Abstract
Truncated-ERBB2 isoforms (t-ERBB2s), resulting from receptor proteolysis or alternative translation of the ERBB2 mRNA, exist in a subset of human breast tumors. t-ERBB2s lack the receptor extracellular domain targeted by therapeutic anti-ERBB2 antibodies and antibody–drug conjugates, including trastuzumab, trastuzumab-DM1 and pertuzumab. In clinical studies, expression of t-ERBB2 in breast tumors correlates with metastasis as well as trastuzumab resistance. By using a novel immuno-microarray method, we detect a significant t-ERBB2 fraction in 18 of 31 (58%) of immunohistochemistry (IHC)3+ ERBB2+ human tumor specimens, and further show that t-ERBB2 isoforms are phosphorylated in a subset of IHC3+ samples (10 of 31, 32%). We investigated t-ERBB2 biological activity via engineered expression of full-length and truncated ERBB2 isoforms in human mammary epithelial cells (HMECs), including HMEC and MCF10A cells. Expression of p110 t-ERBB2, but not p95m (m=membrane, also 648CTF) or intracellular ERBB2s, significantly enhanced cell migration and invasion in multiple cell types. In addition, only expression of the p110 isoform led to human breast epithelial cell (HMLE) xenograft formation in vivo. Expression of t-ERBB2s did not result in hyperactivation of the phosphoinositide kinase-3/AKT or mitogen-activated protein kinase signaling pathways in these cells; rather, phosphoproteomic array profiling revealed attenuation of phosphorylated signal transducer and activator of transcription 5 (STAT5) in p110-t-ERBB2-expressing cells compared to controls. Short hairpin-mediated silencing of STAT5 phenocopied p110-t-ERBB2-driven cell migration and invasion, while expression of constitutively active STAT5 reversed these effects. Thus, we provide novel evidence that (1) expression of p110 t-ERBB2 is sufficient for full transformation of HMEC, yielding in vivo xenograft formation, and (2) truncated p110 t-ERBB2 expression is associated with decreased phosphorylation of STAT5.
Keywords: ERBB2, truncated, p95, p110, breast cancer, STAT5
Introduction
Approximately 20% of breast cancers (BCs) harbor amplification of the ERBB2 proto-oncogene, yielding overexpression of ERBB2 (HER2) receptor. Before advent of ERBB2-targeted therapies, patients with ERBB2+ tumors experienced poor clinical outcome.1, 2 The humanized monoclonal antibody trastuzumab (Herceptin) targets the extracellular domain (ECD) of full-length p185-ERBB2 receptor, and has improved prognosis for many patients with ERBB2+ BC.3, 4, 5, 6, 7 However, only subsets of ERBB2+ patients respond to first-line trastuzumab, and resistance to trastuzumab therapy occurs frequently.5, 8, 9, 10
Several truncated ERBB2 isoforms have been described in human BC, arising via alternative mRNA translation and metalloproteinase cleavage.11, 12 Membrane-localized t-ERBB2 isoforms (t-ERBB2s) can activate AKT and mitogen-activated protein signaling in BC cells;13, 14 however, they lack the bulk of receptor ECD (including the target epitope of trastuzumab) and may confer trastuzumab resistance;15 indeed, patients with t-ERBB2+ BC exhibit impaired trastuzumab response.13, 16 Furthermore, t-ERBB2 expression correlates with increased nodal involvement, and t-ERBB2s are more frequently expressed in metastases than primary tumors.17, 18, 19 Importantly, t-ERBB2 expression is associated with shorter progression-free and overall survival of metastatic BC patients, including those treated with trastuzumab.16, 18, 20
Three t-ERBB2s have been described in clinical specimens and ERBB2-amplified cell lines (Figure 1a): p110 (also referred to as 611CTF17), which arises by alternative translation of ERBB2 mRNA; p95m (m=membrane, also 648CTF), arising via proteolytic cleavage of full-length receptor;21 importantly, both p110 and p95m isoforms contain receptor transmembrane (TM) domain. p95cyto (cytoplasmic, 687CTF), an isoform lacking TM domain, is expressed in the cytoplasm.11, 12 Finally, roles for ERBB2 isoforms in cell nuclei have also been described.22, 23, 24
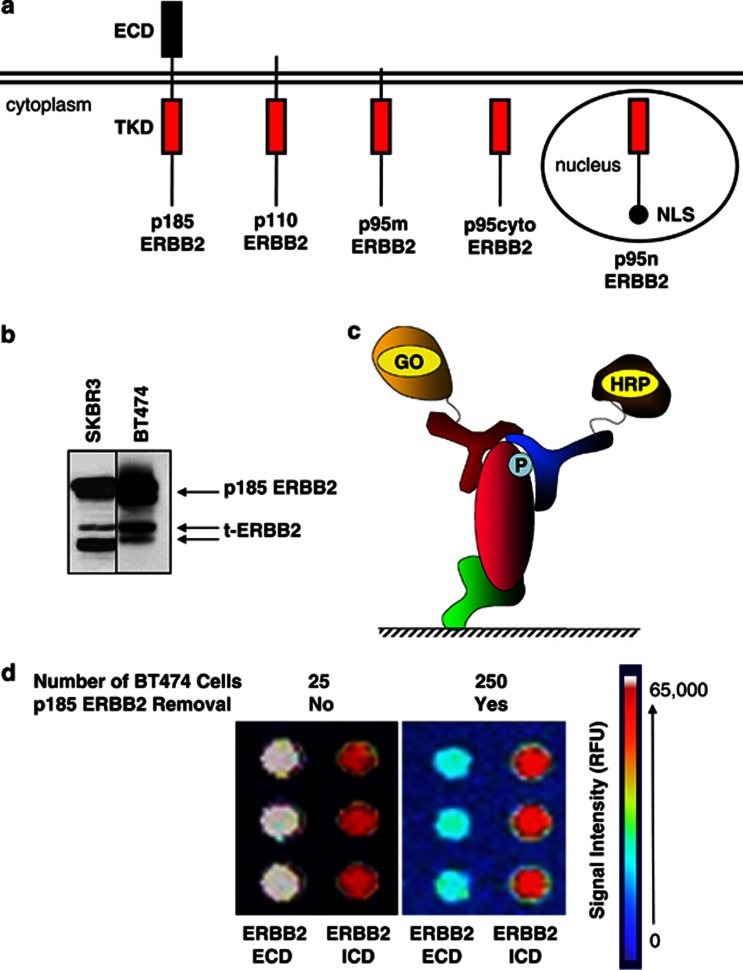
Figure 1.
Detection of ERBB2 isoforms in human breast cancer cell lines. (a) Schematic representation of full-length and truncated ERBB2 isoforms. p185, p110 and p95m isoforms contain transmembrane domain, whereas p95cyto lacks this domain. p95n is targeted to the nucleus by the addition of two tandem nuclear localization sequence motifs on the C terminus. ECD, extracellular domain; TKD, tyrosine kinase domain; NLS, nuclear localization sequence. (b) Western blots of SKBR3 and BT474 lysates reveal detectable expression of truncated ERBB2 isoforms in human ERBB2+ breast cancer cell lines. (c) Schematic of the CEER detection method. Target substrate is immobilized with a capture antibody (green). The first detector antibody (red) conjugated to GO binds to the captured target substrate using a different epitope than the capture antibody. The second detector antibody (blue) conjugated to HRP binds to a third epitope and completes the formation of the immuno-complex necessary for signal generation. GO, glucose oxidase; HRP, horseradish peroxidase; P, phosphorylated residue. (d) Detection of t-ERBB2 isoforms in BT474 cells using the CEER assay. Total protein lysate from 25 BT474 cells (left panel) or protein lysate from 250 BT474 cells from which p185 ERBB2 was removed with an ECD targeting antibody (right panel) were tested for the expression of ERBB2 using antibodies targeting both the ERBB2-ECD and ICD. Following removal of p185 ERBB2 (right panel), primarily t-ERBB2 isoforms are present, which are detected with the ERBB2-ICD targeting antibody (red signal is near maximal signal), but not with an ECD targeting antibody (light blue is close to the background signal). The image for post-p185 ERBB2 removed BT474 profile is shown at a higher photomultiplier tube (Photo Multiplier Tube, hence higher background signal) gain set as the signal was almost non-detectable with the ERBB2-ECD antibody. Utilizing BT474 lysates with and without p185ERBB2 removal and ICD capture configuration, the number of t-ERBB2 per cell were determined to be ∼5.3 × 104 receptors per cell, compared to 1.2 × 106 total ERBB2 receptors per cell. Therefore, the percentage of t-ERBB2 in BT474 cells is determined to be approximately 4.3% (13/300).
