of μ23 for urea (component 3)-Na2NMP (component 2) interaction at constant NaCl (component 4) concentration were obtained from osmolality data on four, three and two component solutions as follows:
| (4) |
| (5) |
This generalization of the three component ΔOsm equation to the situation where a constant background of a fourth component (here NaCl) is present was previously used for DNA osmometry by Hong et al.25 Error propagation in Δ23,4Osm used eq 3.13 in Bevington and Robinson26 with standard errors and covariances of correlated parameters from multiple linear regression of the Osm(m2,m3,m4) and (Osm(m3,m4)) data (eqs 7.14–7.17, 7.19, 7.25 in Bevington and Robinson26 ). To check if the effect of urea on NaCl activity has a significant effect, μ34 was calculated as described in ref23 using urea-salt data from ref 4 and found to be negligibly small (<4% of μ23,4/RT).
Nucleic Acid Base and Base Analog Distribution Assay
Series of 10 mL aqueous urea solutions with concentrations ranging from 0–4 molal were prepared gravimetrically. The nucleobase or base analog was dissolved in hexanol, and 2 mL of this solution were added to each urea solution. Samples were incubated in a 25°C shaking water bath for up to 24 hours; the same concentration ratio was observed if the samples were read anywhere between 1–24 hours after preparation. Absorbances of the water and hexanol phases were measured in a Cary 1 UV-visible spectrophotometer.
From these data, we determine the equilibrium constant (distribution coefficient) KDWH (Eq. 2) characterizing the distribution of the nucleobase between aqueous urea solutions and hexanol; absorbances (directly related to molar concentration) are converted to molal concentration to determine this molal scale concentration ratio as described in supplemental. Values of μ23/RT were determined from initial slopes of plots of lnKDWH vs urea molality (Eq. 2). See Fig. S1 for sample plots of ln(m/m0 ) vs aqueous urea concentration for the hexanol and water phases and the resulting lnKDWH vs aqueous urea concentration plot. As controls for the validity of this assay and the approximation in Eq. 2, results of the distribution assay are compared with results with results of solubility assays for nucleobase-salt interactions and with VPO results for nucleoside-urea interactions in supplemental.
DNA and RNA Dodecamer Thermal Denaturation and Urea Titration
Details of DNA and RNA dodecamer sample preparation are given in supplemental. For thermal melts and urea titrations, dodecamer duplex transitions were monitored at 260 nm using a Cary 100 UV-visible spectrophotometer (Varian) equipped with a Peltier temperature controller. For thermal melts, dodecamer duplex samples were heated at a rate of 0.3 °C/min and absorbance readings were collected every 0.2 °C. For urea titrations, absorbance readings were collected as urea was added and the absorbance was normalized to molar DNA concentration. Dodecamer duplex and single-stranded baseline regions in the absorbance melting profiles were fit by linear regression (slopes were less than 1% of the overall absorbance change between duplex and single stranded states); the fraction of single stranded (fss) dodecamer was determined from the difference in absorbance between the experimentally measured values and the extrapolated baseline values.
The observed equilibrium constant Kobs for duplex formation from the single strands S1 and S2 (S1 + S2→duplex) can be determined from the total strand concentration ([str]tot=[S1]+[S2]+[duplex]) and fss:
| (6) |
For titrations, Kobs is determined over the range of urea concentrations where 0.2<fss<0.8. For thermal melts, van’t Hoff plots were made of ln(Kobs) vs 1/T in the T range where 0.2<fss<0.8 (the slope is ΔH°obs/R). The value of Kobs used in m-value analysis is interpolated from these plots at the temperature where fss=0.2 in 0 m urea (this temperature is in the 0.2<fss<0.8 transition region for all urea concentrations studied). Linear regression of the natural log of Kobs with urea molarity was used to calculate the m-value (eq. 1). Buffer and salt molalities were held constant in these experiments; urea has no effect on the salt dependence of DNA melting (supplemental).
ASA Calculations
5'-NMP, nucleobase/base analog and nucleoside solvent-accessible surface areas (ASA) were calculated using Surface Racer27 with a probe radius of 1.4 Å and the set of van der Waals radii from Richards.28 Coordinate files for the compounds were obtained from NMR solution structures from the Biological Magnetic Resonance Data Bank. The 5’-NMP nucleobases were in the anti conformation, although the difference in ASA between 5'-NMPs in either syn or anti conformations was less than 5%.
The DNA surface area newly exposed in unfolding (ΔASA) for each dodecamer duplex in Table 1 was calculated for three models of the single stranded oligomers, assuming nucleobases were stacked, half-stacked, and fully-unstacked.25 The xleap module in AMBER 1029 was used to construct the B- and A-forms of the DNA and RNA dodecamer duplexes, respectively. The ASA of the duplex and two single strands in either the B- (DNA) or A-form (RNA) conformation were calculated using naccess30 with the same parameters used to calculate 5'-NMP ASA. Single strands stripped out of helices in the B- or A-form have stacked nucleobases. To make unstacked strands, the torsion angles about the O3' – P bonds were rotated 120 degrees in PyMol31 to break up any nucleobase stacking starting at the 5' end of the single strands. The ASA for a single strand in the half-stacked model was obtained by averaging the ASA for stacked and unstacked single strands. The ΔASA for unfolding a duplex was then calculated by summing the ASA of the two single strands and subtracting the ASA of the duplex.
Table 1.
Values of μ23/RT for interactions of urea with model compounds.
| Model Compound | Experimental μ23/RT (m−1) ± SD | Predicted μ23/RT (m−1) ± SD |
|---|---|---|
| Nucleobases | ||
| Cytosine | −0.17 ± 0.04 | −0.19 ± 0.02 |
| Uracil | −0.22 ± 0.02 | −0.22 ± 0.02 |
| Thymine | −0.23 ± 0.01 | −0.26 ± 0.02 |
| Purine | −0.26 ± 0.01 | −0.28 ± 0.02 |
| Adenine | −0.27 ± 0.01 | −0.25 ± 0.02 |
| Hypoxanthine | −0.28 ± 0.01 | −0.27 ± 0.02 |
| Theobromine | −0.37 ± 0.02 | −0.35 ± 0.02 |
| Theophylline | −0.37 ± 0.02 | −0.35 ± 0.02 |
| Caffeine | −0.40 ± 0.02 | −0.40 ± 0.03 |
| Mononucleotides (Na2NMP) | ||
| 2Na+ + 5’-CMP2− | −0.102 ± 0.014 | −0.10 ± 0.04 |
| 5’-CMP2− | −0.299 ± 0.031a | −0.30 ± 0.05 |
| 2Na+ + 5’-AMP2− | −0.126 ± 0.012 | −0.16 ± 0.04 |
| 5’-AMP2− | −0.323 ± 0.030a | −0.36 ± 0.05 |
| 2Na+ + 5’-GMP2− | −0.140 ± 0.016 | −0.13 ± 0.04 |
| 5’-GMP2− | −0.337 ± 0.032a | −0.33 ± 0.05 |
| 2Na+ + 5’-dTMP2− | −0.148 ± 0.013 | −0.17 ± 0.04 |
| 5’-dTMP2− | −0.345 ± 0.031a | −0.37 ± 0.05 |
| 2Na+ + 5’-UMP2− | −0.166 ± 0.012 | −0.12 ± 0.04 |
| 5’-UMP2− | −0.363 ± 0.057a | −0.32 ± 0.05 |
| Nucleosides | ||
| Guanosine | −0.31 ± 0.03 | −0.31 ± 0.03 |
| Uridine | −0.31 ± 0.02b | −0.31 ± 0.03 |
| deoxyThymidine | −0.31 ± 0.02c | −0.33 ± 0.02 |
| Adenosine | −0.35 ± 0.02 | −0.34 ± 0.03 |
Determined by subtracting contribution of interaction with 2Na+ (2βNa+ = 0.197 ± 0.028 m−1) predicted from model compound data in ref4 from experimental value.
