SUMMARY
Assembly of SAS-6 dimers to form the centriolar cartwheel requires the ZYG-1/Plk4 kinase. Here we show that ZYG-1 recruits SAS-6 to the mother centriole independently of its kinase activity; kinase activity is subsequently required for cartwheel assembly. We identify a direct interaction between ZYG-1 and the SAS-6 coiled-coil that explains its kinase activity-independent function in SAS-6 recruitment. Perturbing this interaction, or the interaction between an adjacent segment of the SAS-6 coiled-coil and SAS-5, prevented SAS-6 recruitment and cartwheel assembly. SAS-6 mutants with alanine substitutions in a previously described ZYG-1 target site or in 37 other residues, either phosphorylated by ZYG-1 in vitro or conserved in closely related nematodes, all supported cartwheel assembly. We propose that ZYG-1 binding to the SAS-6 coiled-coil recruits the SAS-6—SAS-5 complex to the mother centriole, where a ZYG-1 kinase activity-dependent step, whose target is unlikely to be SAS-6, triggers cartwheel assembly.
Keywords: Centriole, Plk4, Centrosome, Mitosis, Spindle
INTRODUCTION
Centrioles are barrel-shaped organelles whose structural features include a cartwheel, composed of a central hub with nine radially symmetric spokes, and an outer wall containing a nine-fold symmetric array of stabilized microtubules (Loncarek and Khodjakov, 2009; Azimzadeh and Marshall, 2010; Carvalho-Santos et al., 2011). Centrioles serve two primary functions: they direct the formation of cilia and recruit pericentriolar material (PCM) to form centrosomes that nucleate and organize microtubules (Azimzadeh and Marshall, 2010; Carvalho-Santos et al., 2011; Nigg and Stearns, 2011). In animal cells, centrioles duplicate concurrently with DNA replication (Loncarek and Khodjakov, 2009; Azimzadeh and Marshall, 2010; Nigg and Stearns, 2011; Brito et al., 2012). Centriole duplication is tightly regulated to ensure that mitotic cells have precisely two centrosomes, as extra centrosomes lead to aberrant chromosome-microtubule attachments and chromosomal instability (Nigg and Raff, 2009; Thompson et al., 2010).
SAS-6 and SAS-4/CPAP, initially discovered in C. elegans (Strnad and Gönczy, 2008), are components of a universally conserved eukaryotic module for centriole assembly (Carvalho-Santos et al., 2010; Hodges et al., 2010). In C. elegans, centriole formation also requires SAS-5 (Dammermann et al., 2004; Delattre et al., 2004), a divergent member of the STIL/Ana2 family in mammals and Drosophila (Stevens et al., 2010; Castiel et al., 2011; Tang et al., 2011; Kitagawa et al., 2011b; Arquint et al., 2012; Vulprecht et al., 2012). In metazoans, centriole duplication is controlled by the Polo family kinase Plk4 (Bettencourt-Dias et al., 2005; Habedanck et al., 2005), whose functional C. elegans analog is ZYG-1 (O'Connell et al., 2001; Carvalho-Santos et al., 2010; Hodges et al., 2010).
Analysis of protein recruitment during new centriole formation in C. elegans has shown that ZYG-1 targets independently of SAS-5 and SAS-6 and is necessary for their recruitment (Delattre et al., 2006; Pelletier et al., 2006). SAS-5 and SAS-6 are in turn required to recruit SAS-4 (Dammermann et al., 2004; Delattre et al., 2004; Leidel et al., 2005). Ultrastructural work in C. elegans has shown that centriole assembly occurs in two genetically separable steps (Pelletier et al., 2006). During the first step, which occurs in S-phase, a central tube forms adjacent to the parent centriole through a process that requires ZYG-1, SAS-5, and SAS-6, but not SAS-4 (Pelletier et al., 2006). The C. elegans central tube is analogous to the cartwheel in other systems, as its formation requires interactions between SAS-6 dimers (Kitagawa et al., 2011c, see also Fig. 2); we therefore refer to the central tube as the cartwheel throughout the manuscript for simplicity. Unlike cartwheels in other systems, a 9-fold symmetric array of spokes has not been observed in C. elegans (Pelletier et al., 2006). However, it remains unclear if this is due to technical limitations in the EM or to a difference in cartwheel structure between species. During the second step of centriole assembly, which requires SAS-4 (Pelletier et al., 2006) and mitotic entry (Dammermann et al., 2004), the outer centriole wall containing the 9-fold symmetric microtubule array forms around the cartwheel.
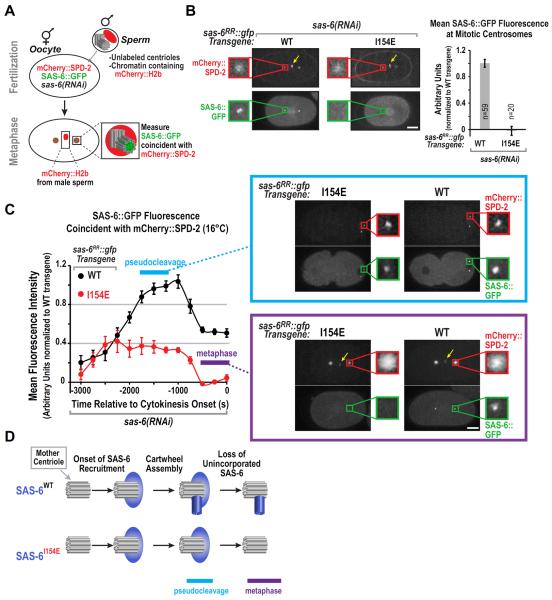
Figure 2. The assembly-defective I154E SAS-6 mutant is recruited to the mother centriole but fails subsequent cartwheel assembly.
(A) Schematic of the method used to monitor SAS-6 recruitment during daughter centriole assembly, while rendering the SAS-6 already present in the mother (sperm-derived) centrioles invisible. Oocytes expressing mCherry∷SPD-2 and SAS-6∷GFP and depleted of endogenous SAS-6 were fertilized by male sperm containing unlabeled centrioles and mCherry∷H2b-labeled chromatin. Presence of mCherry∷H2b chromatin confirmed that each monitored embryo was cross-fertilized. mCherry∷SPD-2 is recruited to sperm centrioles immediately following fertilization, providing a marker for the mother centrioles. The amount of SAS-6∷GFP coincident with the mCherry∷SPD-2 labeled mother centrioles was monitored over time. (B) (left) Maximum intensity projection confocal images of metaphase embryos derived using the experimental scheme in (A). Sperm-derived mCherry∷H2b weakly labels chromatin (yellow arrows). Insets are magnified 3.8-fold. (right) Quantification of SAS-6∷GFP fluorescence coincident with mCherry∷SPD-2 during the 300s interval prior to cytokinesis, when the amount of centriolar SAS-6 is nearly constant. Measured values were normalized by dividing by the mean value for WT SAS-6∷GFP. Error bars are the SEM, n=number of measurements. (C) Kinetic analysis at 16°C of SAS-6∷GFP (WT or I154E) recruitment during daughter centriole assembly using the experimental scheme shown in (A). Data points are the mean normalized value for the 500 second interval centered around the plotted point. Error bars=SEM. Representative confocal projection images at indicated time points are shown. Insets are magnified 5-fold. (D) Schematic summarizing localization dynamics of WT and assembly-defective I154E mutant SAS-6. Bars, 10μm. See also Figure S2.
SAS-6, the key cartwheel structural component across systems (Dammermann et al., 2004; Leidel et al., 2005; Pelletier et al., 2006; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007; Yabe et al., 2007; Culver et al., 2009; Jerka-Dziadosz et al., 2010), has an N-terminal head domain, a central coiled-coil, and a C-terminal tail that is predicted to be unstructured. SAS-6 dimerizes through a parallel coiled-coil interaction, resulting in two opposite-facing N-terminal domains at one end of the coiled-coil (van Breugel et al., 2011; Kitagawa et al., 2011c). Atomic structures revealed that the SAS-6 N-terminal domains interact between dimers via a peg-into-hole mechanism involving insertion of a critical hydrophobic residue (I154 in C. elegans) from the loop of one N-terminal domain into a pocket in the interacting N-terminal domain (van Breugel et al., 2011; Kitagawa et al., 2011c). Models of the observed packing between the SAS-6 N-terminal head domains can explain the nine-fold symmetric hub-and-spoke architecture of the cartwheel.