Clinical testing for ERBB2 overexpression frequently relies on an intracellular domain-specific antibody (HercepTest; Dako, Carpenteria, CA, USA),25, 26 and thus cannot distinguish between ERBB2 isoforms. If t-ERBB2 expression impacts trastuzumab response, accurate measurement of t-ERBB2s may influence choice of targeted therapy, for example, therapeutic antibodies vs kinase inhibitors.13, 16 Therefore, methods distinguishing full-length from t-ERBB2s in a sensitive and specific manner are needed. Herein, we demonstrate a novel antibody-microarray format, Collaborative Enzyme Enhanced Reactive Immunoassay (CEER),27 to quantify total and phosphorylated t-ERBB2s in human BC, using frozen primary specimens and fine-needle aspirate (FNA) metastatic biopsies. This method is able to detect and discern full length vs t-ERBB2s with a high degree of sensitivity, using minute amounts of biopsied tumor material.
Having confirmed t-ERBB2 expression in a clinical cohort, we next investigated functional effects of t-ERBB2s in HMLEs.28, 29 cDNAs encoding ERBB2 isoforms were stably expressed in human mammary epithelial cell lines, HMLE, HME and MCF10A. t-ERBB2 effects on cells were investigated in vitro using soft agar colony formation, and invasion and migration assays. In addition, in vivo engraftment of HMLE cells expressing t-ERBB2s was investigated using orthotopic xenografts in non-obese diabetic/severe combined immune deficiency mice.
The signal transducer and activator of transcription 5 (STAT5) transcription factor has an important role in directing differentiation of both normal and transformed breast epithelial cells,30 and ERBB receptors (including ERBB2) can regulate phosphorylation of STAT5.31 Levels of phosphorylated STAT5 (pSTAT5) are inversely correlated with BC tumor stage, and pSTAT5 predicts favorable outcome in patients.30, 32 In vitro, pSTAT5 inhibits BC cell migration and invasion, potentially explaining its association with favorable prognosis.33 Herein, we report attenuation of pSTAT5 accompanying p110-t-ERBB2 expression in multiple BC cell types.
Results
Detection of t-ERBB2s in BC
Full-length and t-ERBB2s are present in cultured BC cell lines SKBR3 and BT474 (Figure 1b). In addition, t-ERBB2s exist in a subset of ERBB2-amplified human BC.16, 17, 18 Thus, we sought to confirm the expression of t-ERBB2s in a cohort of BC specimens, using a novel antibody-capture, proximity-based immune-microarray platform34 (CEER; Figure 1c). This assay requires immunocomplex formation between target-specific capture antibody and two detector antibodies (one conjugated to glucose oxidase and one to horseradish peroxidase (HRP)) by binding to a single target protein.27, 35, 36 As shown in Figure 1d, the assay detects total and phosphorylated t-ERBB2 using lysates from as few as 250 BT474 cells (see Materials and methods and Supplementary Figure 1A). Comparison of differential ERBB2 profiling (with ERBB2-ECD and ERBB2-intracellular domain (ICD) captures) of BT474 cells with and without the removal of full-length p185-ERBB2 demonstrated that approximately 4.3% of the total ERBB2 receptors were t-ERBB2s (or ∼5.3 × 104 t-ERBB2 receptors per cell) in this ERBB2-amplified cell line, with about 1.2 × 106 total ERBB2 receptors per cell. The enrichment of t-ERBB2 was efficient, as removal of p185-ERBB2 from lysates prepared from 250 BT474 cells yielded substantially lower signal with ERBB2-ECD capture antibody, indicating that the signal in ERBB2-ICD antibody capture is predominantly from t-ERBB2. Levels of expression and phosphorylation of t-ERBB2s were determined by comparing values of both capture antibodies on samples before and after p185-ERBB2 clearance by immunoprecipitation. Standard curves of levels of total ERBB2 and phosphorylated ERBB2 were generated as references, using CEER analysis of the BT474 cell line (Supplementary Figure 2).
Frozen BC specimens from 74 patients were scored for ERBB2 levels using standard immunohistochemical analysis (HercepTest) and then analyzed using CEER (Table 1 and Supplementary Table S1). In all, 24 samples were ERBB2 low/negative (score=0–1 by immunohistochemistry (IHC)), 19 were IHC score=2 and 31 had high-level expression of ERBB2 (IHC score=3+). The numbers and percentages of samples harboring t-ERBB2 receptors in each IHC group are summarized in Table 1. In this cohort, two IHC=0/1 specimens had detectable full-length ERBB2 above background, but none had t-ERBB2 above background cutoff (p185 level below 300 000 RTK ng−1 was considered background based on CEER measurement of ERBB2 in T47D non-amplified cell line). In all, 15.7% (3 of 19) of IHC2 ERBB2+ tumors expressed t-ERBB2, whereas 58% (18 of 31) of IHC3+ ERBB2+ tumors expressed t-ERBB2. The association of t-ERBB2 expression with IHC3+ score was statistically significant (IHC3+ cohort compared to others, P<0.001). Furthermore, detection via pan-antiphosphotyrosine antibody revealed that t-ERBB2 isoforms were phosphorylated in subsets of ERBB2 IHC2 tumors (11%, 2 of 19 samples) and IHC3+ tumors (32%, 10 of 31). IHC3+ score was significantly associated with detectable phosphorylated t-ERBB2 (IHC3+ cohort compared to others, P<0.005). CEER-ERBB2 profiling and immunoprecipitation-western blot analyses of ERBB2 isoforms in three individual tumor samples are provided in Figures 2a and b, respectively (values for these samples are highlighted in red; Supplementary Table S1). Supplementary Table S1 provides t-ERBB2 quantification for all 74 samples. Lastly, FNAs of metastatic lesions from eight BC patients were analyzed. Three samples of ERBB2 IHC+ metastases showed varying degrees of total and phosphorylated t-ERBB2 (Table S2), whereas t-ERBB2 were not detected in IHC− metastases (not shown).
Table 1. Expression and phosphorylation of truncated ERBB2 receptors in human breast cancer specimens.
| IHC | ERBB2 score 0/1 | ERBB2 score 2 | ERBB2 score 3 |
|---|---|---|---|
| Sample no. (n) | 24 | 19 | 31 |
| CEER t-ERBB2+ | 0 | 3 | 18 |
| CEER t-ERBB2-P+ | 0 | 2 | 10 |
| % t-ERBB2+ | 0 | 15.7 | 58 |
| % t-ERBB2-P+ | 0 | 11 | 32 |
Abbreviations: CEER, collaborative enzyme enhanced reactive immunoassay; IHC, immunohistochemistry; t-ERBB2, truncated-ERBB2 isoforms.
Tumor samples scored by IHC for ERBB2 expression were measured by CEER for t-ERBB2 and phosphorylated t-ERBB2. The number and percentage positive for t-ERBB2 and phospho-t-ERBB2 are provided. IHC3+ score significantly associates with the presence of total (P<0.0001) and phosphorylated (P<0.005) t-ERBB2 (two-tailed Fisher's exact test).
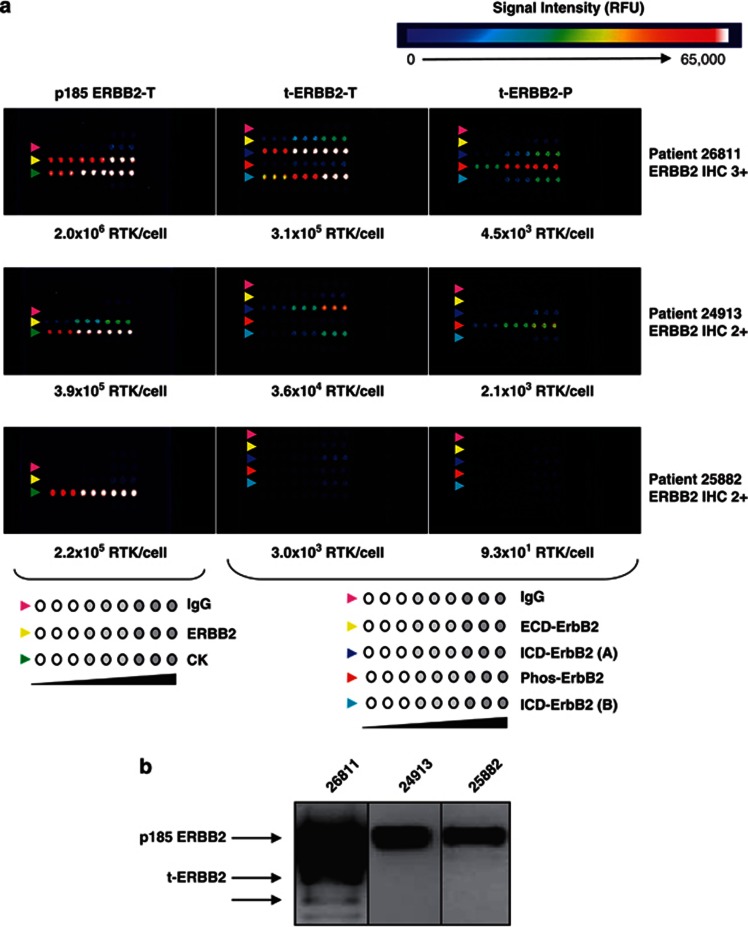
Figure 2.
