Summary
Cytotoxic chemotherapy for acute myeloid leukemia (AML) usually produces only temporary remissions, at the cost of significant toxicity and risk for death. One fundamental reason for treatment failure is that it is designed to activate apoptosis genes (eg., TP53) that may be unavailable because of mutation or deletion. Unlike deletion of apoptosis genes, genes that mediate cell cycle exit by differentiation are present in myelodysplastic syndrome (MDS) and AML cells but are epigenetically repressed: MDS/AML cells express high levels of key lineage-specifying transcription factors (TF). Mutation in these TF (eg., CEBPA) or their cofactors (eg., RUNX1) affect transactivation function and produce epigenetic repression of late-differentiation genes that antagonize MYC. Importantly, this aberrant epigenetic repression can be redressed clinically by depleting DNA methyltransferase 1 (DNMT1, a central component of the epigenetic network that mediates transcription repression) using the deoxycytidine analogue decitabine (DAC) at non-cytotoxic concentrations. The DNMT1 depletion is sufficient to trigger upregulation of late-differentiation genes and irreversible cell cycle exit by p53-independent differentiation mechanisms. Fortuitously, the same treatment maintains or increases self-renewal of normal hematopoietic stem cells (HSC), which do not express high levels of lineage-specifying TF. The biological rationale for this approach to therapy appears to apply to cancers other than MDS/AML also. DAC or 5-azacytidine dose and schedule can be rationalized to emphasize this mechanism of action, as an alternative or complement to conventional apoptosis-based oncotherapy.
Keywords: Decitabine, differentiation, p53, p16, p27, CDKN2A, CDKN1B, therapy, chromatin modifying enzymes, cancer, leukemia
The lineage and maturation context of cancer cells
A major goal of cancer research is to find differences between normal stem cells and cancer cells that can be used to selectively destroy cancer cells. One difference is frequent mutation or deletion of key apoptosis genes (e.g., TP53, p16/CDKN2A) in cancer cells1-7. However, this feature of cancer cells works against the objectives of conventional apoptosis-based chemo- or radiation therapy, contributing to treatment resistance and toxicity.
Instead of increasing cancer cell apoptosis/death as the primary treatment objective, an alternative is to identify and target pathways of cancer cell proliferation8. Since these pathways may be differentiation-context dependent, it is useful to understand the lineage and maturation stage of cancer cells. Examination of cancer cell morphology and surface phenotype usually reveals lineage-commitment. Indeed, lineage-markers (morphologic and immunohistochemical) underpin classification of malignancy, and lineage-commitment is an implicit component of some oncotherapy, for example, hormonal blockade to treat breast and prostate cancer.
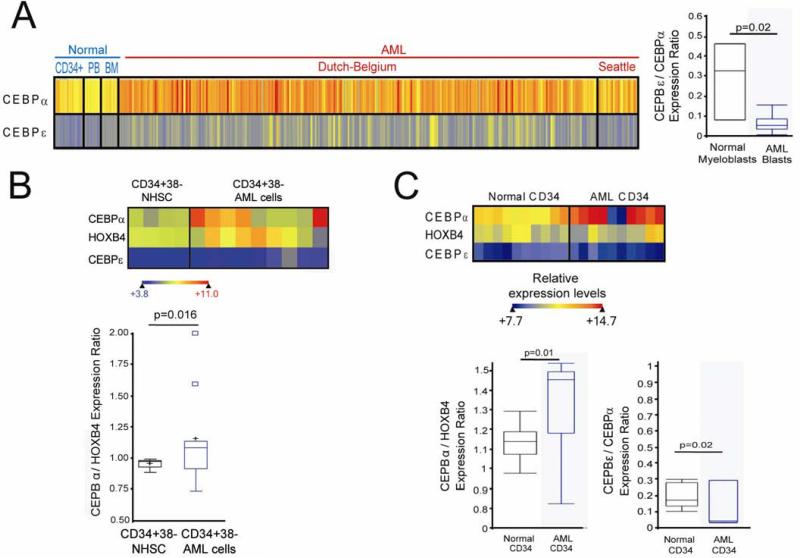
The expression pattern of key DNA binding transcription factors that drive lineage-commitment and progressive maturation provide further valuable insight into differentiation context: lineage-commitment and progressive maturation absolutely requires and is driven by coordinated expression of key transcription factors9-11. Missense mutations in genes for these factors, for example CEBPA and GATA1, are known initiating events in acute myeloid leukemia (AML) pathogenesis12;13. These genes are expressed at high levels only with lineage-commitment. Indeed, AML cells express high levels of these factors (Figure 1)14. Despite the high expression of CEBPA, AML cells express relatively low levels of the key late-differentiation driver transcription factor CEBPE (Figure 1)15;16 (expression levels of CEBPE increase during the transition from proliferating pro-myelocytes to non-proliferating myelocytes11, and CEBPE terminates proliferation in myeloid and AML cells10;17-22).