Average of μ23/RT values obtained from VPO (−0.34 ± 0.02 m−1) and hexanol-water distribution assays (−0.28 ± 0.02 m−1).
Average of μ23/RT values obtained from VPO (−0.33 ± 0.03 m−1) and hexanol-water distribution assays (−0.29 ± 0.02 m−1).
Determining αi Values
The ASA of the 5’-NMPs, nucleobases/base analogs and nucleosides is divided into six distinct surface types: anionic phosphate O, sugar C and O, heterocyclic aromatic ring C and N, and the functional groups on the heterocyclic rings (carbonyl O, amino N, aliphatic C). Even though the heterocyclic aromatic ring contains both C and N atoms, of necessity it is treated as one surface type because the ASA ratio of aromatic C/N is similar in all the model compounds we studied. Likewise, the ASA ratio of sugar C and O in all the nucleosides and nucleotides studied is similar so is treated as one surface type. We determine the ASA of each type of surface on the model compounds from structural data as described in above.
Values of μ23/RT for model compounds in Table 1 and model compound ASA data from Table S1 were globally fit to eq. 3 using multiple linear regression in Igor 5.04B to determine αi values for interaction of urea with individual nucleic acid functional groups. For the urea-Na+ interaction, the value of βNa+ (0.099±0.014 m−1) was determined from model compound data in ref 4 reanalyzed as described in supplemental. Nucleobase and base analog partitioning data were also fit to eq. 3 as above without the nucleoside partitioning data or 5’-NMP VPO data. The resulting interaction potentials for the interactions of urea with surface types found on these nucleobases and analogs (ring methyl, aromatic ring, carbonyl O, amino N) were the same within error as those determined from the fit including the nucleoside and 5’-NMP data.
Results and Discussion
Interactions of Urea with Model Compounds; Distribution Studies
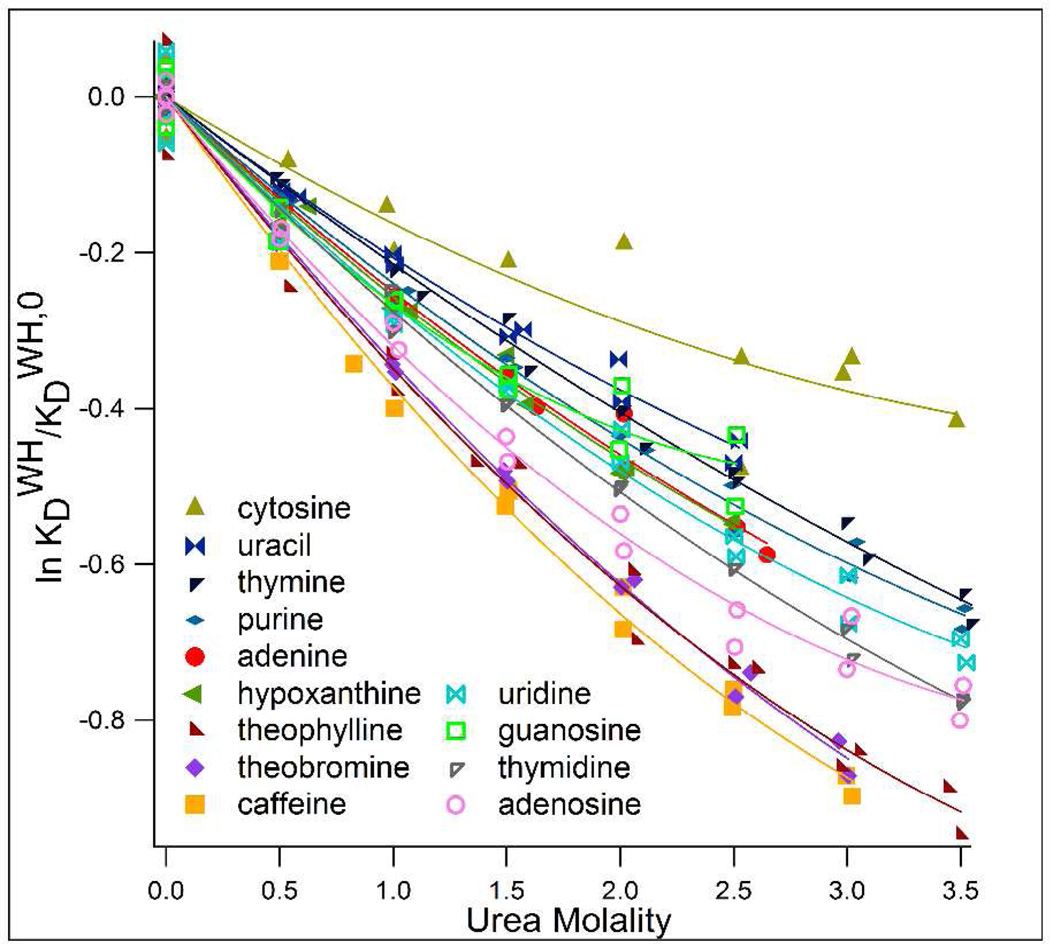
Interactions of urea with the sparingly soluble nucleosides and nucleic acid bases and base analogs were determined from the effect of urea concentration on the distribution of the nucleobase between a hexanol-rich phase and predominantly aqueous urea solutions. In the absence of urea, all nucleobases and base analogs concentrate in the hexanol-rich phase; the distribution of the nucleobase is quantified by the waterhexanol distribution coefficient KDWH. Fig. 1 plots the natural logarithm of KDWH (normalized to KDWH in the absence of urea) against urea concentration (Eq. 2). The negative slopes here indicate that urea (which is concentrated in the aqueous phase, see supplemental) increases the concentration of the nucleobase, base analog or nucleoside in the aqueous phase and therefore has a favorable interaction with these compounds, as is the case for interactions of urea with other nonelectrolytes studied4. Values of μ23/RT were determined from initial slopes of quadratic fits to these plots (see supplemental for details) and are summarized in Table 1. The distribution assay was adopted because most nucleosides and nucleobases/base analogs are too insoluble for VPO and too slow (~2 weeks) to achieve solubility equilibrium before artifacts (perhaps arising from degradation of urea32) become significant. Long and McDevit33 described the use of distribution assays to determine the effect of Hofmeister salts on nonpolar solute activity, which have been used to quantify the interaction of Hofmeister salts with formamide34 and with nitrobenzoic acids, where the same results were obtained from solubility and distribution experiments35. Distribution assays of this type have also been used to quantify the hydrophobicity of carboxylic acids and nucleic acid bases from their distribution between heptane and water,36,37 and to estimate the solubility of various compounds in water from the correlation of solubility with octanol-water distribution coefficients38,39.
Figure 1.
Plots of the logarithm of the macroscopic distribution coefficient ratio KDWH/KDWH,0 vs urea molality where KDWH is the molal scale distribution coefficient (m2hex/m2aq)eq for a nucleobase or base analog between hexanol-rich and water-rich phases in the presence of urea and KDWH,0 is the distribution coefficient for the nucleobase between hexanol-rich and water-rich phases in the absence of urea; initial slopes are used to determine μ23/RT (Eq. 2).