Detection of ERBB2 isoforms in primary human breast tumor samples. (a) CEER analysis of human tumor samples reveals the presence and phosphorylation of t-ERBB2s. Lysates of tumors from three patients with high (no. 26811) or intermediate (nos. 24913 and 25882) ERBB2 expression as scored by IHC were tested. Assay configuration is illustrated below the CEER panels. Antibodies were arrayed in triplicate at three concentrations. Left panel antibodies for total p185 ERBB2 assay: control IgG (pink arrows), ECD-ERBB2 (yellow arrows) and cytokeratin (CK, green arrows). Center and right panel antibodies for total and phosphorylated t-ERBB2 assay: control IgG (pink arrows), ECD-ERBB2 (yellow arrows), ICD-ERBB2 (dark and light blue arrows) and phosphorylated ERBB2 (red arrows). Before t-ERBB2 CEER assay (center and right panels), full-length p185 ERBB2 was removed with an N-terminal ECD-directed antibody. (b) Western blot analysis of ERBB2 isoforms in human breast tumor samples with varying levels of ERBB2 (by IHC) assayed in part (a). Patient specimen lysates 26811, 24913 and 25882 were probed with antibody against ICD-ERBB2. Patient 26811 with positive western analysis also showed high level of ERBB2 as well as t-ERBB2 expression by CEER, with the use of much less cell lysate. Using CEER configuration, a quantitative detection of full-length ERBB2 and t-ERBB2 was achieved in samples (24913 and 25882) with IHC2+ status. Of note, t-ERBB2 was not detected by western blot analysis in these samples. Furthermore, a robust phosphorylation of t-ERBB2 was observed in patient 26811, whereas lower level was detected in patient 24913.
Engineered expression of t-ERBB2s
Having confirmed t-ERBB2 expression and phosphorylation in human BC specimens, we sought to determine their biological effects in HMLEs. cDNAs encoding p185, p110, p95m and p95cyto were stably expressed using pLXSN retroviral vector (Figure 3a). In addition, a nuclear isoform (p95n) was constructed by adding two tandem nuclear localization sequences to the p95cyto C terminus (see Figure 1a). Importantly, parental HMLEs lack detectable endogenous t-ERBB2s (Figure 3a). Expression of p185-ERBB2 was used as positive control, while non-transduced and empty-vector-transduced cells served as negative controls in experiments. As shown in Figure 3a, western blotting of HMLE lysates confirmed the expression of ERBB2 isoforms in each stable pool of cells. While protein expression levels varied slightly, comparable levels of p110 compared to p95m and p95cyto compared to p95n were achieved in these pooled populations. HMLE-p110 cells also express the p95cyto isoform due to alternate translation from the downstream AUG codon corresponding to methionine 687 in the ERBB2 peptide sequence (see Figure 3b, upper left panel); however, we have also engineered a version of p110 t-ERBB2 with a mutated 687 methionine codon (M687G) to prevent translation of p95cyto (see western blot; Supplementary Figure 1B). This construct has yielded the same phenotypic effects in cells as the original (data not shown). Although an unknown lower molecular weight band does appear in the p185 lysates, repeated blots indicate that p110 and p95 isoforms are not expressed in the p185-transduced population (see Supplementary Figure 1B).
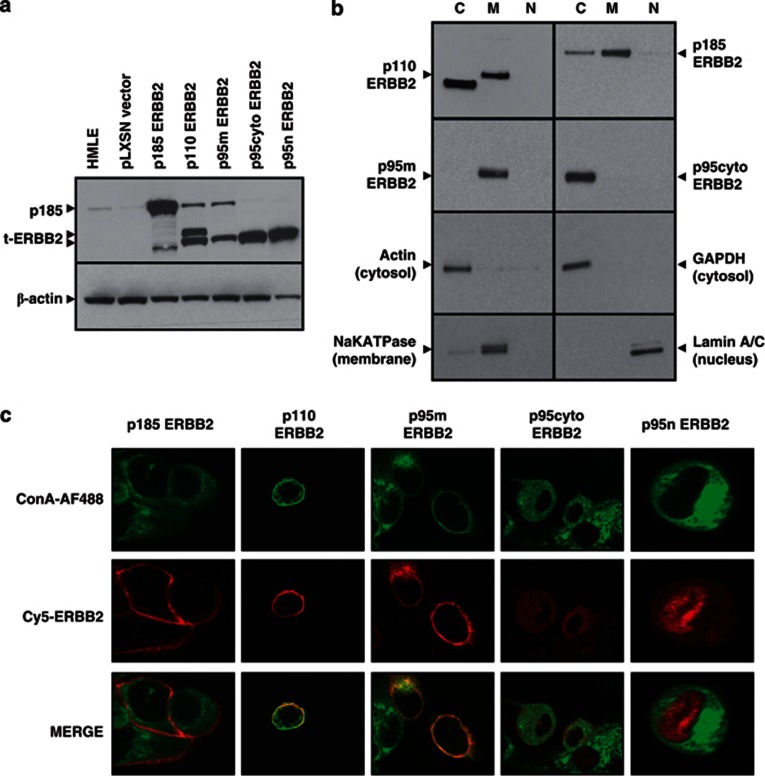
Figure 3.
Expression and localization of ERBB2 receptor isoforms in HMLE cells. (a) Lysates from parental or recombinant ERBB2-transduced HMLE cells were probed with anti-ERBB2 antibody in a western blot assay. Actin was blotted as loading control. (b) Recombinant HMLE cells were fractionated into cytoplasmic ‘C', membrane ‘M' and nuclear ‘N' fractions, followed by western blotting for ERBB2. Control antibodies identify proteins restricted to the cytoplasm (GAPDH, actin), the plasma membrane (NaK-ATPase) and the nucleus (Lamin A/C). p110 HMLE also express the p95cyto isoform due to translation from the AUG codon corresponding to methionine 687. (c) Confocal microscopy of recombinant HMLE cells. Plasma membranes were stained with concanavalin A (green), followed by a mouse anti-ERBB2 antibody and anti-mouse Cy5 (red) for receptor labeling. p185, p110 and p95m reside primarily in the plasma membrane, whereas p95cyto and p95n reside in the cytoplasm and nucleus, respectively. Images are representative of the population at large and were taken at × 80 magnification on a Zeiss confocal microscope. Cells expressing p95n were analyzed at higher magnification to confirm nuclear localization.
Subcellular localization of ERBB2 isoforms
Alternative translation of ERBB2 mRNA is responsible for the generation of p110 (CTF611) t-ERBB2.12 This isoform lacks an N-terminal leader peptide to facilitate canonical membrane protein trafficking.17 We investigated subcellular localization of this isoform using two complementary methods, western blots of cell protein fractions (Figure 3b) and immunofluorescence confocal microscopy (Figure 3c). p185, p110 and p95m receptors (those harboring TM domain) were localized to cell membranes. The p95m cDNA used in this study also lacks a canonical N-terminal leader peptide, yet this isoform also resides in the plasma membrane. Thus, it is apparent that the TM domain is sufficient to drive plasma membrane localization of p110 t-ERBB2. In contrast, p95cyto existed predominantly in the cytoplasm, whereas engineered nuclear isoform p95n was predominantly localized to the nucleus (this image is at × 2 higher magnification to enhance detail). Because of the loss of protein during the extraction method for fractionation, some signal is lost for p185 ERBB2 in the p110 and p95m lanes.
p110-t-ERBB2 enhances cell migration and invasion in vitro
Expression of t-ERBB2 has been previously associated with increased nodal involvement and metastasis,18 suggesting a role for t-ERBB2s in the pathophysiology of BC migration and invasion. Therefore, we analyzed effects of t-ERBB2s using in vitro migration and invasion assays. As shown in Figure 4a, p110 expression enhanced migration of HMLEs compared to p185 ERBB2 (p110 compared to p185, P=0.04). While p95m increased migration compared to parental and empty-vector HMLEs, its effect was not significant compared to p185. Neither p95cyto nor p95n significantly affected cell migration. To rule out cell type- or population-specific effects, we also determined effects of p185 and p110 expression in HME and MCF10A cells. p110 t-ERBB2 also significantly increased migration of HME cells (Figure 4c) compared to p185. MCF10A migration was also enhanced by the expression of p110 compared to p185 (mean number of cells per field=464 (p110) vs 182 (p185), P=0.004 (Student's t-test)).