Figure 1.
The pattern of expression of key lineage-specifying and late differentiation factors in AML cells suggests impaired differentiation in lineage-committed cells. A) CEBPA (lineage-specifying factor) and CEBPE (late differentiation factor) expression in AML myeloblasts (n=318) compared with normal CD34+ cells, bone marrow and peripheral blood (n=38), and normal myeloblasts (n=3) (p-values Wilcoxon Two-sample test. Raw data extracted from GEO Datasets 148;149, gene expression measured by microarray). B) CD34+CD38- cells from AML patient bone marrow (n=9) express higher levels of CEBPA, and a higher CEBPA/HOXB4 ratio, than CD34+CD38- cells from normal bone marrow (n=4). p-values Median Two-Sample test. Raw data extracted from GEO Datasets 150. C) Similar findings in CD34+ AML cells (n=10) and normal CD34+ cells (n=11) analyzed by gene-expression microarray. Expression levels represented by heat-map. p-values Wilcoxon Two-sample test. Raw data extracted from GEO Datasets 151;152D) CD34+CD38- cells from AML patient bone marrow (n=9) express higher levels of CEBPα, and a higher CEBPα/HOXB4 ratio, than CD34+CD38- cells from normal bone marrow (n=4) 150. p-values Median Two-Sample test. Raw data extracted from GEO Datasets 150.
The identity and gene expression profiles of key transcription factors that drive progressive maturation of solid tissues are not as well characterized as for hematopoiesis23. Nonetheless, where the identity of these transcription factors is known, the solid tumors that arise from these tissues express high levels of lineage-commitment transcription factors, and low levels of key late-differentiation genes: malignant melanoma cells express high levels of the melanocyte commitment factor MITF, and point mutations in melanoma target MITF and another key driver of melanocyte commitment and early differentiation, SOX1024;25. However, melanocyte late-differentiation driver genes, eg., SOX9, are epigenetically repressed26;27. Medulloblastoma cells express high levels of genes that are expressed early in cerebellar development/differentiation, but relatively low levels of late cerebellar differentiation genes28 (the medulloblastoma gene expression profile corresponds to the normal maturation stage with the highest rate of proliferation and migration28). Squamous cell lung carcinoma cells demonstrate a gene expression profile of an intermediate stage of normal lung development/differentiation28. Rhabdomyosarcomas express high levels of the lineage-specifying transcription factor MYOD29, however, disruptions to the usual interactions between MYOD and E-proteins results in repression rather than activation of late-differentiation target genes29. Chromosome translocations target the master regulator of differentiation ETV6 in breast cancer and sarcoma30;31, PAX8 in follicular thyroid cancer32, PAX3 and PAX7 in rhabdomyosarcoma33;34 and TFE3 in papillary renal cell cancer35. Evidence for lineage-commitment and lineage-dependency in solid tumors has also been reviewed elsewhere36.
Is lineage-commitment a feature of cancer ‘stem cells’ or cancer initiating cells?
Surface phenotype can be used to sort cancer cell populations into subsets. These subsets can then be xeno-transplanted into immunocompromised mice for evaluation of cancer initiating efficiency (as a measurement of self-renewal capacity). The earliest studies suggested that AML cells with leukemia-initiating capacity had a surface phenotype resembling that of normal hematopoietic stem cells (CD34+38-)37;38. This suggested that AML cell populations might recapitulate the hierarchical structure of normal hematopoiesis, with only cells with a stem cell phenotype having the self-renewal capacity to sustain the bulk AML cell population38. Recently, it has been reported that technical factors may have biased results from the earliest studies39. Accordingly, in numerous recent studies, AML initiating cells had a surface phenotype suggesting lineage-commitment (progenitor phenotype) (CD34+38+, CLL-1+, CD71+, CD90 -, c-Kit -)39-46. Also, with use of more immunocompromised mice as recipients, AML initiating cell surface-phenotypes are not stem cell restricted47-50. Even AML cells with a stem cell surface phenotype (CD34+CD38-) express high levels of the lineage-specifying transcription factor CEBPA, low levels of the late-differentiation driver CEBPE, and low levels of stem cell genes such as HOXB4, when compared to normal CD34+38- cells (Figure 1)15.
In solid cancers, surface markers that identify cancer cell subsets with the highest cancer initiating efficiency, such as CD133, are surface markers of both progenitors and stem cells51-54, and therefore, provide limited information regarding stem versus progenitor context. Similar to the experience with AML cells, use of more immunocompromised mice in these assays suggests cancer-initiating capacity is less restricted than early estimates, with a similar frequency of cancer initiating capacity in the CD133 positive and negative compartments55.