Vapor Pressure Osmometry Studies
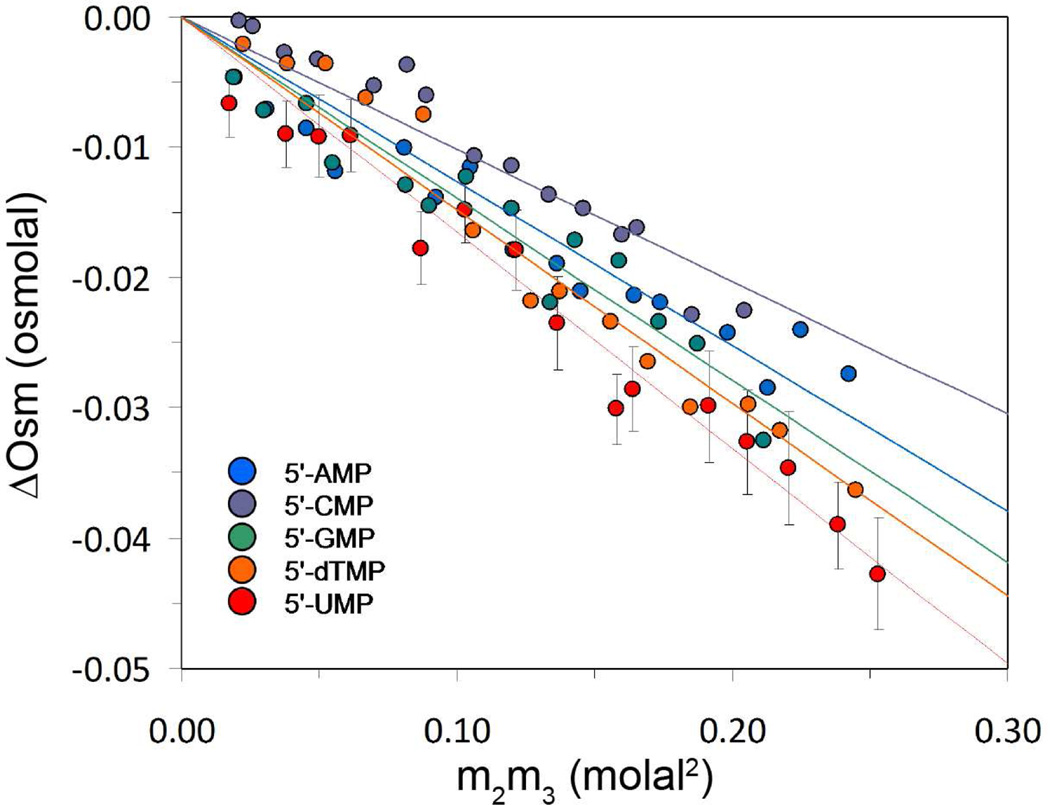
Interactions of urea with the modestly soluble disodium salts of the 5'-NMPs were quantified from VPO measurements by determining the osmolality difference ΔOsm (Eq. 4) as a function of m2m3, the product of the 5'-NMP and urea molalities (Figure 2). Linear regression of the data in Figure 3 with intercepts fixed at zero yields values of μ23/RT (Eq. 4) summarized in Table 1. The negative slopes in Figure 2 indicate a net favorable preferential interaction of urea with the 5'-NMP anion and the 2 Na+ ions.
Figure 2.
Excess osmolality ΔOsm (Eq. 5) of disodium salts of 5'-NMPs in aqueous urea solutions as a function of m2m3, the product of molalities of 5'-NMP (m2) and urea (m3); slopes are μ23/RT (Eq. 4).
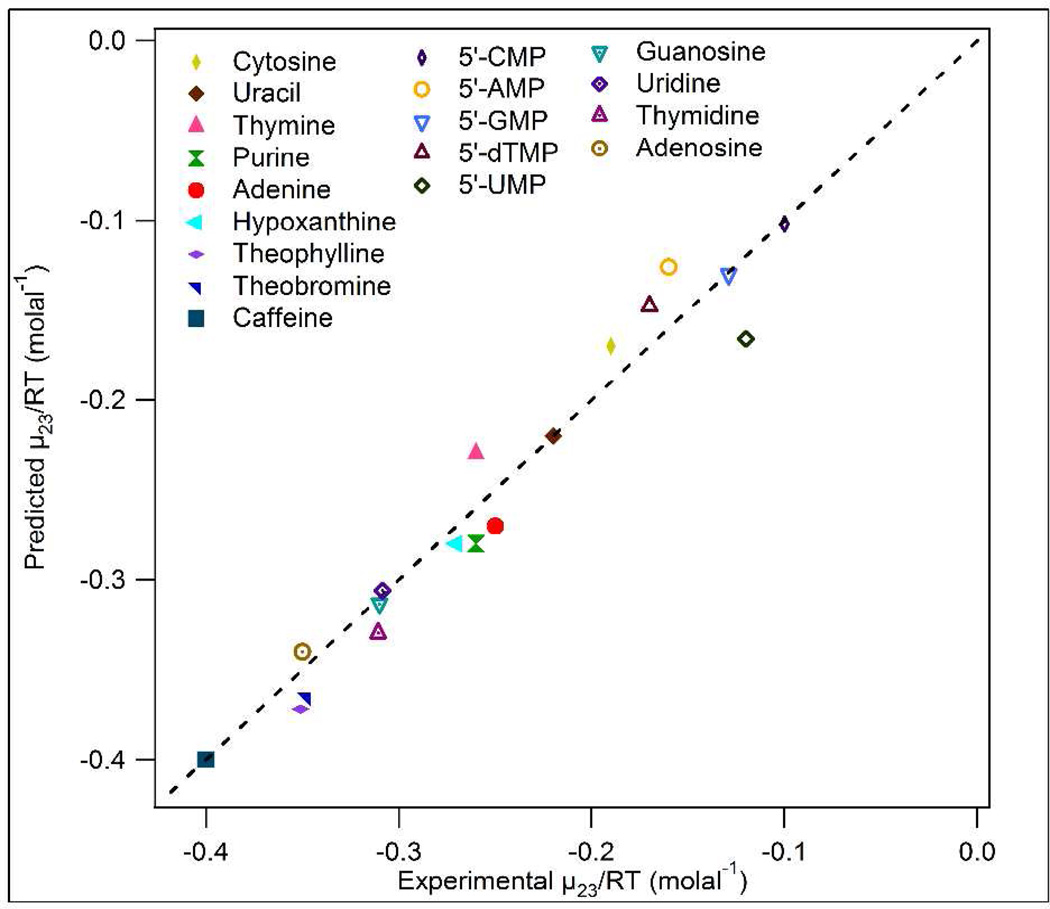
Figure 3.
Plot of predicted vs observed values of μ23/RT for interactions of urea with model compounds (see Table 1). Line represents equality of predicted and observed values.
Interpreting Preferential Interactions (μ23/RT values) Between Urea and Nucleic Acids
Values of μ23/RT for interactions between urea and nucleobases/base analogs/nucleosides/nucleotides obtained from the slopes of plots of VPO and partitioning data (Figs 2 and 3) are summarized in Table 1. The groups in Table 1 (nucleobases, nucleosides, mononucleotides) are arranged in order of increasing molecular weight and ASA (Table S1). Since interactions of urea with all but one type of organic surface investigated to date (the exception being cationic N)4 are favorable, we expect urea to have a more favorable interaction (more negative μ23/RT value) with larger compounds. Indeed, uridine, deoxythymidine and adenosine μ23/RT values are more favorable (more negative) than uracil, thymine and adenine μ23/RT values by approximately 0.1 m−1, presumably due to a favorable interaction with the sugar ASA on nucleosides. However, urea exhibits less favorable interactions with the 5’-NMP disodium salts than with corresponding nucleobases and nucleosides. This is consistent with the strong unfavorable interaction of urea with Na+ ions deduced from model compound data4 (2βNa+ = 0.197 ± 0.028 m−1). Correction for this effect shows that μ23/RT values for interactions of urea with the 5’-UMP dianion (−0.363 ± 0.057 m−1) and 5’-dTMP dianion (−0.345 ± 0.031 m−1) are more negative than for the nucleosides uridine (−0.31 ± 0.02 m−1) and deoxythymidine (−0.31 ± 0.02 m−1). While the μ23/RT value calculated for the 5’-AMP dianion (−0.323 ± 0.030 m−1) is less negative than for the nucleoside adenosine (−0.35 ± 0.02 m−1 ), both are more negative than μ23/RT for the nucleobase adenine (−0.27 m−1 ± 0.01).