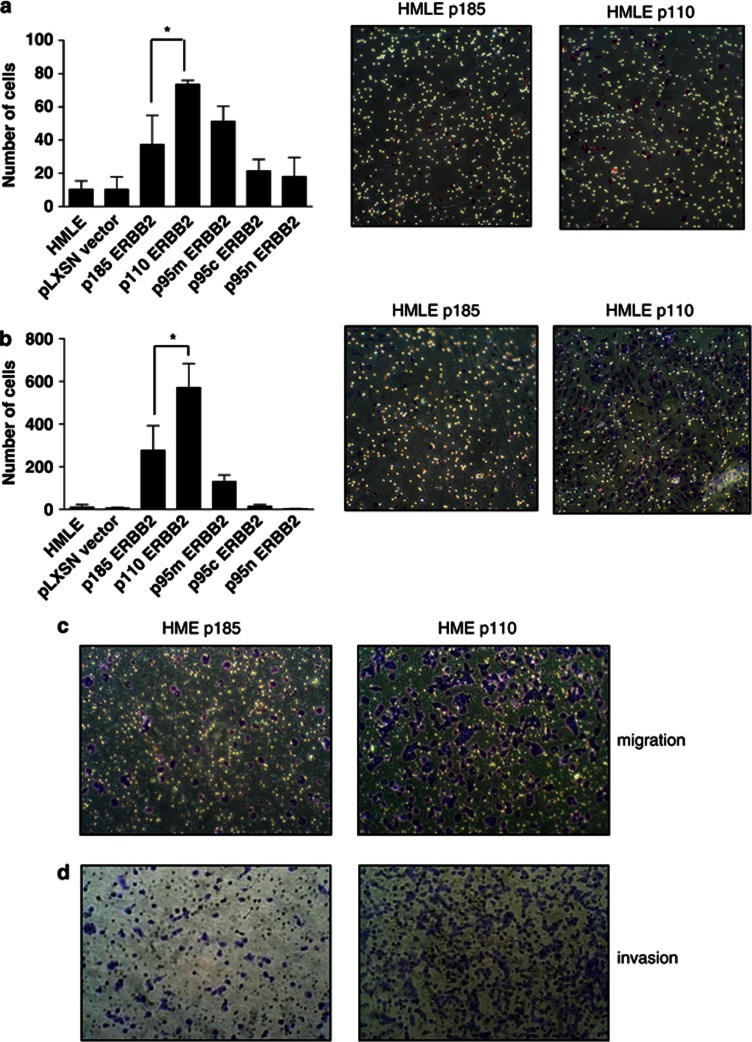
Figure 4.
p110 t-ERBB2 increases migration and invasion of HMLE and HME cells in vitro. (a) Migration of parental and recombinant HMLE cells. Numbers of cells in nine random fields on the lower sides of triplicate 8 μm filters were counted after 24 h. *P=0.04 (Student's t-test). Shown are representative images of HMLE p185 and HMLE p110 migration transwells after staining with crystal violet. (b) Invasion through Matrigel of parental and recombinant HMLE cells. Numbers of cells on the lower sides of triplicate 8 μm filters were counted after 36 h. *P=0.03 (Student's t-test). Shown are representative images of HMLE p185 and HMLE p110 invasion transwells after staining with crystal violet. (c) HME cells expressing p185 and p110 isoforms of t-ERBB2 were used in migration assays, and representative images of transwell membranes are shown. (d) HME cells expressing p185 and p110 isoforms of t-ERBB2 were used in invasion assays, and representative images of transwell membranes are shown.
Invasion assays using Matrigel-coated filters (Figure 4b) revealed that p110 and p95m isoforms increased HMLE invasion compared to parental and vector control cells (p110 vs controls, P=0.001; p95m vs controls, P=0.003); however, only p110 significantly increased invasion compared to p185-expressing cells (P=0.03). p110-expressing HME cells were also more invasive than p185-expressing cells (Figure 4d). Importantly, the differences seen in these assays were not due to enhanced proliferation by subsets of cells, as there was no significant difference in the proliferation rate of HMLE cells expressing the different isoforms within the 36 h window of this assay (Supplementary Figure 3C).
In addition, the ability of HMLEs expressing different isoforms of ERBB2 to form colonies in soft agar was assayed. The number of colonies formed by HMLEs significantly increased with the expression of membrane-bound isoforms p185 (P=0.0001), p95m (P=0.003) and p110-ERBB2 (P=0.0001) when compared to vector control cells; however, this effect was not elicited by intracellular t-ERBB2 species (Supplementary Figure 3A).
p110 t-ERBB2 promotes HMLE xenograft formation
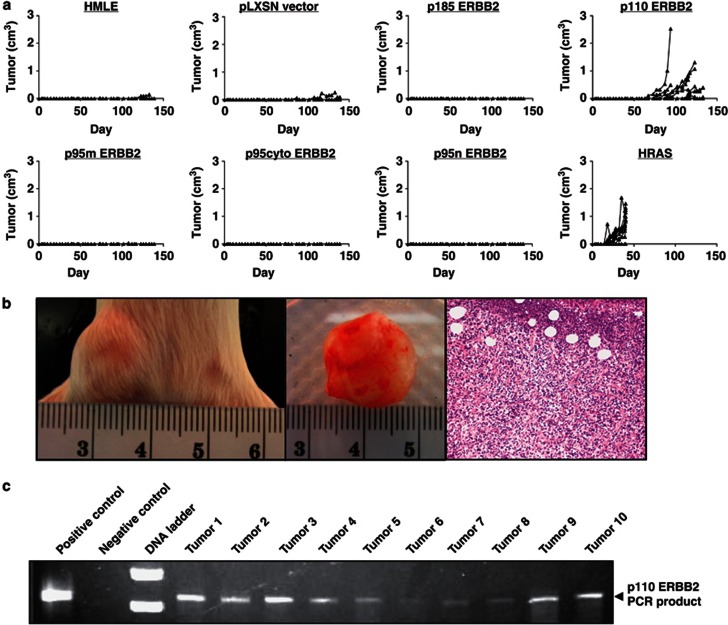
Transgenic mouse models have implicated p110 (611CTF) in promoting primary breast tumor formation.17 Thus, we investigated t-ERBB2 effects on orthotopic xenograft formation by HMLE cells. Recombinant HMLE cells were grafted into mammary fat pads of non-obese diabetic/severe combined immune deficient mice. Cells expressing oncogenic HRAS were used as positive controls for xenograft formation, whereas parental and empty vector cells served as negative controls. As shown in Figure 5a, only HMLEs expressing HRAS or p110-t-ERBB2 formed detectable xenografts within 150 days. HMLE-HRAS xenografts exhibited a shorter latency period (mean time to xenograft detection=19.5 days; 95% confidence interval:±10.0 days) than p110 t-ERBB2 tumors (mean time to detection=98 days; 95% confidence interval: ±41.3 days). Images of a p110 HMLE xenograft in situ, ex vivo and a hematoxylin/eosin-stained section are shown in Figure 5b. Polymerase chain reaction using ERBB2-specific forward primer and vector-specific reverse primer demonstrated that genomic DNA from HMLE-p110 tumors was positive for pLXSN-p110 construct (Figure 5c). Expression of other ERBB2s, including p185, did not lead to xenograft outgrowth within 150 days.