Gene expression profiles of cancer cells and embryonic stem cells can overlap42. Recently, it has been shown that the overlapping gene expression signatures can be attributed, in large part, to the activity of MYC, a driver of cell proliferation56. Importantly, MYC upregulation is a feature of, and required for, the active proliferation that occurs with lineage-commitment by stem cells57;58. In other words, the MYC module that is associated with cancer is a normal feature of early progenitors.
Progressive epigenetic repression of late-differentiation genes during neoplastic evolution
Coactivator protein complexes recruited by DNA binding transcription factors contain chromatin modifying enzymes that create activation marks on histones, and also assist in recruitment of the basal transcription factor complex. Conversely, corepressor protein complexes that can be alternatively recruited by transcription factors contain chromatin-modifying enzymes that create repression marks on histones and DNA. As discussed earlier, mutations or translocations in lineage-commitment/early-differentiation transcription factors (or their cofactors) affect corepressor/coactivator recruitment decisions59 and produce epigenetic repression of late-differentiation genes that would otherwise terminate active MYC-driven proliferation10;16-22;59;60.
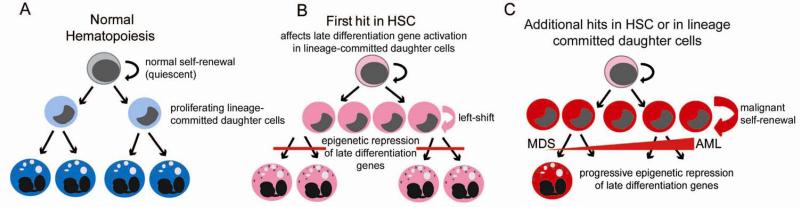
Typically, evolution and progression of cancer is accompanied by increasing impairment of maturation, illustrated for example during myelodysplastic syndrome (MDS) progression into AML (Figure 2). Conceivably, this progressive impairment of maturation is caused by further imbalance in corepressor/coactivator recruitment by transcription factors that regulate expression of late-differentiation genes. Supporting this possibility, genetic abnormalities that target chromatin modifying enzymes (e.g., ASXL1) accompany MDS progression to AML61-63 (Figure 2). A high rate of mutation or amplification in chromatin modifying enzymes is also observed in solid tumors64 (comprehensively reviewed elsewhere65-69). One net consequence of these abnormalities is an increase in repression marks on histones and in gene promoters that have correlated with disease aggression in multi-variate analyses of multiple malignancies including AML, ALL, and multiple solid tumors70-76. Indeed, repressive chromosome marks can be observed over large chromosome regions in cancer cells (reviewed in77).
Figure 2.
Progression of myelodysplastic syndrome (MDS) into acute myeloid leukemia (AML). A) Normal hematopoiesis: Hematopoietic stem cells (HSC) can self-renew or give rise to lineage-committed daughter cells (progenitors). Progenitors actively divide (transit-amplification) until mature cells are formed. B) The initial abnormality (first hit) in multi-hit neoplastic evolution may occur in an HSC (the cell of origin is an HSC). However, the growth advantage is conferred to progenitors, by epigenetic repression of late differentiation genes. Early in the disease process, the differentiation impairment produced by the initiating abnormality may not be severe enough to decrease mature cell numbers. However, a left-shift in the marrow compartment may be noted, as differentiation impaired precursors accumulate. C) Mature cell numbers decrease, and there is a progressive left-shift, with additional hits that cause progressive epigenetic repression of late differentiation genes in the progenitor compartment, conferring the property of self-renewal to lineage-committed cells (producing leukemia-initiating cells, LIC, and evolution of MDS into AML).
The model suggested by these observations
The preceding observations suggest the following model of MDS/AML and possibly other cancers:
Adult stem cells are quiescent (reviewed in78). However, daughter cells that lineage-commit proliferate actively (a MYC-driven process57;58). This proliferation is usually self-limited by the activation of late-differentiation genes that antagonize MYC16;22;60;79. However, mutation or translocation in early-differentiation driving transcription factors produces aberrant epigenetic repression of these late-differentiation genes. In other words, the MYC activation may be physiologic, having occurred as a consequence of lineage-commitment by stem cells; the pathologic event is failure to activate late-differentiation genes that antagonize MYC. Since the repression of late-differentiation genes is epigenetic and not genetic, it is potentially reversible. Furthermore, the key DNA-binding factors that usually drive expression of the late-differentiation genes are expressed at high levels in the lineage-committed malignant cells. The fundamental problem is an imbalance in corepressor versus coactivator recruitment at late-differentiation genes59.