A current focus of investigation is to understand how SAS-6 oligomerization is regulated to ensure that cartwheels form only during S-phase adjacent to the mother centriole. The affinity of the SAS-6 N-terminal head domains for each other is low (Kd ~50–100 μM; van Breugel et al., 2011; Kitagawa et al., 2011c), suggesting that cartwheel formation is controlled by regulatory mechanisms that promote SAS-6 polymerization. SAS-5/Ana2/STIL and the Plk4/ZYG-1 kinase, which are also required for cartwheel formation, are key candidates to regulate SAS-6 assembly (Azimzadeh and Marshall, 2010; Nigg and Stearns, 2011; Brito et al., 2012). The kinase activities of Plk4 and ZYG-1 are required for centriole assembly (O'Connell et al., 2001; Habedanck et al., 2005), and C. elegans centriole assembly has been proposed to require phosphorylation of SAS-6 serine 123 by ZYG-1 (Kitagawa et al., 2009). Centriole assembly has also been proposed to be regulated by the phosphatase PP2A, which interacts with the SAS-5/6 complex and promotes cartwheel formation (Song et al., 2011; Kitagawa et al., 2011a).
Here, we show that SAS-6 recruitment to the mother centriole and cartwheel assembly are separable events in vivo. ZYG-1 is required for SAS-6 recruitment and cartwheel assembly, but its kinase activity is only required for the latter. This finding is explained by a direct ZYG-1—SAS-6 interaction that recruits SAS-6 to the mother centriole. Surprisingly, phosphorylation of the previously reported ZYG-1 target site on SAS-6 (serine 123; Kitagawa et al., 2009) is not important for centriole assembly. The same was true for 37 additional candidate serines and threonines in SAS-6, identified either because they are phosphorylated by ZYG-1 in vitro or because they are conserved among closely related nematode species. We propose that ZYG-1 binding to the SAS-6 coiled-coil recruits the SAS-6—SAS-5 complex to the mother centriole, where a ZYG-1 kinase activity-dependent step, whose target is unlikely to be SAS-6, triggers cartwheel assembly.
RESULTS
A Single-Copy Transgene Insertion System for SAS-6 in the C. elegans Embryo
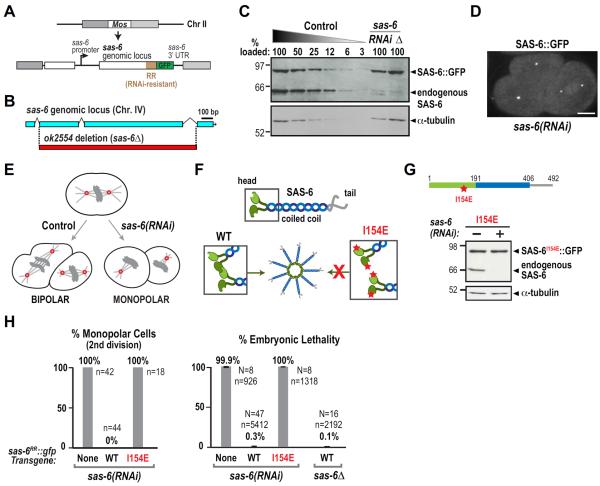
To investigate the regulation of SAS-6 by ZYG-1, we used Mos1-mediated single-copy insertion (MosSCI; Frøkjaer-Jensen et al., 2008) to generate a single-copy sas-6∷gfp transgene integrated into a specific site on Chromosome II (Fig. 1A). The transgene was engineered to be resistant to RNA interference (RR: RNAi-resistant) by altering a region of nucleotide sequence without affecting protein coding (Fig. 1A; Fig. S1A). Transgene-encoded SAS-6∷GFP was expressed at endogenous levels (Fig. 1C), resistant to a dsRNA targeting the altered region (Fig. 1C), and localized to centrioles (Fig. 1D).
Figure 1. A single-copy transgene insertion system for SAS-6 in the C. elegans embryo.
(A) Schematic of the sas-6RR∷gfp single-copy transgene insertion. (B) Schematic of the sas-6(ok2554) allele (sas-6Δ). (C) Immunoblot of sas-6RR∷gfp transgenic worm lysates probed with antibodies to SAS-6 (top) and α-tubulin as a loading control (bottom). Serial dilutions of control lysate were used to assess endogenous SAS-6 depletion (RNAi). Numbers above each lane are the percentage loaded relative to 100% control. Lysate from sas-6Δ worms rescued by sas-6RR∷gfp is in the last lane (Δ). (D) Image of a two-cell embryo expressing SAS-6∷GFP after depletion of endogenous SAS-6. Bar, 10 μm. (E) Schematic illustrating the fate of embryos unable to duplicate centrioles due to SAS-6 depletion. (F) Schematic illustrating the effect of the I154E mutation (red star) on SAS-6 inter-dimer interactions and cartwheel assembly. (G) Immunoblot of lysates prepared from worms expressing I154E SAS-6∷GFP. (H) Plots of the frequency of second division monopolar spindles (left) and embryonic lethality (right) following endogenous SAS-6 depletion for the indicated conditions. WT= wild-type sas-6RR∷gfp transgene. Error bars = SD of percent lethality per worm; N = number of worms, n = total number of embryos scored. See also Figure S1.
SAS-6 depletion by RNAi prevents centriole assembly, resulting in a normal first mitotic division followed by monopolar spindles during the second division (Fig. 1E; O'Connell et al., 2001). This phenotype arises because the sperm that fertilize SAS-6-depleted oocytes carry two normal centrioles, as sperm production occurs prior to introduction of the dsRNA. The sperm-derived centrioles organize the two spindle poles during the first embryonic division. If no new centrioles form, each daughter cell inherits only one sperm-derived centriole leading to monopolar spindles in the second division and 100% embryonic lethality. The wild-type sas-6RR∷gfp transgene fully rescued the second division monopolar spindle phenotype and embryonic lethality caused by SAS-6 depletion (Fig. 1H), as well as the inviability of an sas-6Δ mutant (Fig. 1B,H). The absence of endogenous SAS-6 was confirmed in the transgene-rescued sas-6Δ strain by immunoblotting (Fig. 1C).
To validate the use of this transgenic system, we generated a transgene expressing SAS-6∷GFP with an I154E substitution, which blocks the interaction between SAS-6 N-termini (van Breugel et al., 2011, Kitagawa et al., 2011; Fig. 1F,G). Transgene-encoded I154E mutant SAS-6 was expressed at wild-type levels after endogenous SAS-6 depletion (Fig. 1G). As expected from prior work (van Breugel et al., 2011; Kitagawa et al., 2011c), the I154E mutation led to a fully penetrant second division monopolar spindle phenotype and 100% embryonic lethality following endogenous SAS-6 depletion (Fig. 1H). Thus, the functional transgene-encoded SAS-6∷GFP facilitates analysis of SAS-6 mutations in vivo.
SAS-6 Recruitment to the Mother Centriole and Cartwheel Incorporation are Distinct Events that can be Monitored In Vivo
We previously developed a method to monitor SAS-6 recruitment during daughter centriole assembly, while rendering the SAS-6 already incorporated into the mother (sperm-derived) centrioles invisible (Dammermann et al., 2008). This method revealed that SAS-6 reaches its maximal levels during cartwheel formation in S-phase. Subsequently, as cells enter mitosis, SAS-6 levels decrease by ~50% (Dammermann et al., 2008; Fig. 2C). One explanation for this behavior is that SAS-6 recruitment to the mother centriole and its incorporation into the cartwheel are separable events, with SAS-6 that is recruited to the mother, but not incorporated into the cartwheel, being lost following completion of cartwheel assembly (Fig. 2D). To test this idea, we used the SAS-6 I154E mutant, which is defective for the N-terminal inter-dimer interactions underlying cartwheel formation.