Figure 5.
p110 t-ERBB2 promotes HMLE orthotopic xenograft formation. (a) Growth curves of parental and recombinant HMLE xenografts over 150 days. (b) Photographs and hematoxylin and eosin staining of a representative p110-HMLE tumor. (c) Polymerase chain reaction using vector and ERBB2 targeted primers and genomic DNA from p110-HMLE tumors as template. Polymerase chain reaction lacking template was used for the negative control reaction.
STAT5 phosphorylation is attenuated by p110-t-ERBB2 expression
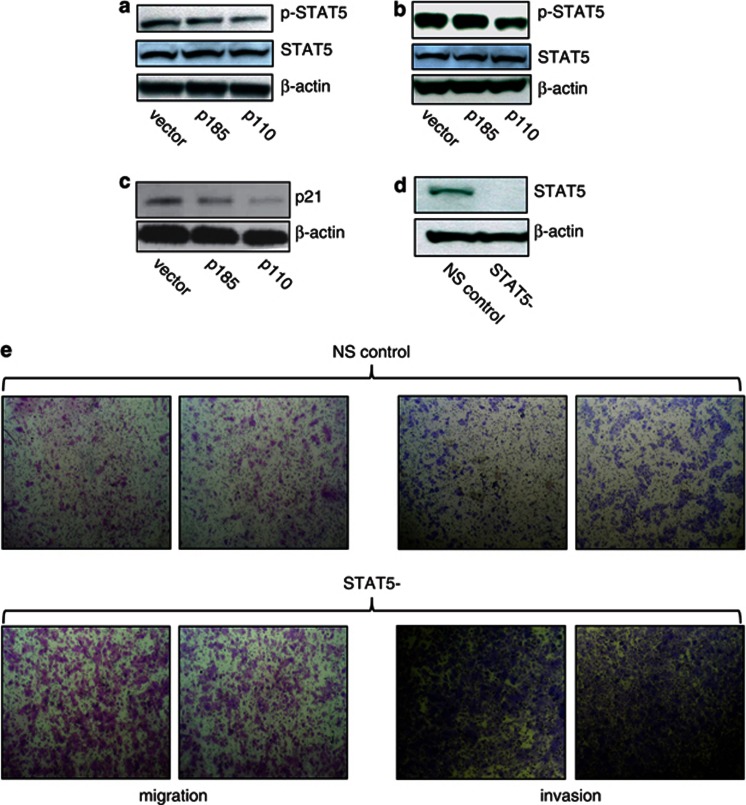
On the basis of our in vitro and in vivo observations, we sought a biological basis for p110-driven phenotypes. Unexpectedly, we did not observe significant changes in phosphorylation of MEK1/2, ERK1/2 or AKT signaling proteins in p110- compared to p185-expressing HMLE cells when examined by western blotting or CEER (data not shown), in contrast to a published study using engineered expression in MCF7 cells.14 In addition, there was no significant proliferation rate difference between the HMLE populations (Supplementary Figure 3b). To determine potential mechanisms by which p110 t-ERBB2 elicits phenotypes distinct from p185, we used the Proteome Profiler array (R&D Systems, Minneapolis, MN, USA; no. ARY003) to examine phosphorylation of multiple signaling proteins in HMLE cells. Analysis revealed decreased phosphorylation of STAT5 in HMLE cells expressing p110 isoform, compared to p185-expressing and control HMLE cells (Supplementary Figure 3B). Densitometric analysis revealed approximately twofold reduction of pSTAT5 by this method. Moreover, western blotting verified loss of pSTAT5 in p110-expressing HMLE cells (Figure 6a), and CEER measurement confirmed approximate twofold decrease of pSTAT5 in p110-HMLE compared to p185-HMLE (data not shown). pSTAT5 was also decreased in an independent p110-expressing cell line, HME, when compared to expression of p185 ERBB2 (Figure 6b), ruling out population- and cell-type-specific effects. Loss of pSTAT5 has previously been associated with increased tumor grade and poor clinical outcome in human BC30, 32 (see Discussion). To confirm that the decrease in pSTAT5 was sufficient to effect STAT5-dependent gene transcription, we analyzed levels of the STAT5-responsive gene p21WAF1/CIP1. p21 protein was decreased in p110-expressing HMLE cells compared to p185 and control HMLE cells (Figure 6c).
Figure 6.
STAT5 phosphorylation is attenuated in p110 t-ERBB2-expressing cells, and elicits migration and invasion of HMLE cells. (a) Western blot of phospho-STAT5 in lysates from recombinant HMLE cells expressing ERBB2 isoforms. (b) Western blot of phospho-STAT5 in lysates from recombinant HME cells expressing ERBB2 isoforms. (c) Western blot of p21 in lysates from recombinant HMLE cells expressing ERBB2 isoforms. (d) Western blot of total STAT5 protein in lysates from HMLE transduced with non-silencing control (NSC) shRNA or STAT5-targeted shRNA. (e) Migration (left panels) and invasion (right panels) of control HMLE (top rows) and HMLE with silenced STAT5 expression (bottom rows) from (d).
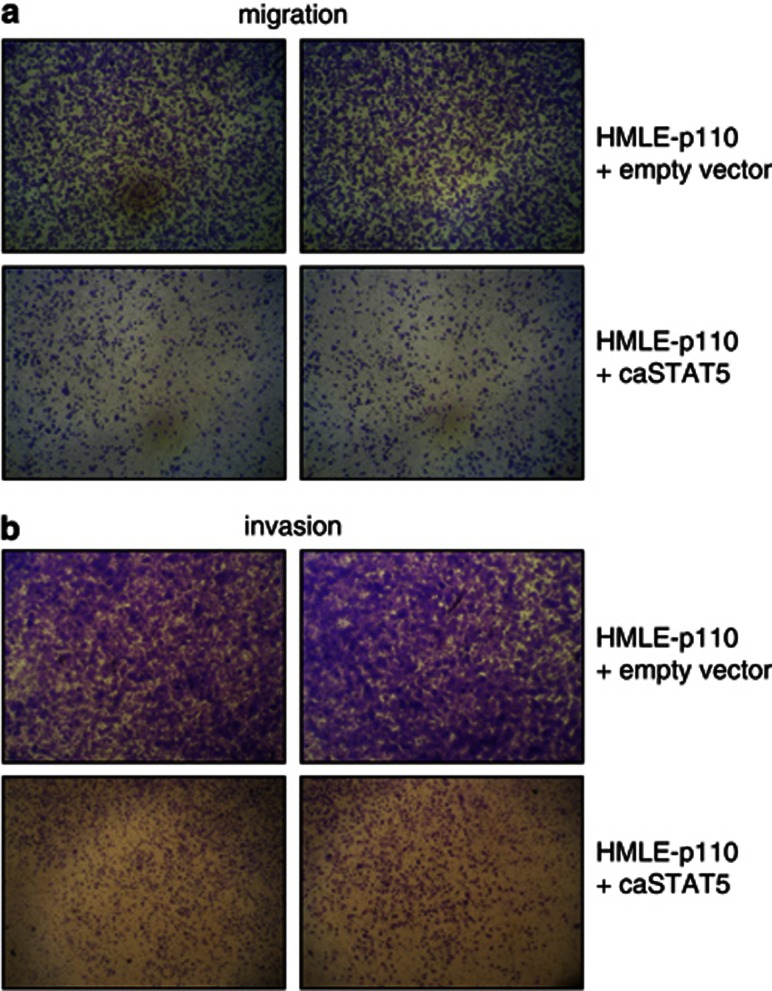
To determine if decreased pSTAT5 is sufficient to elicit phenotypes similar to those elicited by p110, we transduced HMLE cells with short hairpin (shRNA) lentivirus (Open Biosystems, Hunstville, AL, USA) to silence STAT5 expression. As shown in Figure 6d, HMLE cells transduced with shRNA virus have reduced expression of STAT5 protein, compared to control cells with transduced with non-silencing control shRNA. Importantly, STAT5-silenced HMLE cells exhibited enhanced migration (Figure 6e, left panels) and invasion (Figure 6e, right panels) when compared to control cells using transwell assays in vitro, mimicking phenotypes observed in p110-t-ERBB2-expressing cells. To confirm the role of pSTAT5 loss in the migration and invasion driven by p110 expression, we expressed a constitutively active mutant form of STAT5b (STAT5b1*6) in HMLE cells also expressing p110 t-ERBB2. Expression of constitutively active STAT5b in p110 HMLE reversed the p110-driven migratory (Figure 7a) and invasive phenotypes (Figure 7b).
Figure 7.