This model is readily tested: treating MDS/AML or other cancer cells with conditions or drugs that antagonize corepressor function should restore late-differentiation gene expression and terminate proliferation. This has indeed been observed, with a number of different strategies to relax chromatin, and in a spectrum of cancer histologies and genotypes: aggressive, differentiation-impaired melanoma and breast cancer cells resumed differentiation and exited cell cycle when exposed to an embryonic cell micro-environment that opens chromatin80;81. Similarly, oocyte extracts, another micro-environment that induces DNA hypomethylation and removes repressive histone marks, terminated tumorigenicity of breast cancer cells82. Drugs that inhibit histone deacetylases (chromatin modifying enzymes that create repressive histone marks) induce terminal differentiation in a spectrum of leukemia and cancer primary cells and cell lines83-89. Similarly, the deoxycytidine analogue decitabine, which depletes DNA methyl-transferase 1 (DNMT1) (DNMT1 creates the methyl-CpG DNA repression mark) and relaxes chromatin90;91 terminates proliferation of various AML and cancer primary cells and cell lines85;88;92-95.
Irreversible cell cycle exit by epigenetic-differentiation does not require functional p53 or p16/CDKN2A
p53 and p16/CDKN2A-null mice, although cancer-prone, demonstrate essentially normal development96;97, suggesting that cell cycle exit by differentiation may not require these master regulators of apoptosis. Evaluating this possibility using clinically available drugs is particularly relevant from a translational perspective. One confounding factor in the interpretation of such studies is that drug therapy can have non-epigenetic effects, including antimetabolite or DNA damaging effects that cause apoptosis, that may contribute to cell cycle exit98. To address this issue, we have conducted experiments focused on the drug decitabine14-16;27: unlike the cytidine analogues cytarabine or gemcitabine, the sugar moiety of decitabine is unmodified. Therefore, at low concentrations, DNA-incorporated decitabine does not terminate DNA chain elongation99;100. Accordingly, decitabine can be administered at doses that deplete DNMT1 without causing significant DNA damage or cytotoxicity, both in vitro and in vivo92;99-103.
Treatment of AML, renal cell cancer and melanoma cells with these concentrations of decitabine allowed one or two cell divisions, upregulated key drivers of late myeloid (CEBPE), epithelial (HNF4A) and melanocyte (SOX9) differentiation respectively, and induced cell cycle exit accompanied by upregulation of p27/CDKN1B, the cyclin dependent kinase inhibitor that mediates cell cycle exit by differentiation104-107. Further underlining the p53-independence of the differentiation-mediated cell cycle exit, many of the cells used in these experiments were p16/CDKN2A and/or p53 null15;16;27;108. In vitro observations were readily recapitulated in murine xenotransplantation models of AML, renal cell cancer and melanoma, by using a dose and schedule of decitabine that depleted DNMT1 without in vivo myelotoxicity15;16;27;108. Fortuitously, p27/CDKN1B is rarely deleted from cancer cells, unlike p16/CDKN2A and p53: in 770 cell lines analyzed by the Cancer Genome Project (Wellcome Trust Sanger Institute), there was homozygous deletion of p16/CDKN2A in 218 cell lines, loss of heterozygosity in 278 cell lines, and mutation in 276 cell lines. There was homozygous deletion of TP53 in 5 cell lines, loss of hetrozygosity in 482 cell lines, and mutation in 482 cell lines. In contrast, there was p27/CDKN1B loss of heterozygosity in 161 cell lines but no mutations or homozygous deletions in any of the 770 cell lines analyzed.