Qualitative analysis of these μ23/RT values leads to the following picture of the preferential interactions of urea with nucleic acid base, sugar, and phosphate functional groups:
Values of μ23/RT for nucleobases, base analog, nucleosides, and Na2NMPs are all negative, showing that preferential interactions of urea with all model compounds studied are favorable. Therefore, urea must interact favorably with most if not all types of nucleic acid surface.
Values of μ23/RT for purine and pyrimidine bases and base analogs with similar functional groups (e.g. adenine vs. cytosine, 5’-AMP vs. 5’-CMP or hypoxanthine vs. uracil) reveal a more favorable μ23/RT of the purines (with two fused heterocyclic aromatic rings) than the pyrimidines (with one heterocyclic aromatic ring), indicating a favorable interaction of urea with ring surface.
Values of μ23/RT for the nucleic acid bases adenine, thymine and uracil are less negative (less favorable) than the corresponding values for the nucleosides adenosine, thymidine, guanosine and uridine, indicating that urea has a favorable interaction with the nucleoside sugar.
In general, values (Table 3) of μ23/RT become more negative (more favorable) as more functional groups are added to the heterocyclic ring. Comparing purines, μ23/RT values for caffeine, theobromine and theophylline, with 4–5 groups attached to the ring, are significantly more negative than μ23/RT values for hypoxanthine, adenine and purine (which have 0–1 group attached). This indicates a favorable preferential interaction of urea with these functional groups that is large enough to compensate for the loss of favorable urea-ring interaction resulting from the steric effect of these groups.
Values of μ23/RT for uracil (also 5’-UMP) are more negative than for cytosine (also 5’-CMP). Here the chemical difference is between a carbonyl oxygen (uracil) and an amino nitrogen (cytosine); both nucleobases display similar amounts of ring ASA. Therefore urea must interact more favorably with carbonyl oxygen than amino nitrogen, analogous to the more favorable interaction of urea with amide O than with amide N on protein model compounds.4
The value of μ23/RT for thymine is slightly more negative than for uracil. The addition of the methyl group to the C5 position of uracil to give thymine adds aliphatic ASA but eliminates aromatic ring ASA. The elimination of the favorable interaction of this aromatic ASA with urea is found to be compensated completely by a favorable interaction of urea with the added methyl ASA.
Table 3.
Experimentally determined urea m-values/RT for DNA and RNA duplex formationa
| Sequence | % GC |
Temperature (°C)a |
van’t Hoff ΔHobs (kcal mol−1)b |
m-value (kcal mol−1 molal−1)c |
Predicted m- value for 75% stacked single strands (kcal mol−1 molal−1)d |
|---|---|---|---|---|---|
| DNA duplexes | |||||
| 5'-d(GAAATTATAAAC)-3' | 17 | 15 | 0.75 ± 0.07 | 0.81 ± 0.09 | |
| 5'-d(GAAAGTATAAAC)-3' | 25 | 18 | 0.85 ± 0.02 | 0.81 ± 0.09 | |
| 5'-d(GAAAGTAGAAAC)-3' | 33 | 18 | 0.72 ± 0.04 | 0.79 ± 0.09 | |
| 5'-d(GCAAAGTAAACG)-3' | 42 | 30 | 0.81 ± 0.04 | 0.80 ± 0.10 | |
| 5'-d(GCAAAGCAAACG)-3' | 50 | 41 | −18.7 ± 0.7 | 0.73 ± 0.06 | 0.79 ± 0.10 |
| 5'-d(GCATAGCATACG)-3' | 50 | 41 | −17.5 ± 1.0 | 0.58 ± 0.05 | 0.80 ± 0.10 |
| RNA duplexes | |||||
| 5'-r(GAAAGUAGAAAC)-3' | 33 | 41 | −23.1 ± 0.1 | 0.98 ± 0.03 | 0.78 ± 0.09 |
| 5'-r(GCAAAGCAAACG)-3' | 50 | 52 | −25.0 ± 0.1 | 0.93 ± 0.03 | 0.79 ± 0.09 |
| 5'-r(GCAUAGCAUACG)-3' | 50 | 52 | −24.1 ± 0.2 | 0.87 ± 0.04 | 0.79 ± 0.09 |
| 5'-r(GCGAAGCCAACG)-3' | 67 | 63 | −26.4 ± 0.5 | 0.84 ± 0.03 | 0.77 ± 0.10 |
| 5'-r(GCGCCGCCGGCG)-3' | 100 | 81 | −27.4 ± 0.5 | 0.59 ± 0.04 | 0.77 ± 0.11 |
m-values for duplex formation from thermal denaturation experiments determined at the temperature with 0.80 fraction duplex intact in 0 m urea. m-values from isothermal titration experiments determined at temperature with 1.0 fraction duplex intact in 0m urea.
van’t Hoff enthalpies of duplex formation calculated as described in methods from thermal melts in the absence of urea.
First four m-values determined from isothermal urea titrations. All other m-values determined from thermal melts. For all experiments, concentration of DNA and RNA strands used is ~2.5µM.
Because in general each functional group affects the accessible surface area of neighboring groups, a quantitative analysis of the interactions of urea with nucleobases cannot be based solely on functional groups, but must incorporate the accessibility of these groups, as in the next section.
Analysis of μ23/RT: interactions of urea per unit area of each nucleic acid functional group
Using ASA data and the experimentally determined μ23/RT values we globally fit the model compound data to Eq. 3 as described in methods to determine interaction potentials αi for the interaction of urea with each type of nucleic acid surface (Table 2). The fits for other surface types are unaffected by grouping ring C and N and sugar C and O together. Urea interacts favorably (negative αi values) with all types of nucleic acid surface studied here, interacting most favorably with the aromatic ring and the methyl group attached to it.
Table 2.
Values of the interaction potential αi for the interaction of urea with nucleic acid and protein surface types.
| Nucleic Acid Surface Type |
αi* 104 (m−1Å−2) |
Protein Surface Typea |
αi * 104 (m−1Å−2) |
|---|---|---|---|
| Aromatic Ring (C,N) | −10.9 ± 0.7 | Aromatic C | −8.9 ± 0.5 |
| Ring Methyl | −12.0 ± 1.0 | Aliphatic C | −1.1 ± 0.5 |
| Carbonyl O | −6.2 ± 1.9 | Amide O | −8.5 ± 1.8 |
| Amino N | −2.8 ± 1.9 | Amide N | −3.7 ± 2.2 |
| Phosphate O | −5.8 ± 1.2 | Carboxylate O | −3.7 ± 1.6 |
| Sugar | −6.6 ± 0.7 |
From ref4 with minor revision (see supplemental)
The αi values in Table 2 were used to predict μ23/RT values for interactions of urea with all model compounds in the training set. Experimental and calculated values are given in Table 1 and compared in Figure 3; agreement is very good. The average deviation of calculated values from experimental values is ±8%, indicating that we have successfully developed the ability to predict interactions of urea with compounds displaying nucleic acid surface types. A next step is to use urea αi values to predict effects of urea on nucleic acid processes like RNA and DNA duplex formation (see last section of Results).