To compare the recruitment profiles of wild-type and I154E mutant SAS-6∷GFP, centriole assembly was monitored in embryos expressing mCherry∷SPD-2 and SAS-6∷GFP (Fig. 2A). As C. elegans undergoes self-fertilization, the centrioles derived from the sperm in these worms contain SAS-6∷GFP, which complicates analysis of SAS-6∷GFP recruitment during daughter centriole formation. To circumvent this problem, hermaphrodites expressing mCherry∷SPD-2 and SAS-6∷GFP were mated to males containing mCherry∷H2b-labeled chromatin and unlabeled centrioles. As centriolar SAS-6 does not turn over (Leidel et al., 2005; Dammermann et al., 2008), the male-derived sperm centrioles remain unlabeled after fertilization; the presence of mCherry∷H2b chromatin (yellow arrows in Fig. 2B,C) confirmed that each analyzed embryo was cross-fertilized with male sperm. The mCherry∷SPD-2, which is recruited to sperm centrioles immediately after fertilization and independently of cartwheel assembly (Delattre et al., 2006; Pelletier et al., 2006), marked the location of the sperm centrioles. Finally, to selectively monitor transgene-encoded SAS-6∷GFP (WT or I154E mutant), endogenous SAS-6 was depleted from oocytes by RNAi.
We first monitored the endpoint of cartwheel formation by examining the centrosomes of the mitotic spindle. WT SAS-6∷GFP formed a bright focus, representing the cartwheel of one daughter centriole at each mitotic centrosome. No signal was detected at mitotic centrosomes in the SAS-6∷GFP I154E mutant (Fig. 2B). As expected from prior work, formation of the SAS-6∷GFP focus was not affected by depletion of SAS-4 (Fig. S2A,B), which is required for outer wall assembly (Kirkham et al., 2003; Leidel and Gönczy, 2003; Pelletier et al., 2006, Dammermann, 2008).
Next, we performed a kinetic analysis of WT and I154E mutant SAS-6∷GFP. This analysis was performed at 16°C for consistency with subsequent experiments monitoring SAS-6 recruitment in strains expressing temperature-sensitive ZYG-1 (next section). The dynamic changes in WT SAS-6∷GFP during daughter centriole assembly were similar to those documented previously at 21°C (Dammermann et al., 2008); SAS-6∷GFP reached its maximal level during S-phase and decreased by ~50% as cells progressed into mitosis. The I154E mutant SAS-6∷GFP was also recruited to the site of new centriole assembly, reaching ~40% of the maximum observed for WT SAS-6; this localized I154E mutant SAS-6 pool subsequently declined as cells progressed into mitosis, resulting in mitotic centrosomes with no GFP focus (Fig. 2C). Thus, SAS-6 dimers are recruited to the mother centriole independently of their ability to assemble through their N-terminal domains to form the cartwheel. One model to explain why I154E SAS-6∷GFP reaches only 40% of the maximum level of WT SAS-6∷GFP is that this reflects the amount of SAS-6 recruited to the the mother centriole to provide a local reservoir for cartwheel assembly. WT SAS-6 lost from the reservoir due to incorporation into the cartwheel would be replenished by recruiting more SAS-6 (Fig. 2D). We conclude that recruitment of SAS-6 to the mother centriole and its incorporation into the cartwheel are separable events that can be monitored in vivo.
ZYG-1 Kinase Activity is Required for Cartwheel Assembly but Not for Recruitment of SAS-6 to the Mother Centriole
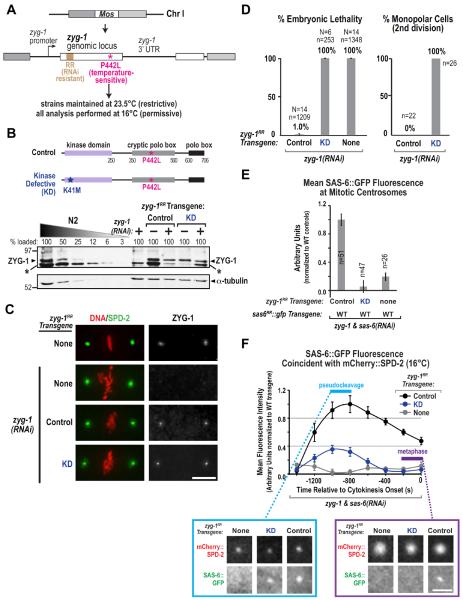
ZYG-1 is required for SAS-6 recruitment during daughter centriole assembly (Delattre et al., 2006; Pelletier et al., 2006) and its kinase activity is important for embryonic viability (O'Connell et al., 2001). However, it is not known which step in centriole assembly— recruitment of SAS-6 to the vicinity of the mother, cartwheel assembly, or outer wall assembly—is the first to require ZYG-1 kinase activity. To determine this, we generated a zyg-1 transgenic system (Figs. 3A, S3A) to compare the consequences of depleting ZYG-1 versus inhibiting its kinase activity. Despite extensive efforts, we were unable to generate transgenic worms carrying the wild-type zyg-1 locus, as injection of DNA containing this locus was toxic, possibly due to transient overexpression. To circumvent this problem, we introduced a temperature-sensitive point mutation (P442L; Kemphues et al., 1988; Kemp et al., 2007) into the transgene. Strains harboring transgenes with this mutation alone (Control) or with an additional Kinase-Defective (KD) mutation were generated and maintained at the non-permissive temperature to keep the transgenic protein inactive (23.5°C; Fig. 3A). After injection of dsRNA to deplete endogenous ZYG-1, strains were shifted to the permissive temperature of 16°C. This approach enabled replacing endogenous ZYG-1 with transgene-encoded Control- or KD-ZYG-1, with all phenotypic analysis being performed at 16°C. As a zyg-1 transgene containing a GFP tag failed to rescue the embryonic lethality associated with endogenous ZYG-1 depletion (not shown), we generated and used untagged Control and KD zyg-1 transgenes (Fig. 3A,B).
Figure 3. The kinase activity of ZYG-1 is required for cartwheel assembly but not for recruitment of SAS-6 to the mother centriole.
(A) Schematic of the zyg-1RR single-copy transgene. (B) Schematics of transgene-encoded Control and Kinase-Defective (KD) ZYG-1 and immunoblot of lysates prepared from the indicated strains and conditions probed with antibodies to ZYG-1 (top) and α-tubulin (bottom). The asterisk (*) marks a variable background band likely derived from the E. coli bacteria that worms eat. (C) Immunofluorescence analysis of ZYG-1 and SPD-2 in embryos derived from the indicated strains and conditions. (D) Plots of embryonic lethality (left) and frequency of monopolar second division (right), as in Fig. 1H, for the indicated strains and conditions. (E) SAS-6∷GFP fluorescence at mitotic centrosomes following mating as in Fig. 2A and plotted as in Fig. 2B, in embryos expressing Control, KD, or no transgenic ZYG-1 and depleted of endogenous ZYG-1 and SAS-6. (F) Kinetic analysis at 16°C of SAS-6∷GFP recruitment during daughter centriole assembly using the experimental scheme shown in Fig. 2A in embryos expressing the indicated ZYG-1 variants and depleted of both endogenous ZYG-1 and SAS-6. Data points are the mean normalized value for the 400 second interval centered around the plotted point. Error bars=SEM. Representative confocal projection images of one centrosome at early and late time points in each strain are shown. Bars, 5 μm. See also Figure S3.
Immunoblotting and immunofluorescence confirmed that transgene-encoded Control- and KD-ZYG-1 were expressed (Fig. 3B,C). ZYG-1, like Plk4, is exceedingly rare and difficult to detect by immunoblotting. However, an ultrasensitive detection method revealed a band of the expected size that was lost following RNAi-mediated depletion (Fig. 3B; the variable intensity band under ZYG-1 is likely a contaminant from the E. coli that worms eat). Whereas the ZYG-1 band was lost completely following zyg-1(RNAi) in the absence of a transgene, the signal was reduced but not lost in the transgenic strains. This conclusion was confirmed by immunofluorescence. ZYG-1 forms a focus at each mitotic spindle pole in 1-cell embryos; this localization was lost following zyg-1(RNAi) in the absence of a transgene but was still detected in the two transgene-containing strains (Fig. 3C). Thus, both Control- and KD-ZYG-1 are expressed at near normal levels and localize to centrioles, indicating that the kinase activity of ZYG-1 is dispensable for its localization to the mother centriole.