Constitutively active STAT5 reverses p110-driven migration and invasion of HMLE cells. HMLE cells stably expressing p110 t-ERBB2 were transduced with empty vector or constitutively active STAT5b vector. (a) Migration of p110-HMLE cells expressing empty vector (top panels) or caSTAT5b (lower panels). Representative photographs of transwell membranes are shown. (b) Invasion through Matrigel by p110-HMLE cells expressing empty vector (top panels) or caSTAT5b (lower panels). Representative photographs of transwell membranes are shown.
Discussion
Routine clinical testing of BC does not currently assess the expression of t-ERBB2 vs p185-ERBB2.37 Although t-ERBB2s may confer resistance to trastuzumab therapy, ERBB2 kinase inhibitors including lapatinib inhibit the activity of t-ERBB2s.38 Therefore, sensitive methods to accurately and reliably measure expression of t-ERBB2s in BC are needed, and could potentially impact choice of anti-ERBB2 therapy.
Recently, t-ERBB2 levels in clinical samples have been assessed by antibody-based VeraTag assay.16 This method requires formalin-fixed and paraffin-embedded tissue that may alter antigenicity due to fixation and antigen retrieval procedures. Therefore, detection of minor t-ERBB2 variants against the background of predominant full-length ERBB2 proteins can be challenging.
In contrast, CEER utilizes tissues (FNA or biopsy) without fixation, thus minimizing the probability of antigen modification during processing. Full-length ERBB2 proteins present in sample lysate are removed immunomagnetically using an antibody against the ECD of ERBB2 (as shown in Supplementary Figure 1A), followed by detection of t-ERBB2 variants using three different ICD antibodies. This configuration combined with the superior sensitivity of the proximity-mediated platform allows minute amounts of t-ERBB2 to be efficiently detected and quantified.
In a cohort of ERBB2+ BC, 11% (2 of 19) of IHC2+ tumors expressed t-ERBB2 and 52% (16 of 31) of IHC3+ tumors expressed t-ERBB2. Furthermore, t-ERBB2 isoforms were phosphorylated in IHC3+ tumors (32%, 10 of 31) and a minority of IHC2 tumors (11%, 2 of 19 samples). An IHC 3+ score was significantly associated with expression (P<0.0001) and phosphorylation (P<0.005) of t-ERBB2s when compared to IHC 0/1 and 2 groups. In several samples, t-ERBB2s accounted for 10–20% of total ERBB2 receptors per cell, revealing a significant fraction of t-ERBB2s in these specimens (Table 1 and Supplementary Table S1).17 Moreover, t-ERBB2s were detectable in FNA samples of metastatic lesions from several patients (Supplementary Table S2), suggesting potential clinical application of this technology in monitoring therapeutic response or progression of metastatic disease.
To examine effects of t-ERBB2 expression in human breast cells, isoforms were stably expressed in HMLE, HME and MCF10A cells. We confirm association of p110 t-ERBB2 with the plasma membrane, despite lack of a leader peptide sequence. Only the membrane-bound isoforms of ERBB2 (p185, p110 and p95m) elicited significant phenotypes in the invasion, migration and soft agar colony formation assays used in this study. It is important to note that we chose to analyze the effect(s) of each t-ERBB2 alone; it is possible that in ERBB2-amplified BC, there exists a mixed population of different isoforms of the receptors that yield other phenotypes. Although p185, p110 and p95m had similar effects on soft agar colony formation by HMLEs, p110 consistently and significantly enhanced HMLE and HME migration and invasion when compared to p185.17, 39 To rule out cell-type-specific effects, we also confirmed increased migration driven by p110 in MCF10A cells. Finally, only the expression of p110-t-ERBB2 led to HMLE xenograft formation in vivo. Consistent with our data, p110-ERBB2 transgenic mice develop mammary adenocarcinomas with a shorter latency than those in p185-ERBB2 transgenic animals.17 Combined, these data indicate that p110 t-ERBB2 is highly tumorigenic compared to p185 and other isoforms,40 identifying p110 as a potential therapeutic target in ERBB2+ BC.
Unexpectedly, expression of t-ERBB2s did not result in hyperactivation of mitogen-activated protein kinase or AKT signaling pathways, as was described in MCF7 cells.14 Importantly, MCF7 are a fully transformed human cell line with an activating mutation in PI3KCA (among other genetic alterations), possibly explaining the differences in our observations using partially transformed HMLE cells. Rather, p110 t-ERBB2 led to decreased pSTAT5 when compared to p185 ERBB2, in both HME and HMLE cells.
The role of STAT5 in human BC is currently controversial and not fully understood. Activated STAT5 may promote survival and antiapoptotic signaling in mammary cells, contributing to initial neoplastic transformation. However, the presence of pSTAT5 is inversely correlated with BC tumor stage.32 STAT5 is involved in differentiation of breast epithelium; thus, tumors with high pSTAT5 tend to be well-differentiated and have more favorable prognostic characteristics compared to those lacking pSTAT5,30 implying a role for STAT5 in the pathophysiology of BC. Accordingly, silencing of STAT5 in HMLE cells led to increased cell migration and invasion, mimicking p110 t-ERBB2 phenotypes, while expression of constitutively active STAT5 reversed p110-driven migration and invasion. Deregulation of STAT5 signaling has been implicated in motility, invasion and metastasis of BC cells.33, 41 Taken together, our data offer a mechanism by which p110 t-ERBB2 may augment malignant phenotypes such as migration and invasion, namely via perturbation of STAT5 signaling. Further elucidation of the molecular mechanism by which p110 t-ERBB2 exerts effects on STAT5 phosphorylation is an active area of research in our laboratory.
In summary, these data implicate p110-t-ERBB2 as a potential driver of tumorigenesis, migration and invasion of human mammary epithelial cells (HMECs), thus extending observations from fully transformed BC cell lines and transgenic mice. Perturbation of STAT5 signaling by p110 t-ERBB2 represents a newly identified mechanism of t-ERBB2 activity in human mammary cells. t-ERBB2s may be drivers of ERBB2+ BC pathogenesis, and could serve as biomarkers identifying patients that may differentially benefit from anti-ERBB2 therapies (monoclonal antibodies (Abs) vs TKIs).16, 42 As shown here, the specific and sensitive CEER detection method is able to detect and quantify t-ERBB2 isoforms in human breast tumor samples using minute amounts of clinical material, including metastasis samples. Accurate clinical measurement of t-ERBB2 expression in human tumors may help to guide appropriate ERBB2-targeted therapeutic approaches.
Materials and methods
Multiplexed-microarray printing
Capture Abs were printed on nitrocellulose-coated glass slides (ONCYTE; Grace Bio-Labs, Bend, OR, USA) using non-contact printers (Nanoplotter, GeSiM); spot diameters were approximately 175 µm. Slides were kept in desiccated chambers at 4 °C. Spots included tracking dye and specific capture Abs. Approximately 500 pl of capture Abs were printed in triplicate and serial dilution concentrations of 1, 0.5 and 0.25 mg ml−1. Purified mouse-IgGs served as negative controls.
Antibody conjugation
Target-specific Abs and detector enzymes, glucose oxidase or HRP, were activated with bi-functional crosslinker, succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate, and coupled making antibody–enzyme conjugates.35, 36 Conjugates were high performance liquid chromatography purified. Ab activities in purified conjugates were determined by competition enzyme-linked immunosorbent assay; enzyme activities were detected by functional assays specific for detector enzymes.
CEER34
Immuno-microarray slides were rinsed twice with TBST (50 mℳ Tris/150 mℳ NaCl/0.1% Tween-20, pH 7.2–7.4), blocked with 80 µl Whatman blocking buffer for 1 h at room temperature (RT), and then washed 2 × with TBST.34 Serially diluted lysate controls in 80 µl dilution buffer (2% bovine serum albumin/0.1% Triton X-100/TBS, pH 7.2–7.4) and samples were added to designated subarrays on slides, and then incubated for 1 h at RT. Slides were washed 4 × for 3 min, detector Abs were added in 80 µl reaction buffer and incubated for 2 h at RT. After TBST wash to remove unbound detector Abs, 80 µl of biotin-tyramide solution (5 µg ml−1 in 50 mℳ glucose/phosphate-buffered saline (PBS)) prepared from 400 µg ml−1 in ethanol solution (Perkin-Elmer, Waltham, MA, USA) was added and incubated for 15 min in darkness. The covalent coupling of the highly reactive, short-lived biotin-tyramide radicals to nucleophilic redisues in the proximity of interaction sites causes local deposition of biotinylated tyramide. Subsequent incubation with fluorescently labeled streptavidin generates signals that can be captured by fluorescence microarray scanner for visualization and quantitation. Glucose-oxidase/HRP-mediated tyramide signal amplification process was terminated by washing with TBST 4 × , 3 min each. Local deposition of biotin-tyramide was detected by incubation with streptavidin-Alexa Fluor 647 (Invitrogen, Grand Island, NY, USA) at 0.5 µg ml−1 in 2% bovine serum albumin/0.1% Triton/TBS for 40 min. Upon completion of incubation, slides were washed 4 × with TBST, dried and kept in darkness until imaged via microarray scanner.
t-ERBB2 enrichment
Full-length p185-ERBB2 receptors were cleared from lysates using magnetic-bead-coupled antibodies specific to ECD of ERBB2. Resulting p185-ERBB2 depleted lysates, which contained t-ERBB2 receptor proteins lacking the ECD, were used for subsequent quantification of t-ERBB2 expression and phosphorylation.