To gain a molecular perspective of the interaction of urea with each functional group, these αi values are interpreted using the solute partitioning model in terms of a microscopic partition coefficient defined as the ratio of the concentration of solute in the local hydration water and in bulk water (KpSPM = m3loc/m3bulk) as described in detail in refs 4,5,40,41. A positive αi* 104 value of approximately 30 m−1Å−2 was obtained for Na2SO4, corresponding to complete exclusion of both Na+ and SO42− ions from approximately two layers of water hydrating hydrocarbon surface (Kp=0), while the most negative value obtained to date for chemical interactions is −68 m−1Å−2 for the accumulation of the 3 ions of Na2SO4 at amide surface (Kp=3.49)7; compared to these large αi-values, interactions of urea are relatively subtle.
Lambert and Draper18 obtained VPO data for urea- potassium dimethylphosphate interactions and combined this with literature solubility and VPO data to determine contributions to μ23/ASA (equivalent to RT*αi values) for the interaction of urea with a unit area of nucleic acid base, sugar and phosphate surface. These can be compared with our αi values determined from a different data set.
Their αi value for anionic phosphate O (−6.1*10−4 m−1Å−2, obtained from potassium dimethylphosphate VPO data by correcting for the interaction of urea with K+ ion, ester O and methyl groups using αi values from ref 4),18 is the same within uncertainty as that determined here (Table 2). Since very different compounds and analysis are used, this agreement is very encouraging.
Analysis of uncorrected solubility data for adenine and cytosine in aqueous urea solutions42 yields αi values for adenine of −8.9*10−4 m−1Å−2 and for cytosine of −7.6*10−4 m−1Å−2. Lambert and Draper18 corrected these solubility data for nucleobase self-interaction due to stacking (estimated from purine and cytidine VPO data because adenine and cytosine are too insoluble for VPO) to obtain nucleobase αi values of −11*10−4 m−1Å−2 and −10*10−4 m−1Å−2 respectively. Our nucleobase αi values for adenine (− 9.5*10−4 m−1Å−2) and cytosine (−6.5*10−4 m−1Å−2) determined from the μ23/RT values in Table 1 (obtained using the hexanol-water distribution assay at low nucleobase concentrations where stacking is negligible) are closer to the values determined from the uncorrected solubility data indicating that the correction for stacking may be too large. Lambert and Draper18 determined an average αi value for nucleic acid bases (−11*10−4 m−1Å−2) from this adenine and cytosine data. Because we quantified interactions of urea with 9 nucleobases and base analogs we obtain values for the individual ring functional groups and the ring ASA.
The sugar αi value (−2.0*10−4) predicted18 from sugar ASA and model compound VPO data for glycerol and sucrose (ref 4) is only 1/3 as large in magnitude as the sugar αi value in Table 2, which is determined from nucleosides and mononucleotides. This is surprising since it indicates a failure of the assumption of additivity, which generally is found to be valid1,4–7 as Fig 3 (above) indicates. This context dependence indicates that the neighboring nucleic acid base affects how the sugar interacts with urea, as is observed with the ring methyl (see below). This does not affect the use of Eq. 3 to analyze the data here as the context of the sugar is always the same.
Comparison of αi values and their molecular interpretation for interactions of urea with protein and nucleic acid functional groups
Proteins and nucleic acids contain similar types of functional groups; how do interaction potentials αi for the interaction of urea with each of these similar groups compare (Table 2)? Can these nucleic acid αi values be interpreted in terms of noncovalent interactions of urea with the nucleic acid functional groups, as has been reported for interactions of urea and GB with protein functional groups4?
Aromatic ring (C,N): Table 2 shows that urea has a more favorable preferential interaction with the heterocyclic rings found on nucleic acid bases (Kp=1.34), which can accept hydrogen bonds to the π-system as well as hydrogen bonds to or from the ring N’s, than with homocyclic aromatic rings of tyrosine and phenylalanine residues on proteins (Kp=1.28) suggesting that urea may have a preference for ring N over ring C. The interaction of urea with both ring types may involve a hydrogen bond donated from the urea amide N to the π-system of the aromatic ring,43 a partial cation-π interaction with the urea amide N44 or a π-π stacking interaction of urea with the ring (observed in simulations of urea with a RNA hairpin45 and with 5’-NMPs, detailed in supplemental), all allowing urea to interact more favorably with the ring than water does.
Carbonyl O: The urea-carbonyl O preferential interaction (Kp=1.19) is slightly less favorable than that of urea with amide O (Kp=1.28). As with amide O, the urea-carbonyl O interaction can be interpreted as a hydrogen bond donated from the urea NH2. Moreover, we would expect the carbonyl O groups attached to the heterocyclic aromatic rings N to behave similarly to amide O because they can participate in resonance structures with the ring in the same way that amide O can participate in resonance structures with amide N.
Amino N: Urea can act as a hydrogen bond donor and acceptor with amino N (Kp=1.09), like with amide N (Kp=1.10). Amino N groups can also participate in resonance structures with the ring as amide N groups do with amide O and we find a similar weak favorable interaction of urea with these two groups. Urea interacts less favorably with amino N than carbonyl O. We interpreted the less favorable interaction of urea with amide N than amide O as a favorable urea O-amide N hydrogen bond counteracted by an unfavorable urea N-amide N hydrogen bond. Likewise, we expect a favorable interaction between urea amide O and amino N similar in magnitude to the urea-carbonyl O interaction, indicating that while urea O-amino N hydrogen bonds are favorable relative to water, urea NH2-amino N hydrogen bonds are disfavored in comparison to hydrogen bonds with water.
Aliphatic C: Urea has a moderate favorable preferential interaction with the methyl group attached to nucleic acid heterocyclic rings (Kp=1.37, similar to the interaction with the ring itself). Surprisingly, the αi value for the interaction of urea with aliphatic C surface on the nucleobase aromatic ring (proportional to Kp-1) is almost an order of magnitude larger than for ordinary aliphatic C (Kp=1.03). Urea also has a larger than expected interaction with the methyl group on toluene and the tetramethylbutyl group on the phenyl ring of triton X-100 micelles (see supplemental for details) indicating that urea has a more favorable interaction with aliphatic hydrocarbon that is a substituent on an aromatic ring, which could affect the electron distribution of the attached methyl groups through both inductive and resonance effects. Additionally, urea could be involved in a π-π interaction with the ring45 while interacting with the methyl group attached to the ring, making the interaction more favorable than that with water, which lacks a π-system. Since the only aliphatic groups considered explicitly here are these ring methyls, this deviation from additivity does not affect the application of Eq. 3 to the nucleobase/base analog data set.
Examples of non-additivity have been found for certain Hofmeister salt ions as well. Consistent with the stacking hypothesis, Hofmeister ions with a π-system that can stack on the ring (e. g. GuH+) also have a more favorable interaction with the ring methyl than with aliphatic C while ions lacking a π-system do not (see supplemental). We do not see the same effect of the ring on the preferential interaction of urea or GuH+ salts with ring moieties with which they can hydrogen bond (amino N, carbonyl O), suggesting that stacking only significantly affects ring moiety interactions relative to water when stronger hydrogen bonding interactions (that water can also participate in) are not possible. In another example of context dependence involving a π-system, spectroscopic studies and MD simulations46 indicate that interactions of Hofmeister anion SCN− (also I−) with the methylene C adjacent to the amide π-system of the peptide backbone are more favorable than their interactions with side chain aliphatic C.
Phosphate O: Phosphate O and carboxylate O are both anionic O functional groups but the interaction of urea with phosphate O is somewhat more favorable (Kp=1.18). We have interpreted the favorable interaction of urea with carboxylate O (Kp=1.13) in terms of a hydrogen bond donated from urea -NH2 so perhaps the hydrogen bond with phosphate O is stronger due to its larger partial negative charge (2/3 vs 1/2).