We next compared the consequences of depleting ZYG-1 to those resulting from specifically inhibiting its kinase activity. Control-ZYG-1 fully rescued the embryonic lethality and monopolar second division phenotype resulting from endogenous ZYG-1 depletion (Fig. 3D). In contrast, KD-ZYG-1 failed to rescue either, indicating that ZYG-1 kinase activity is essential for centriole assembly in vivo. We next generated strains homozygous for the zyg-1 transgenes (WT or KD, integrated on Chr. I) along with the wild-type sas-6RR∷gfp transgene (integrated on Chr. II) and a transgene encoding mCherry∷SPD-2, and performed the mating-based assay (Fig. 2A; the only difference was that both endogenous ZYG-1 and SAS-6 were depleted). A clear SAS-6∷GFP focus was present at mitotic spindle poles in embryos expressing Control-ZYG-1 but not KD-ZYG-1 (Fig. 3E). Thus, ZYG-1 kinase activity is required for cartwheel assembly (Fig. 3E). Next, we performed a kinetic analysis as in Fig. 2C. No SAS-6∷GFP was recruited when ZYG-1 was depleted in the absence of a transgene (Fig. 3F). This was not due to SAS-6 destabilization, since SAS-6 levels were not affected by ZYG-1 depletion in the presence or absence of the Control- or KD-zyg-1 transgenes (Fig. S3B). The Control-zyg-1 transgene rescued normal SAS-6∷GFP recruitment, whereas a recruitment profile similar to that for the I154E SAS-6∷GFP mutant (Fig. 2C) was observed when endogenous ZYG-1 was depleted in the presence of the KD-zyg-1 transgene (Fig. 3F). This result indicates that ZYG-1 recruits SAS-6 to the mother centriole independently of its kinase activity; kinase activity is only required for the subsequent incorporation of SAS-6 into the cartwheel.
ZYG-1 Binds to a Region of the SAS-6 Coiled-Coil Distinct from the SAS-5 Interaction Region
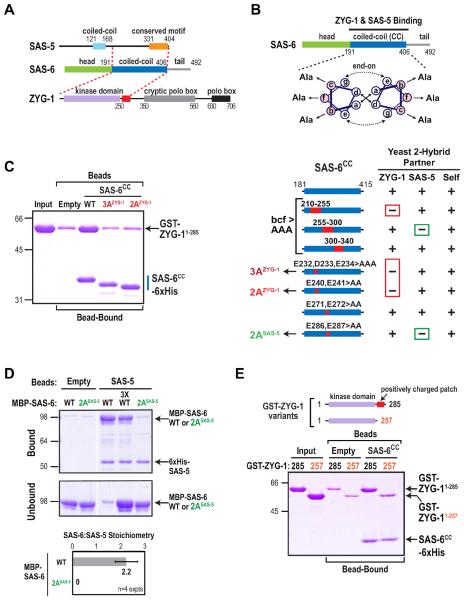
To understand how ZYG-1 recruits SAS-6 to the mother centriole independently of its kinase activity, we tested whether ZYG-1 directly interacts with SAS-6. We observed a robust yeast two-hybrid interaction between the N-terminal region of ZYG-1 and the SAS-6 coiled-coil (Fig. 4A), which also interacts with SAS-5 (Leidel, 2005; Boxem, 2008). The region of ZYG-1 important for the interaction (aa 1–285) includes its kinase domain and an adjacent region enriched in positively charged residues (red box in Fig. 4A). To determine if this interaction helps recruit SAS-6 to the mother centriole, we set out to identify mutations in the SAS-6 coiled-coil that specifically disrupt the interaction with ZYG-1 but not SAS-5, and vice versa.
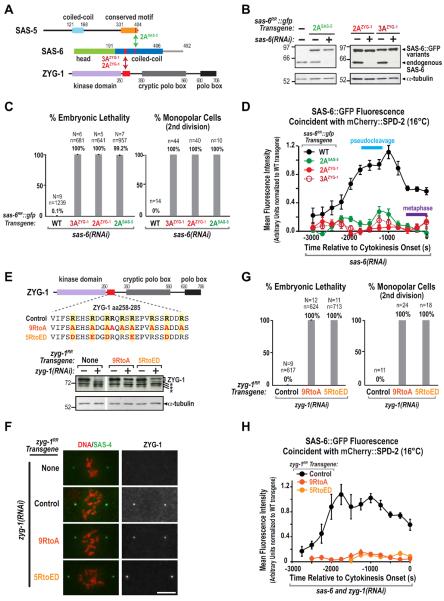
Figure 4. Engineering specific mutations that disrupt direct interactions of adjacent regions of the SAS-6 coiled-coil with ZYG-1 and SAS-5.
(A) Summary of yeast two-hybrid interactions between SAS-6, ZYG-1 and SAS-5. The SAS-6 coiled-coil interacts with the C-terminus of SAS-5 (Leidel et al., 2005; Boxem et al., 2008) and the N-terminus of ZYG-1, which includes its kinase domain and an adjacent region enriched in positively charged residues (red box). (B) (top) Schematic of the method used to identify specific residues in the SAS-6 coiled-coil required for ZYG-1 and SAS-5 binding. Depicted are hydrophobic interactions between a and d position residues (solid arrows) and electrostatic interactions between e and g position residues (dashed arrows). Charged residues in the b, c and f positions were mutated to alanine (Ala). (bottom) Summary of mutational two-hybrid analysis of the SAS-6 coiled-coil. (C–E) Purified protein pull-down assays with the indicated protein-coated beads and soluble input partners. Coomassie-stained gels of the binding assays are shown. Migration of the 3AZYG-1 and 2AZYG-1 mutant SAS-6 coiled-coils in C is accelerated relative to WT likely because the mutations render the proteins less acidic. Under the conditions used, SAS-5-coated beads saturated with SAS-6 at a SAS-6:SAS-5 stoichiometry of 2:1. The stoichiometry was calculated using molecular weight-corrected background-subtracted Coomassie staining intensities (D; bottom panel). Error bars are the SD. See also Figure S4.
Dimeric coiled-coils are composed of two α-helices with a heptad repeat, the positions in which are designated by the letters a to g. Residues in the a, d, e, and g positions typically mediate interactions between the α-helices (Fig. 4B; Mason and Arndt, 2004). We therefore mutated to alanine all of the charged residues in the b, c, and f positions, which typically have outward-facing side chains (Fig. 4B), in non-overlapping segments of 40–45 residues. Mutating 15 bcf positions in the segment containing residues 210–255 eliminated the interaction with ZYG-1 but not SAS-5; by contrast, mutation of 14 bcf residues in the 255–300 segment eliminated the interaction with SAS-5 but not ZYG-1 (Fig. 4B). Mutation of bcf residues in the 300–340 segment did not affect either interaction. All three of the bcf-to-alanine mutants still interacted with themselves, suggesting that the coiled-coil was not disrupted. Additional mutagenesis revealed that mutation of E232, D233 and E234 (referred to as 3AZYG-1) or of E240 and E241 (referred to as 2AZYG-1) specifically disrupted the ZYG-1 interaction, whereas mutation of E286 and E287 (referred to as 2ASAS-5) specifically disrupted the SAS-5 interaction (Fig. 4B, S4A).
Next, we confirmed the results of the two-hybrid analysis using pull-down assays with purified proteins. Purified ZYG-11–285 interacted with the WT but not the 3AZYG-1 or 2AZYG-1 mutant versions of the SAS-6 coiled-coil (Fig. 4C); whereas the 3AZYG-1 or 2AZYG-1 coiled-coil mutants retained the ability to interact with the SAS-5 C-terminus (Fig. S4B). Full-length WT SAS-6 bound to SAS-5 immobilized on beads at a stoichiometry of 2:1 (SAS-6:SAS-5); in contrast, no binding was detected for 2ASAS-5 mutant SAS-6 (Fig. 4D). The SAS-6 residues we identified as important for SAS-5 binding are consistent with a recent structure of the SAS-6 coiled-coil (Qiao et al., 2012). This analysis showed that SAS-5 binds to a negatively charged patch between aa 275 and 288 of the SAS-6 coiled-coil and identified SAS-6 E287 as essential for SAS-5 binding (Qiao et al., 2012). The ZYG-1 binding region we identified (aa 232–241) is just outside the region in the crystal structure (aa 250–407); however, like the adjacent SAS-5 binding region, this region of the SAS-6 coiled-coil is negatively charged. Thus, a negatively charged region of the SAS-6 coiled-coil provides adjacent independent interaction surfaces for ZYG-1 and SAS-5.