Why self-renewal of normal stem cells is maintained with this treatment approach
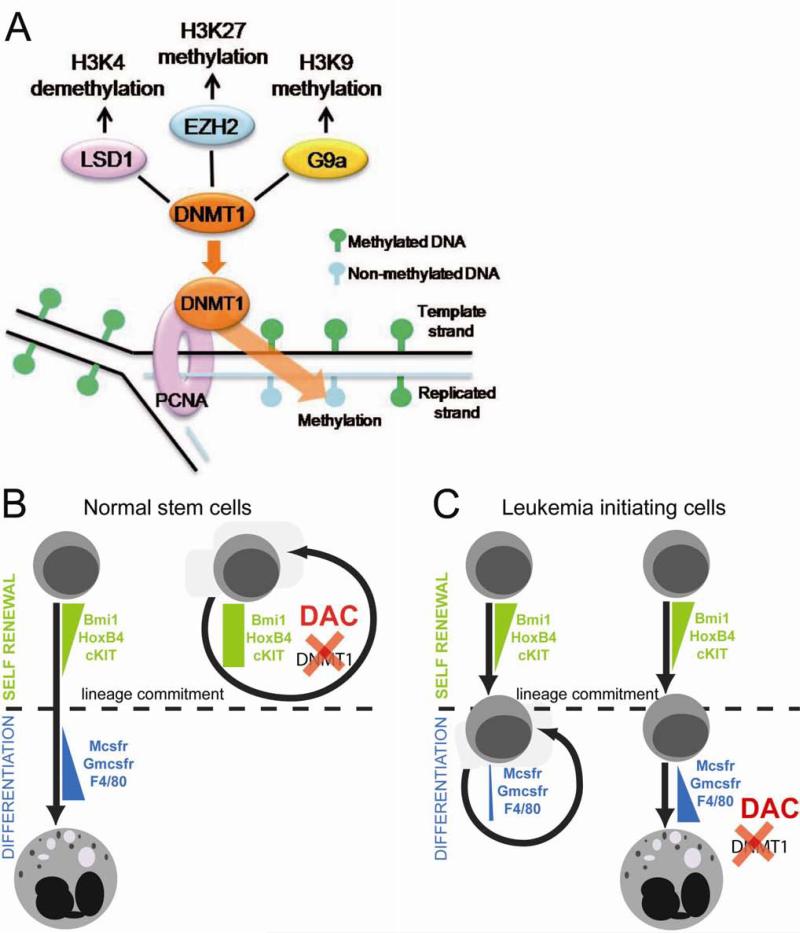
Hematopoietic stem cell genes, such as HOXB4 and c-Kit, are rapidly repressed during the process of hematopoietic lineage commitment and differentiation109. Since DNMT1 is a central component of the epigenetic network that mediates transcription repression110, DNMT1 depletion by shRNA or decitabine prevented stem cell gene repression by differentiation stimuli and maintained stem cell phenotype109. These observations also explain why other drugs that antagonize transcription repression, such as histone deacetylase inhibitors and 5-azacytidine, also increase hematopoietic stem cell self-renewal111-118. However, decitabine treatment after the stem cell gene repression phase of the differentiation process augmented differentiation109. Therefore, the cell fate consequences of depleting DNMT1 with decitabine depend on baseline maturation stage (Figure 3). In other words, differences in maturation stage/lineage-commitment underlie the contrasting effects of chromatin-relaxing drugs on self-renewal and differentiation of malignant and normal stem cells.
Figure 3.
Maturation context explains why non-cytotoxic, DNA methyl-transferase 1 (DNMT1) depleting concentrations of decitabine increase normal hematopoietic stem cell self-renewal but induce terminal differentiation of AML cells. A) DNMT1 plays a central role in the network of chromatin-modifying enzymes that are implicated in transcription repression. B) In normal hematopoietic stem cells (HSC), decitabine (DAC) to deplete DNMT1 and antagonize transcription repression prevents a necessary first step in lineage-commitment, which is repression of stem cell gene expression. Therefore, DAC treatment maintains HSC self-renewal, even in differentiation promoting conditions 109;111-118. If DAC is added shortly after the differentiation-inducing stimulus (after the phase of stem cell gene repression), it does not prevent and may even increase differentiation 109. C) Leukemia-initiating events, such as RUNX1 mutation, can originate in the germ-line or in hematopoietic stem cells, however, RUNX1 deficient stem cells can lineage-commit in response to a differentiation stimulus, with intact repression of stem cell genes. Instead, RUNX1 cooperation with lineage-specifying transcription factor to active late-differentiation genes is impaired, by coactivator/corepressor imbalance at late-differentiation genes, producing aberrant self-renewal (proliferation at the same level of differentiation) in lineage-committed cells. In these cells, primed to differentiate with high levels of lineage-specifying factors, and in which repression of late differentiation genes is by epigenetic means, DAC can resume differentiation and differentiation-mediated cell cycle exit.
Towards effective clinical translation
The idea of using differentiation to terminate malignant cell proliferation (differentiation therapy) was mooted more than 50 years ago119-121. However, as a primary objective of clinical therapy, it is currently limited to all-trans retinoic acid (ATRA) treatment of acute promyelocytic leukemia (APL)122.
One reason has been that the biological model or pathway basis for pursuing epigenetic-differentiation therapy has not been clear. Indeed, the model of cancer as being sustained by self-renewing cancer ‘stem cells’, does not provide a rationale for this mode of therapy, since in the stem cell model, differentiation-impairment is presumably a consequence rather than a cause of malignant self-renewal36;38;123. However, recent data, as outlined above, challenge the stem cell model, and provide a biologic and mechanistic rationale for epigenetic-differentiation therapy.