Sugar: The sugar ring consists of highly correlated C and O groups with an O/C ASA ratio of 1.2–1.5 for all 5’-NMP’s and nucleosides except 5’-dTMP and deoxythymidine which had an O/C ratio of 0.6 (but were still well fit by the same sugar αi value). The interaction of urea with sugar (Kp=1.21) is similarly favorable to the interaction of urea with the other O surface types in the data set (carbonyl O, phosphate O) despite the additional hydrocarbon ASA on the sugar ring. The sugar Kp value predicted from sucrose and glycerol model compound data as described above is less favorable. This could be another situation where the context of a group affects its interactions with urea. Whether the significant context is the constrained geometry of the sugar ring or its proximity to the π system of the base ring remains to be determined.
Molecular dynamics simulations using the Amber force field for urea were performed to visualize the accumulation of urea at the 5’-NMPs studied here and are described in supplemental. They indicate that urea can participate in the hydrogen bonding and stacking interactions that we discuss above. The interaction potentials in Table 2 can be used to tune MD force fields to allow for more quantitative thermodynamic predictions to be made from simulations.
Effect of Urea on DNA and RNA Dodecamer Melting
To test the ability of urea αi values (Table 2) to predict or interpret the effects of urea on nucleic acid processes, we quantified m-values for the effect of urea on DNA and RNA dodecamer duplex formation by urea titration and thermal denaturation. Urea destabilizes all RNA and DNA duplexes, indicating favorable interactions between urea and ΔASA, the surface area of the nucleic acid (mostly of the nucleobases) that becomes water accessible during the melting process. Values of ΔG°obs=-RTlnKobs for each oligomer, interpolated from melting curves to a selected reference temperature in the transition region of the oligomer at all urea concentrations studied (as described in methods) were plotted as a function of urea molality to obtain the urea m-value for duplex formation from the slope (Figure S2). All m-values (Table 3) are positive due to favorable interactions between urea and the surface described by theΔASA. Values of ΔHobs for formation of oligomers studied by thermal denaturation, determined from van’t Hoff plots as described in methods, are also listed in Table 3. Like dΔG°/dm3 (the m-value), dΔH°/dm3 derivatives are positive but are not accurately enough determined to be able to dissect enthalpic and entropic contributions to the urea m-value as was done for protein unfolding.47 RNA Kobs values were determined at higher temperatures because RNA duplex dodecamers had higher Tm values than DNA duplex dodecamers with similar nucleobase sequence, as expected.48
Urea-nucleic acid αi values (Table 2) predict urea m-values for forming these duplexes using the ΔASA for duplex association from single strands (Eq. 3). Previously Guinn et al predicted protein folding m-values using urea-protein functional group interaction potentials, obtaining good agreement using an extended (unstructured) model for the unfolded state.4 Here, because the amount of single stranded stacking is expected to be large, we calculated ΔASA values for DNA and RNA melting determined assuming single strands in fully stacked and half-stacked states to compare with predictions for unstacked states (see Experimental Section, Table S3). We divided ΔASA into the same functional groups as the nucleobases/base analogs and 5’-NMPs. Our ΔASA values calculated for a model with the half-stacked DNA and RNA single strands are in good agreement with Hong et al.25 Most of ΔASA arises from exposure of heterocyclic ring, carbonyl O and amino N groups; a small reduction in phosphate oxygen surface area is also predicted.
Using these ΔASA values and the urea-nucleic acid αi values in Table 2, we predicted m-values for the effect of urea on RNA and DNA duplex formation assuming different extents of residual stacking in the single strands at the temperature where Kobs is determined (Table S4). We compare urea m-values determined at different temperatures here because we expect (and observe) that urea m-values for nucleic acid duplex formation are less temperature dependent than urea m-values/RT (see supplemental). Our αi values are determined at 25°C while the experimental m-values were determined at temperatures ranging from 15–81°C due to the large range of Tm values for the dodecamers studied (Table 3). These predictions are most applicable to the first four entries in Table 2 which are determined at ~25°C.
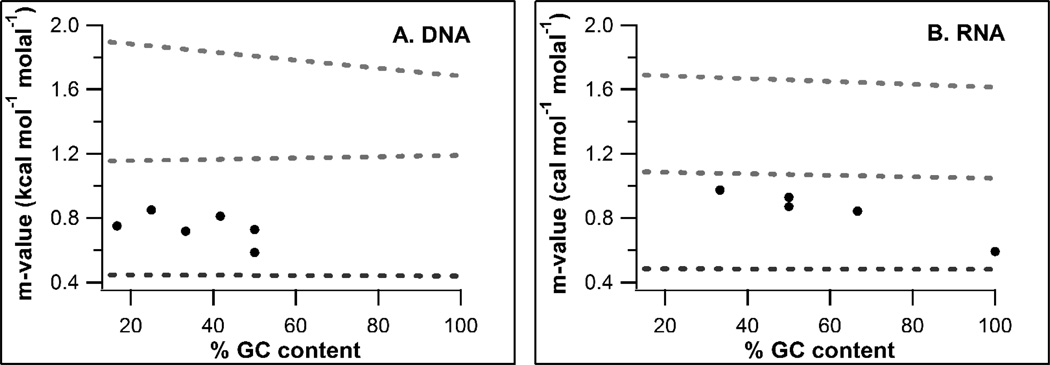
m-Values predicted for DNA and RNA duplex formation assuming completely stacked single strands do not show a significant dependence on nucleobase composition (Fig 4, blue dashed lines). m-Values predicted assuming completely unstacked nucleobases (Fig 4, pink dashed lines) show no nucleobase composition dependence for RNA and a small reduction in the magnitude of the m-value with increasing %GC for DNA due to the smaller contribution to the ΔASA from thymine, the ring methyl of which interacts very favorably with urea. Experimental m-values scatter between the predictions obtained for completely stacked and half-stacked single strands and show little nucleobase composition dependence. m-Values for all DNAs are best predicted assuming 70–90% stacking. m-Values for most RNAs are best predicted assuming 60–70% stacking, while the m-value for the 100% GC RNA oligomer is best predicted assuming almost 100% stacking. The four DNA oligomers studied at 15–30°C (closest to 25°C, the temperature at which the urea interaction potentials used to predict m-values were obtained) have quite similar urea m-values (Table 3), indicating no large effect of nucleobase composition.
Figure 4.
Predicted (dashed lines) and observed (black points) urea m-values for formation of 12 bp duplex DNAs (A) or RNAs (B) to stacked (blue), half stacked (green) or unstacked (pink) single strands vs %GC content of DNA oligomers.
Stacking can be estimated directly from hyperchromicity by assuming the absorbance of duplex DNA represents the absorbance of a completely stacked DNA while the absorbance of NMP’s from a DNA digest represents the absorbance of completely unstacked single strands. Using this technique, Holbrook et al found a 66% GC DNA single strand is 75% stacked at 40°C, consistent with what we see in Fig. 4 for the more GC rich DNAs studied at 41°C, and stacking decreases from 89% at 10°C to 52% at 80°C (the temperature range studied here); all our DNAs and RNAs fall within this range of stacking.49 Lambert and Draper18 predicted m-values for the effect of urea on melting a series of RNA’s, some containing extensive tertiary structure, using their more course-grained model of nucleic acid components. They found that m-values were better predicted when they used an A-form single stranded helix as a model for unfolded RNA than an extended unstacked model, consistent with our prediction of significant single strand stacking.
Different single stranded nucleic acid homopolymers exhibit different degrees of stacking. Poly(T), poly(U) and poly(C) exhibit essentially no stacking at 25°C while poly(A) stacks extensively.50–52 Hence the amount of stacking in our single stranded oligomers may depend in part on nucleobase sequence, perhaps explaining the scatter in Fig. 4. The m-values for RNAs were determined at higher temperature due to their increased thermal stability, which could explain why their m-values are more accurately predicted by invoking a smaller extent of stacking. Because urea promotes exposure of nucleic acid surface, urea is expected to reduce residual stacking in the unfolded form. This has not been observed, and the observation that urea m-values are independent of urea concentration up to at least seven molar urea for RNA duplexes and yeast tRNAPhe indicates that the ΔASA of unfolding is not a significant function of urea concentration.17 Likewise, linearity of protein folding m-values2 indicates no significant urea concentration dependence of the protein denatured state ensemble even though we might expect the denatured state to become more solvent-accessible at higher urea concentrations.