Next, we identified the regions of ZYG-1 and SAS-5 required to bind to SAS-6. ZYG-11–285 includes the kinase domain and a subsequent adjacent stretch enriched in positively charged residues (Fig. 5E, S5C). Whereas purified ZYG-11–285 binds to the WT SAS-6 coiled-coil, binding of a truncated protein lacking the charged patch (ZYG-11–257) was reduced to background levels (Fig. 4E), suggesting that this region is key for the interaction of ZYG-1 with SAS-6. In agreement with other studies (Leidel et al., 2005; Boxem et al., 2008, Qiao et al., 2012), we found that an isoleucine/arginine (IR) motif near the SAS-5 C-terminus is required for interaction with the SAS-6 coiled-coil (Fig. S4C,D). Multi-angle light scattering indicated that the SAS-5 C-terminus is a dimer (Fig. S4E), which suggests that the native SAS-6—SAS-5 complex may be a hexamer composed of two dimers of SAS-6 and one dimer of SAS-5. As we have been unable to generate full-length SAS-5 in sufficient quantities for native molecular weight measurements, this is a preliminary proposal.
Figure 5. Direct interactions of the SAS-6 coiled-coil with ZYG-1 and SAS-5 are required to recruit SAS-6 to the mother centriole.
(A) Summary of the SAS-6 coiled-coil mutations that disrupt its interactions with SAS-5 and ZYG-1. (B) Immunoblots of lysates prepared from worms expressing the indicated SAS-6∷GFP variants with or without depletion of endogenous SAS-6; blots were probed for SAS-6 (top) and α-tubulin (bottom). (C) Plots of percent embryonic lethality (left) and frequency of monopolar second division (right), as in Fig. 1H, for the indicated conditions. (D) Kinetic analysis at 16°C of recruitment of the indicated SAS-6∷GFP variants during daughter centriole assembly using the experimental scheme shown in Fig. 2A and plotted as in Fig. 2C. (E) (top) Amino acid sequence in the region of ZYG-1 required for binding to the SAS-6 coiled-coil (red box) and mutants engineered to alter all 9 arginines to alanine (9RtoA) or charge-reverse 5 arginines that are perfectly conserved among closely related nematodes (5RtoED). (bottom) Immunoblot of lysates from strains carrying the indicated zyg-1RR transgenes, with or without depletion of endogenous ZYG-1, probed for ZYG-1 (top) and α-tubulin (bottom). Asterisks indicate non-specific bands. (F) Immunofluorescence showing that ZYG-1Control, ZYG-19RtoA and ZYG-15RtoED localize to centrioles after depletion of endogenous ZYG-1. Embryos were stained for DNA (red, left panels), ZYG-1 (right panels), and SAS-4 (green, left panels) as a centriole marker. Bar, 5 μm. (G) Plots of percent embryonic lethality (left) and frequency of monopolar second division (right), as in Fig. 1H, for the indicated conditions. (H) Kinetic analysis at 16°C of SAS-6∷GFP recruitment during daughter centriole assembly using the experimental scheme shown in Fig. 2A and plotted as in Fig. 2C in embryos expressing the indicated ZYG-1 variants and depleted of both endogenous ZYG-1 and SAS-6. See also Figure S5.
Direct Interactions of the SAS-6 Coiled-Coil with ZYG-1 and SAS-5 are Required to Recruit SAS-6 to the Mother Centriole
We next tested whether the ZYG-1—SAS-6 and SAS-5—SAS-6 interactions (Fig. 5A) are important for centriole assembly in vivo by generating transgenes that expressed 2AZYG-1, 3AZYG-1 and 2ASAS-5 mutant versions of SAS-6∷GFP at normal levels (Fig. 5B). Following endogenous SAS-6 depletion, all three mutants resulted in 100% second division monopolar spindle formation and embryonic lethality (Fig. 5C). Putting the transgenes in a background expressing mCherry∷SPD-2 and measuring the GFP signal at mitotic spindle poles in the mating-based assay revealed that all three mutants failed to support cartwheel assembly (Fig. S5A). Consistent with the fact that SAS-6 is not required for ZYG-1 to localize to the mother centriole (Delattre et al., 2006; Pelletier et al., 2006), ZYG-1 localized normally following endogenous SAS-6 depletion in the 2AZYG-1 and 3AZYG-1 SAS-6 mutants (Fig. S5B). Following endogenous SAS-6 depletion, the 2AZYG-1 and 3AZYG-1 mutants failed to be recruited to the mother centriole, exhibiting a phenotype analogous to ZYG-1 depletion (Fig. 5D). Thus, selectively disrupting the ZYG-1—SAS-6 interaction prevents SAS-6 recruitment to the mother centriole. The 2ASAS-5 mutant SAS-6∷GFP also failed to be recruited to the mother centriole (Fig. 5D), indicating that selectively mutating the SAS-5 interaction site on SAS-6 phenocopies SAS-5 depletion (Leidel et al., 2005).
To confirm the significance of the ZYG-1—SAS-6 interaction, we also disrupted it by mutating the positively charged arginine residues in the ZYG-1 region required for SAS-6 binding (aa 258–285; Fig. 5E). We changed all 9 arginine residues in this region to alanine (Fig. 5E, 9RtoA) or reversed the charge of the 5 arginines that are perfectly conserved in closely related nematodes (Fig. 5E, S5C, 5RtoED). Immunoblotting and immunofluorescence confirmed that the 9RtoA and 5RtoED ZYG-1 mutants were expressed and localized to centrioles normally in the absence of endogenous ZYG-1 (Fig. 5E,F). Like the SAS-6 mutants that disrupt ZYG-1 binding, the 9RtoA and 5RtoED ZYG-1 mutants resulted in 100% second division monopolar spindle formation and embryonic lethality (Fig. 5G), and both mutants failed to support SAS-6∷GFP recruitment (Fig. 5H). Alignment of this region of ZYG-1 with Plk4 homologs from other species suggests that this charged motif may be conserved (Fig. S5D); however, additional work will be needed to test this idea. We conclude that direct interactions of ZYG-1 and SAS-5 with adjacent segments of the SAS-6 coiled-coil are essential to recruit SAS-6 to the mother centriole.
Phosphorylation of SAS-6 Serine 123 is Not Important for Centriole Assembly or Embryonic Viability
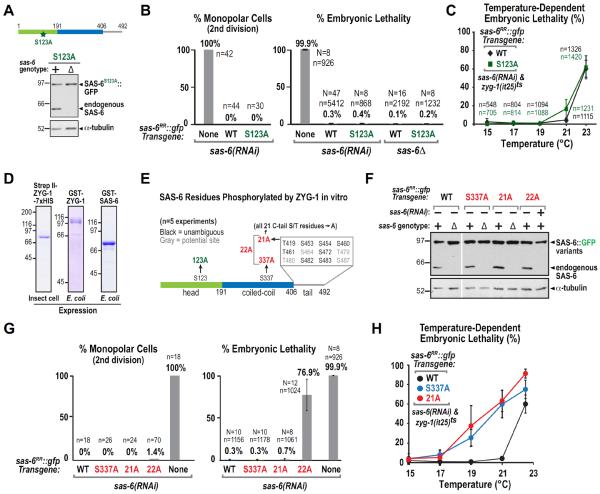
While not required to recruit SAS-6 to the mother centriole, ZYG-1 kinase activity is required for cartwheel assembly. Serine 123 of C. elegans SAS-6 has been proposed as a critical target of ZYG-1 kinase in centriole assembly based on comparisons of wild-type and S123A mutant sas-6 transgenes to rescue depletion of endogenous SAS-6 (Kitagawa et al., 2009). These experiments utilized random transgene insertions generated by ballistic bombardment, which are often expressed at variable levels due to germline silencing and integration at variable copy number in different chromosomal sites (Seydoux and Strome, 1999; Praitis et al., 2001; Green et al., 2008). We therefore reassessed the functional importance of S123 using the single-copy targeted insertion system to generate a transgene expressing S123A mutant SAS-6∷GFP (Fig. 6A). Surprisingly, we observed no significant defects in either second division spindle assembly or embryonic viability with the S123A mutant (Fig. 6B), and the S123A mutant transgene fully rescued the lethality of the sas-6 deletion (Fig. 6A,B).