Another important reason for limited translation is pharmacologic and mechanism of action complexity and limitations of compounds that differentiate AML or cancer cells in vitro: ATRA targets leukemia fusion proteins containing RARA, which are present only in APL. Low dose cytosine arabinoside (cytarabine, AraC) may induce differentiation but is predominantly cytotoxic120. Histone deacetylase inhibitor drugs may induce differentiation but also have cytotoxic effects, and it is difficult to separate epigenetic effects of these drugs from DNA damage and apoptosis induction124-126. Similarly, decitabine that depletes DNMT1 can induce both apoptosis and epigenetic/differentiation effects. Indeed, decitabine was originally developed as a DNA-damaging agent for cytotoxic therapy127. Therefore, doses to treat AML were escalated to maximum tolerated levels in traditional phase 1 studies128. Although the regimen in common use to treat MDS has de-escalated doses with an epigenetic mechanism of action in mind129;130, therapy continues to resemble pulse-cycled cytotoxic therapy, and the potential or actual cytotoxicity of current regimens has resulted in controversy regarding the relative importance of differentiation to the clinical mechanism of action98. Although cytotoxicity can contribute to tumor kill in vitro and in vivo, cytotoxicity also impairs treatment eligibility, tolerance and feasible exposure, and destroys normal hematopoietic stem cells required for relief of cytopenia and durable remission of myeloid cancers. Furthermore, mutagenicity and micro-environmental insult from anti-metabolite actions can potentially accelerate malignant evolution and resistance131;132.
There is ample pre-clinical evidence that decitabine, at non-cytotoxic but DNMT1 depleting concentrations, can induce cancer cell cycle exit by differentiation pathways14-16;27. Indeed, in the earliest cell biology studies the in vitro differentiation modifying effects of decitabine were most potent at low, non-cytotoxic concentrations92. Therefore, a DNMT1 depleting, but not necessarily cytotoxic dose, administered frequently, should be safer and more effective than a higher, cytotoxic dose administered infrequently, since exposure timings and distribution are critical considerations for S-phase specific depletion of DNMT1101. Furthermore, maximizing cell cycle exit by epigenetic-differentiation would offer a true p53/p16-independent alternative or complement to conventional apoptosis-based therapy. We are currently testing this approach in a National Institutes of Health sponsored clinical trial in MDS, administering decitabine 0.1-0.2 mg/kg (3.5-7 mg/m2) (a dose lower than in previous studies130;133-135, since even these low doses are sufficient to deplete DNMT1), by the subcutaneous route (to avoid high peak drug levels that can cause apoptosis), administered from 1-3X/week (to produce greater exposure than with previous clinical trials, and to distribute exposure and capture MDS cells entering cell cycle asynchronously at different points in time). This type of decitabine dose and schedule has been used to treat non-malignant disease103;136, demonstrating its clinical safety and noncytotoxic epigenetic and differentiation modifying actions103;136.
In parallel, we are developing an approach to oral therapy that combines decitabine with tetrahydrouridine, an inhibitor of cytidine deaminase, the enzyme which rapidly metabolizes decitabine in vivo (because of cytidine deaminase activity, the in vivo half-life of decitabine is <20 minutes137 in contrast to an in vitro half-life at 37°C of approximately 9 hours). Oral administration of tetrahydrouridine-decitabine is more likely to produce the desired pharmacologic profile of low peak drug levels (~0.005-0.2 μM) to avoid cytotoxicity, but extended half-life (hours rather than minutes) to increase depletion of DNMT1, and could decrease inter-individual variability in decitabine pharmacokinetics that arises from pharmacogenetic variation in cytidine deamianse (Lavelle et al., submitted manuscript). In addition, an oral formulation will provide major cost, logistical and accessibility advantages for long-term outpatient therapy. Finally, inhibition of cytidine deaminase may address a mechanism of cancer cell resistance, and sanctuary from the effects of decitabine that can occur in organs that express high levels of cytidine deaminase, for example, the liver and intestines138-146.
Currently, decitabine (~10% of the related compound 5-azacytidine is converted to decitabine by ribonucleotide reductase in vivo) is the only drug that can be repurposed clinically for non-cytotoxic, p53/p16-independent epigenetic-differentiation therapy (since it is difficult to separate the chromatin-modifying effects of histone deacetylase inhibitor drugs from cytotoxicity). Other compounds that can inhibit components of the chromatin modifying network without inducing apoptosis have been identified or will be identified69;147. The maturation and epigenetic context of cancer cells suggests that these agents can also be developed for the purpose of p53/p16-independent, normal stem cell sparing epigenetic-differentiation oncotherapy.