Shelton et al measured urea m-values for melting a series of 5 RNA duplexes from 6 to 18 base pairs in length; 4 of their duplexes are approximately 60% GC and one 20% GC.17 They found that the m-value increased in proportion to length (i.e. to ΔASA).2 From estimates of ΔASA of melting of the duplexes studied by Shelton et al, assuming stacked, half-stacked and unstacked nucleobases in the single strands, we predict m-values using the urea alpha-values of Table 2. Comparison of predicted and experimental m-values yields predictions for the amounts of residual stacking in the single strands (Table 4). For all but the shortest duplex, the predicted amount of single-strand stacking is consistent with that determined for the dodecamers investigated here (Fig. 4).
Table 4.
Experimentally determined urea m-values for RNA duplex formation17 and the amount of stacking predicted
| Sequence | Experimental m-value (kcal mol−1 molal−1) |
Temperature (°C) |
Predicted % residual stacking in single strands in transition |
|---|---|---|---|
| 5'-r(GCAGUAGUCGACUACUGC)-3' | 1.17 | 70 | 80 |
| 5'-r(GCAGUAGCUACUGC)-3' | 1.06 | 60 | 70 |
| 5'-r(GCAGUACUGC)-3' | 0.84 | 50 | 60 |
| 5'-r(GUAAUAUUAC)-3' | 0.74 | 20 | 69 |
| 5'-r(GCAUGC)-3' | 0.64 | 20 | 30 |
Concluding Discussion
We demonstrate that urea interacts favorably with all nucleic acid surface types, relative to interactions with water, and that urea destabilizes nucleic acid duplexes by its favorable interactions with both the ring and functional groups of nucleobases which become solvent accessible in the single stranded state. Per unit of accessible surface area, the most favorable interactions of urea are with the heterocyclic aromatic rings of all bases, and with the methyl group of thymine. We use these data to probe the extent of unstacking in formation of DNA and RNA duplexes and find that a large amount of residual stacking (60% to 90%, depending on nucleobase composition and sequence) in the individual single strands is necessary to obtain agreement between observed and predicted urea m-values. Once dissected into individual interactions of amide O and N (by studying interactions of a compound with a different O:N ratio like malonamide with the same set of model compounds)1, these data will provide significant information about interactions between the protein backbone and nucleic acids in proteinnucleic acid complexes.
Now that interactions of urea with both nucleic acid and protein4 functional groups have been quantified, urea will be useful as one of a set of solute probes being developed to detect and characterize large scale conformational changes and formation of new interfaces in the steps of protein and nucleic acid mechanisms. For urea and the osmolyte-stabilizer glycine betaine (GB), thermodynamic m-values for folding of globular proteins and for formation of a repression complex between lac repressor protein and lac operator DNA are well-predicted from α-values for these solutes and structural information.1,4,6 Kinetic and/or thermodynamic m-values have been determined for urea, GB, and KGlutamate (compared with KCl to eliminate polyelectrolyte effects of K+ and thereby compare the physiological anion Glutamate− with the laboratory anion Cl−) for various steps in forming a transcription initiation complex between RNA polymerase and promoter DNA.53,54 The lack of large effects of any of these solutes on the kinetics and thermodynamics of the step in which the 13 base pairs of DNA is opened provides one line of evidence that opening occurs in the cleft of RNA polymerase and not in a solution environment. The very large stabilizing effects of GB and Glutamate (relative to Cl−), and the large destabilizing effect of urea, on the step that converts the initial unstable open complex to a highly stable open complex, increasing its lifetime from 1 s to >105 s at the promoter investigated, provides the best evidence available to date that this step involves folding and assembly of 100–150 residues of RNA polymerase to form a jaw/clamp structure on the downstream duplex DNA after opening.22,54–56 Even before full information about solute interactions with protein and nucleic acid surfaces is available, kinetic m-value studies can provide qualitative answers about steps with large conformational changes or new interfaces in other assembly processes and the operation of other protein machines, such as the spliceosome57. The data obtained here will help to provide quantitative information about the types and amounts of surfaces buried or exposed in these processes.
Supplementary Material
Acknowledgements
The authors thank D. B. Knowles and M. W. Capp for helpful discussions in developing the distribution assay and the reviewers for valuable comments on the manuscript. This research was supported by National Institutes of Health GM 47022 (to M.T.R.) and R15-GM093331 (to J.J.S. and G.W.M.). The work was also supported in part by a grant to St. Olaf College from the Howard Hughes Medical Institute through the Undergraduate Science Education Program. Computer simulations were performed at the University of Minnesota Supercomputing Institute.
Footnotes
Supporting Information Available
Details of materials used, 5’-NMP VPO sample preparation, assumptions in determining μ23/RT from KDWH values, determining molal scale KDWH values, distribution assay data analysis, determining urea in the hexanol phase, controls for the distribution assay, DNA and RNA dodecamer sample preparation, reanalysis of Guinn et al4 data, urea-aliphatic C interactions, determining interactions of urea with triton micelles, molecular dynamics simulations of 5'-NMPs, determining the effect of urea on the salt dependence of melting, the temperature dependence of m-values vs m-values/RT. Tables containing contributions to the ASA of model compounds, pseudorotation phase angle and glycosyl torsion andle distribution in 5’-mononucleotide molecular dynamics simulations, contributions to ΔASA for formation of DNA and RNA duplexes assuming 100%, 50% and 0% stacked nucleobases in the single strands, experimental and predicted m-values for effect of urea on helix formation assuming different amounts of stacking, values of μ23/RT for the interaction of GuHCl with nucleic acid bases and base analogs determined from hexanol/water 2 phase partitioning and solubility assays. Figures showing plots from the 2-phase distribution assay with urea and caffeine, plots of ΔGobs vs urea molality for duplex formation by DNA and RNA, a plot of ΔOsm determined by VPO against m2m3 for urea with thymidine and uridine, plots of the log of the macroscopic distribution coefficient ratio KDWH/KDWH,0 vs GuHCl molality, plots of the negative log of CMC vs urea molality for Triton X-100 and Triton X-305, average urea oxygen and nitrogen density around adenine and uracil from 5'-NMP MD simulations. This content is available free of charge via the internet at http://pubs.acs.org.
Contributor Information
Jeffrey J. Schwinefus, Email: schwinef@stolaf.edu.
Thomas Record, Jr, Email: mtrecord@wisc.edu.