Figure 6. Neither serine 123 nor the predominant in vitro ZYG-1 target sites in the SAS-6 C-Tail are essential for centriole assembly.
(A) Immunoblot of lysates prepared from worms expressing S123A SAS-6∷GFP in wild-type (+) or in the background of an sas-6 deletion (Δ); the blot was probed for SAS-6 (top) and α-tubulin (bottom). (B) Plots of the frequency of monopolar second division (left) and percent embryonic lethality (right) as in Fig. 1H, for the indicated conditions. The WT and No Transgene data are replotted from Fig. 1H. (C) Temperature-dependent embryonic lethality of strains carrying the WT or S123A sas-6∷gfp transgenes and homozygous for the temperature-sensitive zyg-1(it25) allele. (D) Coomassie-stained gels of purified tagged ZYG-1 and SAS-6 used for the in vitro phosphorylation analysis summarized in E. (E) Summary of ZYG-1–phosphorylated sites in SAS-6, identified by mass spectrometry in at least 2 of 5 separate experiments. Mutations engineered to test contributions of these sites to total SAS-6 phosphorylation in vitro or to centriole assembly in vivo are marked in red. Sites in grey could not be unambiguously identified due to the presence of multiple serines/threonines in one peptide. (F) Immunoblots of lysates prepared from worms expressing the indicated SAS-6 variants in the presence of the sas-6 deletion (Δ) or sas-6(RNAi), as indicated; blots were probed for SAS-6 (top) and α-tubulin (bottom). (G) Plots of frequency of monopolar second division (left) and percent embryonic lethality (right), as in Fig. 1H, for the indicated conditions. The No Transgene embryonic lethality data is replotted from Fig. 1H. (H) Temperature-dependent embryonic lethality of strains carrying the S337A or 21A sas-6∷gfp transgenes and homozygous for the temperature-sensitive zyg-1(it25) allele. WT data is reproduced from C. See also Figure S6.
The above data suggest that S123 phosphorylation is not essential for centriole assembly. However, it remained possible that S123 is one of multiple sites phosphorylated by ZYG-1 and that its phosphorylation enhances centriole assembly. To test this, we generated strains homozygous for the S123A or WT sas-6∷gfp transgenes that were also homozygous for a temperature-sensitive zyg-1 mutation (zyg-1(it25); Kemphues et al., 1988; Kemp et al., 2007). After depletion of endogenous SAS-6, embryonic lethality was measured at progressively higher temperatures (Kemp et al., 2007). If the S123A mutation rendered centriole assembly more sensitive to reduction of ZYG-1 activity, higher lethality should be observed for the S123A relative to the WT sas-6∷gfp transgenic strain across the tested temperature range. Instead, no significant enhancement of temperature-dependent lethality was observed (Fig. 6C; for comparison, see other SAS-6 mutants in Fig. 6H that did result in increased lethality). Repeating this analysis in the sas-6Δ background yielded similar results (Fig. S6A). Thus, analysis of the S123A mutant both alone and in the zyg-1ts mutant background indicates that phosphorylation of serine 123 does not contribute significantly to cartwheel assembly in vivo.
The Dominant In Vitro ZYG-1 Phosphorylation Sites In SAS-6 Contribute Only Weakly to Centriole Assembly In Vivo
The above experiments indicate that ZYG-1 kinase activity is required for cartwheel assembly, but that the functionally relevant target is not S123 of SAS-6. We therefore analyzed phosphorylation of SAS-6 by ZYG-1 in vitro to identify other potential regulatory sites. Mass spectrometry of in vitro phosphorylation reactions combining purified full-length ZYG-1, expressed in insect cells or bacteria, and full-length SAS-6 (Fig. 6D) identified a number of different target serines and threonines in the predicted unstructured SAS-6 C-terminus (C-tail), a single serine residue in a predicted break in the coiled-coil (S337), and the previously identified S123 (Fig. 6E). We next mutated S123, S337 or all 21 S/T residues in the C-tail to alanine in full-length SAS-6, or deleted the C-tail (1–389), and analyzed the effect on phosphorylation by ZYG-1. This analysis indicated that the majority (~80%) of ZYG-1 phosphorylation is targeted to the SAS-6 C-tail (Fig. S6B,C).
To test if the newly identified phosphorylation sites are important in vivo, we generated three alanine substitution mutants that were expressed at normal levels (Fig. 6E,F). Mutation of all 21 serine/threonines in the C-tail (21A), S337 alone (S337A), or all 22 newly identified residues (22A) did not lead to penetrant monopolar spindle assembly formation in the second division (Fig. 6G). In addition, the S337A and 21A mutants rescued viability both of endogenous SAS-6 depletion (Fig. 6G) and of the sas-6Δ mutant (data not shown). A single incidence of a monopolar spindle in the second division was observed when endogenous SAS-6 was depleted in the 22A mutant (from an n of 70), and significant embryonic lethality was also observed (Fig. 6G). Consistent with this, filming embryos through the 8-cell stage revealed a minor but appreciable increase in centriole duplication failure in the 22A mutant (Fig. S6D). As C. elegans embryos undergo ~550 cell divisions prior to hatching, this increase likely underlies the observed embryonic lethality. Finally, we generated strains homozygous for the S337A and 21A sas-6∷gfp transgenes that were also homozygous for the zyg-1(it25) temperature-sensitive mutation and measured temperature-dependent embryonic lethality. Both S337A and 21A enhanced the lethality of zyg-1(it25) significantly more than S123A (compare Fig. 6H to 6C), suggesting that these mutations either partially compromise SAS-6 function or that phosphorylation at these sites by ZYG-1 contributes in a non-essential manner to enhancing centriole assembly.
Overall, the analysis of ZYG-1 target sites in SAS-6, despite mutation of 22 new potential phospho-sites, failed to reveal a centriole assembly defect approaching the severity of kinase-defective ZYG-1.
A Serine/Threonine Mutational Scan of SAS-6
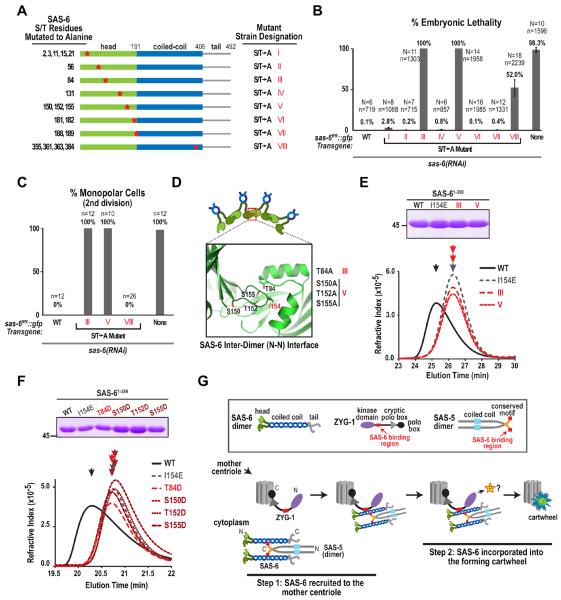
Although phosphorylation of SAS-6 residues targeted by ZYG-1 in vitro was not essential for cartwheel assembly in vivo, it remained possible that the in vitro analysis had failed to identify a critical ZYG-1 target site. We therefore conducted a serine/threonine mutational scan of SAS-6 in vivo. SAS-6 alignments from 5 related Caenorhabditis species were used to identify conserved S/T residues (Fig. S7A). We generated 8 SAS-6∷GFP strains, mutating sets of conserved S/T residues to alanine (Fig. 7A). After depletion of endogenous SAS-6, 2 strains showed penetrant monopolar spindle formation in the second division and embryonic lethality, and a third strain showed moderate embryonic lethality with no significant monopolar spindle formation in the second division (Fig. 7B,C), suggesting compromised SAS-6 function. Strikingly, the residues mutated in the two strains that exhibited the fully penetrant defect were located within the interaction surface between the N-terminal heads (Fig. 7D). The set of residues in strain V (S150, T152, and S155) surround I154 in the loop that inserts into the hydrophobic pocket and T84 (mutated in strain III) lies within the hydrophobic pocket. To test if mutation of these residues to alanine perturbed the inter-dimer interaction (analogous to mutation of I154 to E), as opposed to preventing phosphorylation by ZYG-1, we purified WT and mutant versions of SAS-61-389 (Fig. S7B). WT SAS-61-389 migrated at a higher molecular weight (earlier elution) on a gel filtration column than I154E mutant SAS-61-389, indicative of the I154-dependent interaction of SAS-6 dimers (Fig. 7E). The 2 S/T>A mutants behaved identically to the I154E mutant (Fig. 7E), suggesting that a defect in N-terminal inter-dimer interactions, as opposed to loss of ZYG-1 phosphorylation, underlies their potent in vivo phenotype. Consistent with this, individual alteration of each of the S/T residues in the inter-dimer interface to aspartic acid to mimic phosphorylation also disrupted N-terminal inter-dimer interaction in the gel filtration assay (Fig. 7F). All together, analysis of 12 strains collectively mutating a total of 42 serine/threonine residues in SAS-6 failed to reveal potential phosphorylated residues whose mutation approaches the phenotypic consequences of loss of ZYG-1 kinase activity. These results suggest that SAS-6 is unlikely to be the critical ZYG-1 target during cartwheel assembly.