Clinical samples
Flash-frozen BC tissues were obtained from subjects with invasive ductal carcinoma, stages II or III (ILS Bio, Chestertown, MD, USA). Baseline ERBB2-IHC status was available for all samples. Flash-frozen tissue samples were lysed in 100 µl lysis buffer, kept on ice for 30 min and centrifuged. Protein concentrations of supernatants determined by BCA protein assay kit (Pierce, Rockford, IL, USA) and stored at −80 °C before analysis. FNA samples were collected according to an IRB-approved protocol from subjects with progressive, measurable metastatic stage IIIB or stage IV BC. Subjects had histologically or cytologically confirmed invasive BC. FNA samples were collected using G23-gauge needles. FNA samples were immediately injected into vials containing lysis buffer and shipped overnight for subsequent analysis.
CEER data analysis
Slides were scanned at three photomultiplier tubes gain settings to increase effective dynamic range. Background-corrected signal intensities were averaged from spots printed in triplicate. The relative fluorescence value of the respective reagent blank was subtracted from each sample. Several criteria were used to filter data from further analyses, including limits on spot footprints, coefficient of variation for spot replicates, overall pad background and intensity of reagent blank. For each assay, a standard curve was generated from serially diluted BT474 lysates. Data were fit to a five-parameter equation derived as a function of capture–antibody concentration and photomultiplier tubes. Each curve was plotted as a function of log signal intensity, measured as relative fluorescence unit (RFU) vs log concentration of cell lysates and referenced to the standard cell line, BT474. Individual predictions from each dilution and gain were averaged into a single, final prediction.
Clinical cohort statistical analysis
To determine concordance of IHC score with detection of t-ERBB2 and phospho-t-ERBB2, two-tailed Fisher's exact tests (Graphpad software) were used to determine statistical significance when comparing the number of positive samples in the IHC3 group vs IHC0/1 and IHC2 groups.
IP-western blotting
Cell lysates were incubated with magnetic bead-conjugated antibodies against intracellular domain of ERBB2 overnight at 4 °C. Immunomagnetically enriched lysates were resuspended in sample buffer containing β-mercaptoethanol, boiled 5 min, cooled to RT and loaded onto NuPage (Invitrogen) 4–12% gel. Separated proteins were transferred to nitrocellulose membranes, washed, blocked with 5% milk and incubated with 1° then 2° Abs before detection process using nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate.
Cells and culture
HMLE, HME and MCF10A cells were cultured in mammary epithelial growth medium (Lonza, Walkersville, MD, USA) in 5% CO2 atmosphere at 37 °C. Cells stably transduced with pLXSN vector encoding recombinant ERBB2 isoforms were selected using 0.5 mg ml−1 (HME and HMLE) or 0.75 mg ml−1 G418 (MCF10A), and were kept as pooled populations. HMLE p110 cells with added pLVX-puro constructs were selected by the addition of 0.5 μg ml−1 puromycin. BT474 and SKBR3 cells were cultured in RPMI media supplemented with 10% fetal bovine serum. For proliferation assays, 2500 HMLE cells were seeded in triplicate wells of 24-well plates. Cells were trypsinized and counted 24 and 72 h later using a Coulter Z1 counter.
Cloning and vectors
ERBB2 cDNAs were expressed using pLXSN retroviral vector. p185, p110, p95cyto and p95m were cloned using endogenous start codons from ERBB2 mRNA sequence, whereas an AUG start codon was engineered to initiate translation of p95m. Recombinant pLXSN vectors were packaged using Phoenix cells, virus was harvested, followed by HMLE transduction. Constitutively active STAT5b (STAT5b1*6) was cloned and expressed using pLVX-puro vector (Clontech, Mountain View, CA, USA) via similar techniques. GIPZ shRNA lentiviral vectors targeting STAT5 (Open Biosystems). Lentiviral particles were produced by cotransfection of GIPZ vector DNA with packaging plasmids pMD2.G and psPAX2 in 293 T cells. Viruses were harvested and used to transduce HMLE cells using standard techniques.
Immunofluorescence confocal microscopy
HMLEs were grown on coverslips to ∼50% confluence, washed with PBS and fixed in 4% paraformaldehyde for 20 min at RT. Slides were stained using concanavalin-A-Alexa Fluor 488 (Invitrogen; C11252) diluted in PBS for 30 min at RT, followed by PBS wash. After permeabilization with Triton X-100, anti-ERBB2 antibody (3B5; Abcam, Cambridge, MA, USA; AB16901) was applied in 5% bovine serum albumin-PBS for 1 h at RT, followed by anti-mouse Cy5-conjugated antibody (Millipore, Billerica, MA, USA) for 30 min. Coverslips were washed, mounted on microslides and imaged using Zeiss confocal microscope, followed by the AxioVision software processing.
Subcellular fractionation and western blotting
HMLEs were fractionated using QProteome cell-compartment kit (Qiagen, Valencia, CA, USA; no. 37502) according to the manufacturer's instructions. SKBR3 and BT474 cells were lysed in RIPA buffer. Lysates were run on 4–12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes, probed with anti-ERBB2 antibody (AB16901, 3B5; Abcam), anti-NaK-pump antibody (ab7671; Abcam), anti-lamin A/C antibody (sc-7293; Santa Cruz Biotechnology), anti-actin HRP antibody (Abcam) or anti-GAPDH antibody (ab9482; Abcam) for fraction controls. Blots were developed using standard chemiluminescence techniques.
For STAT5 and p21 western blots, total protein lysates from HMLE and HME cells were used in techniques similar to above, and probed with antiphospho-STAT5 Y699 antibody (antibody 5G4; Santa Cruz Biotechnology, Santa Cruz, CA, USA; no. sc-81524) or anti-p21 antibody (no. sc-397).
Soft agar assays
In all, 5000 HMLEs were seeded in 0.3% Noble agar-media mixture, on a layer of 0.6% agar in six-well plates. Cultures were maintained for 21 days at 37 °C and stained with 0.01% crystal violet in PBS overnight. Following staining, numbers of visible colonies were counted. Experiments were performed in triplicate, and one-way analysis of variance was used to determine significance.
Invasion and migration assays
In all, 50 000 serum-starved HMLEs were seeded into upper chambers of transwells (BD Biosciences, Bedford, MA, USA). For invasion assays, filters were overlaid with Matrigel. Media containing 20% fetal bovine serum was added to lower chambers as chemoattractant. After 24 h (migration) or 36 h (invasion), cells on top of filters were scraped away; filters were fixed with methanol and stained with crystal violet. Filters were mounted on microslides, images were taken at × 10 magnification using light microscopy. Migrating/invading cells were counted for each well. Experiments were performed in triplicate, and one-way analysis of variance and Student's t-tests were used to determine significance.
In vivo methods
Mammary fat pads 4 and 9 of female non-obese diabetic/severe combined immune deficiencymice were injected with 3 × 106 HMLE cells: parental (group 1); vector control (g2); p185-ERBB2 (g3); p110-t-ERBB2 (g4); p95m-t-ERBB2 (g5); p95cyto-t-ERBB2 (g6); p95n-t-ERBB2 (g7); and hRAS (g8). Six mice per group were injected, yielding 12 potential xenografts per group. Xenograft formation and growth was monitored until reaching 5% of bodyweight, or for a maximum of 150 days. Cells were injected in 200 μl of 1:1 (v v−1) mix of Matrigel/CollagenI. Work was performed following Institutional Animal Care and Use Committee guidelines.