References
- 1.Record MTJ, Guinn E, Pegram L, Capp M. Faraday Discussions. 2013;160:9. doi: 10.1039/c2fd20128c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myers JK, Pace CN, Scholtz JM. Protein Sci. 1995;4:2138. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courtenay ES, Capp MW, Record MT. Protein Sci. 2001;10:2485. doi: 10.1110/ps.ps.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT. Proc. Nat. Acad. Sci. U.S.A. 2011;108:16932. doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pegram LM, Record MT. J. Phys. Chem. B. 2008;112:9428. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capp MW, Pegram LM, Saecker RM, Kratz M, Riccardi D, Wendorf T, Cannon JG, Record MT., Jr. Biochemistry. 2009;48:10372. doi: 10.1021/bi901273r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegram LM, Wendorff T, Erdmann R, Shkel I, Bellisimo D, Felitsky DJ, Record MT. Proc. Nat. Acad. Sci. U.S.A. 2010;107:7716. doi: 10.1073/pnas.0913376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong J, Capp MW, Anderson CF, Saecker RM, Felitsky DJ, Anderson MW, Record MT. Biochemistry. 2004;43:14744. doi: 10.1021/bi049096q. [DOI] [PubMed] [Google Scholar]
- 9.Felitsky DJ, Cannon JG, Capp MW, Hong J, Wynsberghe AWV, Anderson CF, Record MT. Biochemistry. 2004;43:14732. doi: 10.1021/bi049115w. [DOI] [PubMed] [Google Scholar]
- 10.Sagle LB, Zhang Y, Litosh VA, Chen X, Cho Y, Cremer PS. J. Am. Chem. Soc. 2009;131:9304. doi: 10.1021/ja9016057. [DOI] [PubMed] [Google Scholar]
- 11.Lim WK, Rosgen J, Englander SW. Proc. Nat. Acad. Sci. U.S.A. 2009;106:2595. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babayan YS. Molek. Biol. 1988;22:1204. [PubMed] [Google Scholar]
- 13.Aslanyan VM, Babayan YS, Arutyunyan SG. Biophysics. 1984;29:410. [Google Scholar]
- 14.Klump H, Burkart W. Biochim. Biophys. Acta. 1977;475:601. doi: 10.1016/0005-2787(77)90320-3. [DOI] [PubMed] [Google Scholar]
- 15.Nordstrom LJ, Clark CA, Andersen B, Champlin SM, Schwinefus JJ. Biochemistry. 2006;45:9604. doi: 10.1021/bi052469i. [DOI] [PubMed] [Google Scholar]
- 16.Schwinefus JJ, Kuprian MJ, Lamppa JW, Merker WE, Dorn KN, Muth GW. Biochemistry. 2007;46:9068. doi: 10.1021/bi602637v. [DOI] [PubMed] [Google Scholar]
- 17.Shelton VM, Sosnick TR, Pan T. Biochemistry. 1999;38:16831. doi: 10.1021/bi991699s. [DOI] [PubMed] [Google Scholar]
- 18.Lambert D, Draper DE. Biochemistry. 2012;51:9014. doi: 10.1021/bi301103j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gluick TC, Wills NM, Gesteland RF, Draper DE. Biochemistry. 1997;36:16173. doi: 10.1021/bi971362v. [DOI] [PubMed] [Google Scholar]
- 20.Gluick TC, Yadav S. J. Am. Chem. Soc. 2003;125:4418. doi: 10.1021/ja0292997. [DOI] [PubMed] [Google Scholar]
- 21.Hong J, Capp MW, Saecker RM, Record MT. Biochemistry. 2005;44:16896. doi: 10.1021/bi0515218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT. Biochemistry. 2006;45:2161. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knowles DB, LaCroix AS, Deines NF, Shkel I, Record MT. Proc. Nat. Acad. Sci. U.S.A. 2011;108:12699. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong J, Capp MW, Anderson CF, Record MT. Biophys. Chem. 2003;105:517. doi: 10.1016/s0301-4622(03)00112-1. [DOI] [PubMed] [Google Scholar]
- 25.Hong J, Capp MW, Anderson CF, Saecker RM, Felitsky DJ, Anderson MW, Record MTJ. Biochemistry. 2004;43:14744. doi: 10.1021/bi049096q. [DOI] [PubMed] [Google Scholar]
- 26.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. New York: McGraw-Hill, Inc.; 1992. [Google Scholar]
- 27.Tsodikov OV, Record MT, Sergeev YV. J. Comp. Chem. 2002;23:600. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 28.Richards FM. Annu. Rev. Biophys. Bioeng. 1977;6:151. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 29.Case DA, Darden TA, Cheatham I TE, Simmerling Cl, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. San Francisco: University of California; 2008. [Google Scholar]
- 30.Hubbard SJ, Thornton JM. Department of Biochemistry and Molecular Biology. London: University College; 1993. [Google Scholar]
- 31.DeLano WL. The PyMOL User's Manual (2002) DeLano Scientific. San Carlos, CA: 2002. [Google Scholar]
- 32.Shaw WHR, Bordeaux JJ. J. Am. Chem. Soc. 1955;77:4729. [Google Scholar]
- 33.Long FA, McDevit WF. Chem. Rev. 1952;51:119. [Google Scholar]
- 34.Nandi PK, Robinson DR. J. Am. Chem. Soc. 1972;94:1308. doi: 10.1021/ja00759a043. [DOI] [PubMed] [Google Scholar]
- 35.Sykes PH, Robertson PW. J. Am. Chem. Soc. 1933;55:2621. [Google Scholar]
- 36.Smith R, Tanford C. Proc. Nat. Acad. Sci. 1973;70:289. doi: 10.1073/pnas.70.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih P, Pederson LG, Gibbs PR, Wolfenden R. J. Mol. Biol. 1998;280:421. doi: 10.1006/jmbi.1998.1880. [DOI] [PubMed] [Google Scholar]
- 38.Meylan WM, Howard PH, Boethling RS. Env. Tox. and Chem. 1995;15:100. [Google Scholar]
- 39.Hansch C, Quinlan JE, Lawrence GL. J. Org. Chem. 1968;33:347. [Google Scholar]
- 40.Capp MW, Pegram LM, Saecker RM, Kratz M, Riccardi D, Wendorff T, Record JGCaMT. Biochemistry. 2009;48:10372. doi: 10.1021/bi901273r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Courtenay ES, Capp MW, Saecker RM, Record MT. Proteins: Struct., Funct., and Genetics. 2000;41:72. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 42.herskovits TT, Bowen JJ. Biochemistry. 1974;13:5474. doi: 10.1021/bi00724a004. [DOI] [PubMed] [Google Scholar]
- 43.Levitt M, Perutz MF. J. Mol. Biol. 1988;201:751. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 44.Scrutton NS, Raine AR. Biochem J. 1996;319(Pt 1):1. doi: 10.1042/bj3190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Priyakumar UD, Hyeon C, Thirumalai D, MacKerell AD. J. Am. Chem. Soc. 2009;131:17759. doi: 10.1021/ja905795v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rembert KB, Paterova J, Heyda J, Hilty C, Jungwirth P, Cremer PS. J. Am. Chem. Soc. 2012;134:10039. doi: 10.1021/ja301297g. [DOI] [PubMed] [Google Scholar]
- 47.Felitsky DJ, Record MT. Biochemistry. 2003;42:2202. doi: 10.1021/bi0270992. [DOI] [PubMed] [Google Scholar]
- 48.Kankia BI, Marky LA. J. Phys. Chem. B. 1999;103:8759. [Google Scholar]
- 49.Holbrook JA, Capp MW, Saecker RM, Record MT. Biochemistry. 1999;38:8409. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 50.Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids: Structure, Properties and Functions. Sausolito, CA: University Science Books; 2000. [Google Scholar]
- 51.Goddard NL, Bonnet G, Krichevsky O, Libchaber A. Phys. Rev. Letters. 2000;85:2400. doi: 10.1103/PhysRevLett.85.2400. [DOI] [PubMed] [Google Scholar]
- 52.Riley M, Maling B, Chamberlin MJ. J. Mol. Biol. 1966;20:359. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- 53.Kontur WS, Saecker RM, Capp MW, Record MT. J. Mol. Biol. 2008;376:1034. doi: 10.1016/j.jmb.2007.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MT. Biochemistry. 2010;49:4361. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drennan A, Kraemer M, Capp M, Gries T, Ruff E, Sheppard C, Wigneshweraraj S, Artsimovitch I, Record MT. Biochemistry. 2012;51:9447. doi: 10.1021/bi301260u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saecker RM, Record MT, Dehaseth PL. J Mol Biol. 2011;412:754. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butcher SE, Brow DA. Biochem Soc Trans. 2005;33:447. doi: 10.1042/BST0330447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.