Figure 7. A serine/threonine mutational scan of SAS-6 and a model for cartwheel assembly.
(A) Serines and threonines conserved in Caenorhabditis species (elegans, briggsae, brenneri, japonica, remanei; Fig. S7A) were mutated as indicated in the schematics, to generate 8 transgenic SAS-6∷GFP strains. (B & C) Plots of percent embryonic lethality (B) and frequency of monopolar second division (C), as in Fig. 1H, for the indicated conditions. (D) Schematic showing the location of serine/threonine residues in the crystal structure of the SAS-6 N-terminus (PDB=3PYI) that when mutated to alanine in strains III and V resulted in 100% embryonic lethality and 100% monopolar second divisions. (E & F) Analysis of SAS-6 inter-dimer interactions by gel filtration chromatography. (top panels) Coomassie-stained gels showing the purified SAS-61–389 variants tested in each experiment (see also Fig S7B). (bottom panels) Elution profiles for the purified proteins on a Superdex 200 (E) or a WTC-030S5 (F) size exclusion column. (G) Model for cartwheel assembly in vivo. A direct interaction between ZYG-1 and the SAS-6 coiled-coil recruits the SAS-6—SAS-5 complex to the mother centriole where ZYG-1 kinase activity promotes cartwheel assembly by phosphorylating a target that is unlikely to be SAS-6. See also Figure S7.
DISCUSSION
The experiments presented here show that centriolar cartwheel assembly occurs in two separable steps (Fig. 7G). In the first step, SAS-6 is recruited to the mother centriole. This recruitment requires direct interaction of adjacent regions on the SAS-6 coiled-coil with ZYG-1 and SAS-5. In the second step, the cartwheel is formed. This step requires the kinase activity of ZYG-1 and SAS-6 inter-dimer interactions. To tackle the question of how ZYG-1 kinase activity promotes cartwheel formation, we analyzed a previously described ZYG-1 target site in SAS-6 and performed an extensive characterization of serine/threonine residues phosphorylated by ZYG-1 in vitro or conserved among closely related nematode SAS-6 homologs. This analysis failed to yield a residue or group of residues whose mutation approached the effect of loss of ZYG-1 kinase activity, suggesting that SAS-6 may not be the critical ZYG-1 target in the cartwheel assembly reaction. We therefore speculate, as discussed below, that the critical target of ZYG-1 may be SAS-5, which is bound close to ZYG-1 on the SAS-6 coiled-coil and is essential for cartwheel assembly in vivo.
Recruitment of SAS-6 to the mother centriole requires its interaction with both ZYG-1 and SAS-5
SAS-6 recruitment during daughter centriole assembly requires its interaction with both ZYG-1 and SAS-5. ZYG-1 targets to centrioles independently of SAS-5 or SAS-6 (Delattre et al., 2006; Pelletier et al., 2006, Fig. S5B) and interacts directly with SAS-6 (Fig. 4), explaining why the interaction with ZYG-1 is required. However, it is not clear why the interaction with SAS-5 is also required for SAS-6 to be recruited to the mother centriole. Our biochemical data suggest that SAS-5 binds to SAS-6 in a 1:2 stoichiometry and that SAS-5 is at least a dimer (the SAS-5 C-terminus is dimeric, Fig. S4E; full length SAS-5 could potentially be a higher-order oligomer). Thus, instead of binding to both available sites in the coiled-coil of a single SAS-6 dimer, dimeric/oligomeric SAS-5 would crosslink two/multiple SAS-6 dimers. Such a hexameric (depicted in Fig. 7G) or multimeric complex may have a higher affinity for the mother centriole as it would contain multiple ZYG-1 binding sites. As SAS-5 binds immediately adjacent to ZYG-1 on the SAS-6 coiled-coil, it is also possible that SAS-5 binding modulates the affinity of the ZYG-1—SAS-6 interaction. Alternatively, the ZYG-1 binding site on SAS-6 could be masked by another protein in vivo, and displacement of this factor by SAS-5 may be necessary for the ZYG-1 interaction. Finally, SAS-5 itself may recognize an element of the mother centriole, such as SPD-2 or ZYG-1, thereby providing a bipartite, multivalent coincidence detection mechanism for the mother centriole. However, to date, we have not observed interactions of SAS-5 with SPD-2 or ZYG-1 that would support this final possibility.
A major current limitation in testing the above ideas is the lack of full-length recombinant SAS-5 that is well-behaved in biochemical assays. Our stoichiometry measurements were performed using full-length SAS-5 concentrated on beads but we were unable to obtain sufficiently concentrated soluble full-length SAS-5 for rigorous complex analysis. Determining the molecular basis for how SAS-5 contributes to recruitment of SAS-6 to the mother centriole will be an important future goal.
SAS-6 is unlikely to be the critical ZYG-1 target during cartwheel assembly
ZYG-1 kinase activity controls cartwheel assembly, but our data suggest that SAS-6 is unlikely to be the critical ZYG-1 target. First, we did not see any significant defect resulting from mutation of the previously described ZYG-1 target site, S123 of SAS-6 (Kitagawa et al., 2009). We suspect the reasons for the difference between our results and the prior work are technical—the single-copy transgene insertions described here allow precise comparisons of wild-type and engineered mutants expressed at equivalent levels. This was difficult to do with the previous transgenesis method of ballistic bombardment, which resulted in random insertions of variable copy number that were susceptible to germline silencing. Although we did observe phosphorylation of S123, the dominant ZYG-1 phosphorylation sites on SAS-6 in vitro are in the C-tail rather than in the N-terminal domain of SAS-6. We believe this difference from the prior study is due to our use of full-length proteins in vitro, whereas SAS-6 fragments were utilized as substrates in the earlier work.
Despite analysis of 22 new potential ZYG-1 phosphorylation sites in SAS-6 as well as 19 additional serine/threonine residues conserved in SAS-6 homologs from closely related nematodes, we did not find SAS-6 target residues of similar importance to ZYG-1 kinase activity in cartwheel assembly. The only critical residues were located within the interaction surface between the N-terminal heads and, when mutated to alanine or to an acidic residue, disrupted inter-dimer interactions independently of phosphorylation in vitro. While it remains possible that our mutagenesis failed to reveal a critical residue set in SAS-6, the data suggest to us that SAS-6 is unlikely to be the key ZYG-1 target during cartwheel assembly.
SAS-5: The Critical Target of ZYG-1?