Phosphoprotein array
R&D Systems Human phosphokinase array kit no. ARY003, protein lysates from parental HMLE cells and those stably transduced with empty vector, p185 ERBB2 or p110-t-ERBB2 were isolated and analyzed according to the manufacturer's instructions. For densitometric analysis, the ImageJ software (NIH, Bethesda, MD, USA) was used.
Acknowledgments
This work was supported by a Young Investigator Award from the Expedition-Inspiration Breast Cancer Research Fund, and by award T32CA119929 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institute of Health.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary Material
References
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Pegram MD, Pienkowski T, Northfelt DW, Eiermann W, Patel R, Fumoleau P, et al. Results of two open-label, multicenter phase II studies of docetaxel, platinum salts, and trastuzumab in HER2-positive advanced breast cancer. J Natl Cancer Inst. 2004;96:759–769. doi: 10.1093/jnci/djh133. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- Mass R.The role of HER-2 expression in predicting response to therapy in breast cancer Semin Oncol 200027(6 Suppl 1146–52., discussion 92–100. [PubMed] [Google Scholar]
- Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin) J Natl Cancer Inst. 2001;93:1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- Nahta R, Esteva FJ. Herceptin: mechanisms of action and resistance. Cancer Lett. 2006;232:123–138. doi: 10.1016/j.canlet.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Pupa SM, Menard S, Morelli D, Pozzi B, De Palo G, Colnaghi MI. The extracellular domain of the c-erbB-2 oncoprotein is released from tumor cells by proteolytic cleavage. Oncogene. 1993;8:2917–2923. [PubMed] [Google Scholar]
- Anido J, Scaltriti M, Bech SJJ, Santiago JB, Todo FR, Baselga J, et al. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. EMBO J. 2006;25:3234–3244. doi: 10.1038/sj.emboj.7601191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- Egeblad M, Mortensen OH, Jaattela M. Truncated ErbB2 receptor enhances ErbB1 signaling and induces reversible, ERK-independent loss of epithelial morphology. Int J Cancer. 2001;94:185–191. doi: 10.1002/ijc.1459. [DOI] [PubMed] [Google Scholar]
- Yuan CX, Lasut AL, Wynn R, Neff NT, Hollis GF, Ramaker ML, et al. Purification of Her-2 extracellular domain and identification of its cleavage site. Protein Expr Purif. 2003;29:217–222. doi: 10.1016/s1046-5928(03)00058-5. [DOI] [PubMed] [Google Scholar]
- Sperinde J, Jin X, Banerjee J, Penuel E, Saha A, Diedrich G, et al. Quantitation of p95HER2 in paraffin sections by using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin Cancer Res. 2010;16:4226–4235. doi: 10.1158/1078-0432.CCR-10-0410. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Angelini PD, Laos S, Bach-Faig A, Cunningham MP, Ferrer-Ramon C, et al. A naturally occurring HER2 carboxy-terminal fragment promotes mammary tumor growth and metastasis. Mol Cell Biol. 2009;29:3319–3331. doi: 10.1128/MCB.01803-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MA, Saez R, Ramsey EE, Garcia-Barchino MJ, Rojo F, Evans AJ, et al. NH(2)-terminal truncated HER-2 protein but not full-length receptor is associated with nodal metastasis in human breast cancer. Clin Cancer Res. 2002;8:347–353. [PubMed] [Google Scholar]
- Saez R, Molina MA, Ramsey EE, Rojo F, Keenan EJ, Albanell J, et al. p95HER-2 predicts worse outcome in patients with HER-2-positive breast cancer. Clin Cancer Res. 2006;12:424–431. doi: 10.1158/1078-0432.CCR-05-1807. [DOI] [PubMed] [Google Scholar]
- Christianson TA, Doherty JK, Lin YJ, Ramsey EE, Holmes R, Keenan EJ, et al. NH2-terminally truncated HER-2/neu protein: relationship with shedding of the extracellular domain and with prognostic factors in breast cancer. Cancer Res. 1998;58:5123–5129. [PubMed] [Google Scholar]
- Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R, et al. Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol Ther. 2006;5:657–664. doi: 10.4161/cbt.5.6.2708. [DOI] [PubMed] [Google Scholar]
- Giri DK, Ali-Seyed M, Li LY, Lee DF, Ling P, Bartholomeusz G, et al. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol Cell Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Lien HC, Xia W, Chen IF, Lo HW, Wang Z, et al. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell. 2004;6:251–261. doi: 10.1016/j.ccr.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Dillon MF, Stafford AT, Kelly G, Redmond AM, McIlroy M, Crotty TB, et al. Cyclooxygenase-2 predicts adverse effects of tamoxifen: a possible mechanism of role for nuclear HER2 in breast cancer patients. Endocr Relat Cancer. 2008;15:745–753. doi: 10.1677/ERC-08-0009. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Hashizume K, Kamihara Y, Itoh H, Tsuda H, Osamura RY, et al. Quantitative immunohistochemical evaluation of HER2/neu expression with HercepTestTM in breast carcinoma by image analysis. Pathol Int. 2001;51:33–36. doi: 10.1046/j.1440-1827.2001.01162.x. [DOI] [PubMed] [Google Scholar]
- Pauletti G, Dandekar S, Rong H, Ramos L, Peng H, Seshadri R, et al. Assessment of methods for tissue-based detection of the HER-2/neu alteration in human breast cancer: a direct comparison of fluorescence in situ hybridization and immunohistochemistry. J Clin Oncol. 2000;18:3651–3664. doi: 10.1200/JCO.2000.18.21.3651. [DOI] [PubMed] [Google Scholar]
- Liu PK, Liu L, Lee T, Barham R, Lin F, Harvey J, et al. Profiling of receptor tyrosine kinases (RTK) activation in circulating tumor cells (CTCs) in metastatic tumors using proximity-mediated microarray immunoassayASCO Annual Meeting.Orlando, FL; 2009(Abstract no. 11024). [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, et al. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7:966–976. doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE. ErbB receptor-induced activation of stat transcription factors is mediated by Src tyrosine kinases. J Biol Chem. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. 2004;22:2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–760. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- Kim P, Liu X, Lee T, Liu L, Barham R, Kirkland R, et al. Highly sensitive proximity mediated immunoassay reveals HER2 status conversion in the circulating tumor cells of metastatic breast cancer patients. Proteome Sci. 2011;9:75. doi: 10.1186/1477-5956-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper MH, Hackett DP. The oxidatic activity of horseradish peroxidase. I. Oxidation of hydro- and naphthohydroquinones. J Biol Chem. 1963;238:3736–42. [PubMed] [Google Scholar]
- Gibson QH, Swoboda BE, Massey V. Kinetics and mechanism of action of glucose oxidase. J Biol Chem. 1964;239:3927–3934. [PubMed] [Google Scholar]
- Xia W, Liu LH, Ho P, Spector NL. Truncated ErbB2 receptor (p95ErbB2) is regulated by heregulin through heterodimer formation with ErbB3 yet remains sensitive to the dual EGFR/ErbB2 kinase inhibitor GW572016. Oncogene. 2004;23:646–653. doi: 10.1038/sj.onc.1207166. [DOI] [PubMed] [Google Scholar]
- Scaltriti M, Chandarlapaty S, Prudkin L, Aura C, Jimenez J, Angelini PD, et al. Clinical benefit of lapatinib-based therapy in patients with human epidermal growth factor receptor 2-positive breast tumors coexpressing the truncated p95HER2 receptor. Clin Cancer Res. 2010;16:2688–2695. doi: 10.1158/1078-0432.CCR-09-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castillo J, Pedersen K, Angelini PD, Bech-Serra JJ, Colome N, Cunningham MP, et al. HER2 carboxyl-terminal fragments regulate cell migration and cortactin phosphorylation. J Biol Chem. 2009;284:25302–25313. doi: 10.1074/jbc.M109.001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Dankort DL, Hardy WR, Muller WJ. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan AS, Brim H, Sherif ZA. Co-overexpression of Janus kinase 2 and signal transducer and activator of transcription 5a promotes differentiation of mammary cancer cells through reversal of epithelial-mesenchymal transition. Cancer Sci. 2008;99:272–279. doi: 10.1111/j.1349-7006.2007.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas J, Parra-Palau JL, Pedersen K. HER2 fragmentation and breast cancer stratification. Clin Cancer Res. 2010;16:4071–4073. doi: 10.1158/1078-0432.CCR-10-1501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.