Conclusion
Conventional therapy focuses on inducing irreversible cell cycle exit in cancer cells by DNA damage or metabolic insult that activates apoptosis pathways (cytotoxicity). This approach to therapy has a major limitation: malignant cells frequently mutate or delete key apoptosis genes. Hence, the goal of activating apoptosis genes may be futile, since these genes may not be present, yet treatment destroys normal stem cells which have intact apoptosis pathways. This manifests clinically as short-term improvement but frequent relapse with more aggressive, apoptosis-resistant disease. Unlike apoptosis genes, genes that mediate cell cycle exit by differentiation are typically present, but are aberrantly repressed by epigenetic means. Furthermore, cancer cells express high levels of lineage-specifying transcription factors. Because of this epigenetic and maturation context of cancer cells, non-cytotoxic antagonism of chromatin-modifying enzymes that mediate transcription repression terminates proliferation by differentiation104-107. Normal stem cells, which do not express high levels of lineage-specifying transcription factors, are spared109-118. Using differentiation to terminate cancer cell proliferation was first described more than 50 years ago119-121, but is not a major component of current clinical therapy. The biological and translational insights that have accrued in the intervening five decades renew the importance and promise of this approach, not just for the myeloid malignancies, but for cancer in general.
Acknowledgements
We would like to thank Soledad Negrotto and Ng Kwok Peng for assistance with figure preparation. YS is supported by NIH (U54HL090513, 1R01CA138858), Dept. of Defense (PR081404), Scott Hamilton CARES Foundation, Mr and Mrs Robert McNeil, and Mr Robert Stein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wattel E, Preudhomme C, Hecquet B, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84(9):3148–3157. [PubMed] [Google Scholar]
- 2.Stirewalt DL, Kopecky KJ, Meshinchi S, et al. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood. 2001;97(11):3589–3595. doi: 10.1182/blood.v97.11.3589. [DOI] [PubMed] [Google Scholar]
- 3.Andersen MK, Christiansen DH, Kirchhoff M, Pedersen-Bjergaard J. Duplication or amplification of chromosome band 11q23, including the unrearranged MLL gene, is a recurrent abnormality in therapy-related MDS and AML, and is closely related to mutation of the TP53 gene and to previous therapy with alkylating agents. Genes Chromosomes.Cancer. 2001;31(1):33–41. doi: 10.1002/gcc.1115. [DOI] [PubMed] [Google Scholar]
- 4.Schoch C, Kern W, Kohlmann A, et al. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosomes.Cancer. 2005;43(3):227–238. doi: 10.1002/gcc.20193. [DOI] [PubMed] [Google Scholar]
- 5.Wendel HG, de Stanchina E, Cepero E, et al. Loss of p53 impedes the antileukemic response to BCR-ABL inhibition. Proc.Natl.Acad.Sci.U.S.A. 2006;103(19):7444–7449. doi: 10.1073/pnas.0602402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suarez L, Vidriales MB, Garcia-Larana J, et al. CD34+ cells from acute myeloid leukemia, myelodysplastic syndromes, and normal bone marrow display different apoptosis and drug resistance-associated phenotypes. Clin.Cancer Res. 2004;10(22):7599–7606. doi: 10.1158/1078-0432.CCR-04-0598. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat.Rev.Drug Discov. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 8.Harris H. Tumour suppression: putting on the brakes. Nature. 2004;427(6971):201. doi: 10.1038/427201a. [DOI] [PubMed] [Google Scholar]
- 9.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka R, Barlow C, Lekstrom-Himes J, et al. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc.Natl.Acad.Sci.U.S.A. 1997;94(24):13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theilgaard-Monch K, Jacobsen LC, Borup R, et al. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105(4):1785–1796. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 12.Klusmann JH, Godinho FJ, Heitmann K, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24(15):1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owen C, Barnett M, Fitzgibbon J. Familial myelodysplasia and acute myeloid leukaemia--a review. Br.J.Haematol. 2008;140(2):123–132. doi: 10.1111/j.1365-2141.2007.06909.x. [DOI] [PubMed] [Google Scholar]
- 14.Negrotto S, Hu Z, Alcazar O, et al. Noncytotoxic differentiation treatment of renal cell cancer. Cancer Res. 2011;71(4):1431–1441. doi: 10.1158/0008-5472.CAN-10-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negrotto S, Hu Z, Link KA, et al. Differentiation-Chronology Specific Function of DNMT1 and Selective Anti-Leukemia Stem-Cell Therapy. Blood. 2008;112(11):81–82. [Google Scholar]