Since ZYG-1 binds immediately adjacent to SAS-5 on the SAS-6 coiled-coil, we speculate that ZYG-1 regulates cartwheel assembly primarily by phosphorylating SAS-5, rather than SAS-6. The notion that ZYG-1/Plk4 kinases regulate cartwheel assembly by phosphorylating SAS-5/Ana2/STIL family proteins is attractive from an evolutionary perspective because, although the basic structural components of centrioles, including SAS-6 and SAS-4, are conserved across a wide range of eukaryotes, SAS-5/Ana2/STIL and ZYG-1/Plk4 are limited to metazoans (Carvalho-Santos et al., 2010; Stevens et al., 2010; Arquint et al., 2012). This evolutionary distribution supports the idea that a regulatory mechanism involving phosphorylation of SAS-5/Ana2/STIL by ZYG-1/Plk4 kinases has been superimposed onto the basal centriole assembly process in metazoans. Future work will be necessary to determine if this speculation bears any merit.
EXPERIMENTAL PROCEDURES
Worm Strains and RNA-mediated Interference
For a list of the C. elegans strains see Supplementary Experimental Procedures. Strains carrying single-copy sas-6 and zyg-1 transgenes were constructed using MosSCI (Frøkjaer-Jensen et al., 2008; Figs. S1, S3). Double-stranded RNAs were generated as described (Oegema et al., 2001) using DNA templates prepared by PCR. L4 hermaphrodites were injected with dsRNA and incubated at 16°C or 20°C depending on the experiment (for details see Supplementary Experimental Procedures).
Western Blotting
Antibodies against SAS-6 and ZYG-1 were generated by injecting GST fusions with SAS-6 aa 2–175 or ZYG-1 aa 250–371 into rabbits. Antibodies were affinity-purified using standard procedures (Harlow, 1988). Worm lysate preparation and blotting were performed as previously described (Lewellyn et al., 2011). ZYG-1 signal was detected using the WesternBright Sirius detection system (Advansta; for details see the Supplementary Experimental Procedures).
Light Microscopy and Immunofluorescence
For detailed information on light microscopy and immunofluorescence see the Supplementary Experimental Procedures. All quantification of fluorescence was performed with MetaMorph software (Molecular Devices) based on previous methods (Dammermann et al., 2008). Immunofluorescence of C. elegans embryos was performed as described (Oegema et al., 2001).
Kinase Reactions and Mass Spectrometry
Strep II-ZYG-1–7xHis and GST-ZYG-1 were purified from Sf9 cells or E. coli as detailed in the Supplementary Experimental Procedures. For analysis by SDS-PAGE and autoradiography, GST-ZYG-1 (0.15 μM) was incubated with GST-SAS-6 (1.6 μM), 0.2mM ATP, and 0.2 μM 32P-ATP for 20 minutes at 30°C. For analysis by mass spectrometry, ZYG-1 (60 nM) was incubated with GST-SAS-6 (1.2 μM) and 0.8mM ATP for 20 minutes at 30°C (similar reactions with GST-ZYG-1 were also analyzed). Resuspended samples were alkylated, trypsinized, and loaded onto a LTQ XL ion trap mass spectrometer (Thermo Scientific). Resulting MS/MS spectral data were searched using the SEQUEST algorithm against a custom database containing human and C.elegans sequences.
Analytical Gel Filtration
For analysis of N-terminal domain-mediated assembly, SAS-6 variants were expressed and purified as detailed in the Supplementary Experimental Procedures. 13–25 μM SAS-6 variants in 20mM Tris pH 7.5, 150 mM NaCl, 1% glycerol, 1 mM DTT were injected onto a Superdex 200 (Fig. 7E, GE Healthcare) or WTC-030S5 size exclusion column (Fig. 7F, Wyatt Technology) connected in-line to an Optilab T-rEX refractive index detector (Wyatt Technology).
Yeast Two-Hybrid and Pull-down assays
Two-hybrid analysis was performed according to the manufacturer's instructions (MATCH-MAKER™, Clontech). For pull down assays, beads coated with purified proteins and incubated with soluble partners were washed three times and resuspended in SDS-PAGE sample buffer before analysis on SDS-PAGE gels. For analysis of ZYG-1 binding to WT, 2AZYG-1, or 3AZYG-1 SAS-6 variants, final concentrations were 12 μM SAS-6 (on 20 μL beads) and 4 μM ZYG-1 in solution. For analysis of SAS-5 binding to WT or 2ASAS-5 SAS-6 variants, final concentrations were 1 μM (for full-length) or 3 μM (for truncations) SAS-5 (on 20 μL beads), and 3 μM SAS-6 in solution. See Supplementary Experimental Procedures for additional details.
Supplementary Material
Highlights
-
-
SAS-6 recruitment to the mother centriole and cartwheel assembly are separable events
-
-
Direct binding of ZYG-1 to the SAS-6 coiled-coil recruits SAS-6 to the assembly site
-
-
ZYG-1 kinase activity is required for cartwheel assembly, but not SAS-6 recruitment
-
-
SAS-6 is unlikely to be a critical ZYG-1 kinase substrate during cartwheel assembly
ACKNOWLEDGEMENTS
We thank the Caenorhabditis Genetics Center and Kevin F. O'Connell for strains, Hayley Pemble for construction of the parental yeast two-hybrid plasmids, Alexander Dammermann for generation of the ZYG-1 antibody used for western blotting. This work was supported by a grant from the National Institutes of Health (R01-GM074207) to K.O. M.M.L. was supported by the University of California, San Diego Genetics (T32 GM008666) and Cancer Cell Biology Training Programs. K.O. and A.D. receive salary and additional support from the Ludwig Institute for Cancer Research.
ABBREVIATIONS
- EM
Electron Microscopy
- KD
Kinase Defective
- RR
RNAi-Resistant
- C-tail
C-terminal tail of SAS-6
- WT
wild-type
- CC
coiled-coil
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arquint C, Sonnen KF, Stierhof Y-D, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125:1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20:R816–R825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Current Biology. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M. Deconstructing the centriole: structure and number control. Curr Opin Cell Biol. 2012;24:4–13. doi: 10.1016/j.ceb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- Castiel A, Danieli MM, David A, Moshkovitz S, Aplan PD, Kirsch IR, Brandeis M, Krämer A, Izraeli S. The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J Cell Sci. 2011;124:532–539. doi: 10.1242/jcs.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver BP, Meehl JB, Giddings TH, Winey M. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol Biol Cell. 2009;20:1865–1877. doi: 10.1091/mbc.E08-08-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Müller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gönczy P. Sequential protein recruitment in C. elegans centriole formation. Current Biology. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gönczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Audhya A, Pozniakovsky A, Dammermann A, Pemble H, Monen J, Portier N, Hyman A, Desai A, Oegema K. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 2008;85:179–218. doi: 10.1016/S0091-679X(08)85009-1. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof Y-D, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton (Hoboken) 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Song MH, Addepalli MK, Hunter G, O'Connell K. Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans. Genetics. 2007;176:95–113. doi: 10.1534/genetics.107.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Kusch M, Wolf N. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics. 1988;120:977–986. doi: 10.1093/genetics/120.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Müller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Busso C, Flückiger I, Gönczy P. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Flückiger I, Polanowska J, Keller D, Reboul J, Gönczy P. PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev Cell. 2011a;20:550–562. doi: 10.1016/j.devcel.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, Flückiger I, Gönczy P. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci. 2011b;124:3884–3893. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011c;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Gönczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Lewellyn L, Carvalho A, Desai A, Maddox AS, Oegema K. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J Cell Biol. 2011;193:155–169. doi: 10.1083/jcb.201008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol. Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Current Biology. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O'Toole E, Schwager A, Hyman AA, Müller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G. SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly. EMBO J. 2012;31:4434–4347. doi: 10.1038/emboj.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Current Biology. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Strome S. Launching the germline in Caenorhabditis elegans: regulation of gene expression in early germ cells. Development. 1999;126:3275–3283. doi: 10.1242/dev.126.15.3275. [DOI] [PubMed] [Google Scholar]
- Song MH, Liu Y, Anderson DE, Jahng WJ, O'Connell KF. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell. 2011;20:563–571. doi: 10.1016/j.devcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Tang C-JC, Lin S-Y, Hsu W-B, Lin Y-N, Wu C-T, Lin Y-C, Chang C-W, Wu K-S, Tang TK. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. The EMBO Journal. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa H-A, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong I-O, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S, et al. STIL is required for centriole duplication in human cells. J Cell Sci. 2012 doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- Yabe T, Ge X, Pelegri F. The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev Biol. 2007;312:44–60. doi: 10.1016/j.ydbio.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.