- 16.Ng KP, Ebrahem Q, Negrotto S, et al. p53 Independent epigenetic-differentiation treatment in xenotransplant models of acute myeloid leukemia. Leukemia. 2011 doi: 10.1038/leu.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima H, Ihle JN. Granulocyte colony-stimulating factor regulates myeloid differentiation through CCAAT/enhancer-binding protein epsilon. Blood. 2001;98(4):897–905. doi: 10.1182/blood.v98.4.897. [DOI] [PubMed] [Google Scholar]
- 18.Truong BT, Lee YJ, Lodie TA, et al. CCAAT/Enhancer binding proteins repress the leukemic phenotype of acute myeloid leukemia. Blood. 2003;101(3):1141–1148. doi: 10.1182/blood-2002-05-1374. [DOI] [PubMed] [Google Scholar]
- 19.Gery S, Gombart AF, Fung YK, Koeffler HP. C/EBPepsilon interacts with retinoblastoma and E2F1 during granulopoiesis. Blood. 2004;103(3):828–835. doi: 10.1182/blood-2003-01-0159. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima H, Watanabe N, Shibata F, et al. N-terminal region of CCAAT/enhancer-binding protein epsilon is critical for cell cycle arrest, apoptosis, and functional maturation during myeloid differentiation. J.Biol.Chem. 2006;281(20):14494–14502. doi: 10.1074/jbc.M600575200. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita H, Nakajima H, Nakamura Y, et al. C/EBPalpha and C/EBPvarepsilon induce the monocytic differentiation of myelomonocytic cells with the MLL-chimeric fusion gene. Oncogene. 2008;27(53):6749–6760. doi: 10.1038/onc.2008.285. [DOI] [PubMed] [Google Scholar]
- 22.Walkley CR, Purton LE, Snelling HJ, et al. Identification of the molecular requirements for an RAR alpha-mediated cell cycle arrest during granulocytic differentiation. Blood. 2004;103(4):1286–1295. doi: 10.1182/blood-2003-07-2391. [DOI] [PubMed] [Google Scholar]
- 23.Warburton D, Schwarz M, Tefft D, et al. The molecular basis of lung morphogenesis. Mech.Dev. 2000;92(1):55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 24.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 25.Cronin JC, Wunderlich J, Loftus SK, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22(4):435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passeron T, Valencia JC, Namiki T, et al. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J.Clin.Invest. 2009;119(4):954–963. doi: 10.1172/JCI34015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcazar O, Achberger S, Aldrich W, et al. Epigenetic regulation by decitabine of melanoma differentiation in vitro and in vivo. Int.J.Cancer. 2011 doi: 10.1002/ijc.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kho AT, Zhao Q, Cai Z, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18(6):629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, MacQuarrie KL, Analau E, et al. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 2009;23(6):694–707. doi: 10.1101/gad.1765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 31.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: congenital (infantile) fibrosarcoma and mesoblastic nephroma. Cancer Genet.Cytogenet. 2002;132(1):1–13. doi: 10.1016/s0165-4608(01)00528-3. [DOI] [PubMed] [Google Scholar]
- 32.McIver B, Grebe SK, Eberhardt NL. The PAX8/PPAR gamma fusion oncogene as a potential therapeutic target in follicular thyroid carcinoma. Curr.Drug Targets.Immune.Endocr.Metabol.Disord. 2004;4(3):221–234. doi: 10.2174/1568008043339802. [DOI] [PubMed] [Google Scholar]
- 33.Galili N, Davis RJ, Fredericks WJ, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat.Genet. 1993;5(3):230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 34.Weber-Hall S, McManus A, Anderson J, et al. Novel formation and amplification of the PAX7-FKHR fusion gene in a case of alveolar rhabdomyosarcoma. Genes Chromosomes.Cancer. 1996;17(1):7–13. doi: 10.1002/(SICI)1098-2264(199609)17:1<7::AID-GCC2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 35.Sinke RJ, de Leeuw B, Janssen HA, et al. Localization of X chromosome short arm markers relative to synovial sarcoma- and renal adenocarcinoma-associated translocation breakpoints. Hum.Genet. 1993;92(3):305–308. doi: 10.1007/BF00244478. [DOI] [PubMed] [Google Scholar]
- 36.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat.Rev.Cancer. 2006;6(8):593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 37.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 38.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 39.Taussig DC, Miraki-Moud F, Anjos-Afonso F, et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112(3):568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 40.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 43.van Rhenen A, Moshaver B, Kelder A, et al. Aberrant marker expression patterns on the CD34+. Leukemia. 2007;21(8):1700–1707. doi: 10.1038/sj.leu.2404754. [DOI] [PubMed] [Google Scholar]
- 44.Blair A, Hogge DE, Ailles LE, Lansdorp PM, Sutherland HJ. Lack of expression of Thy-1 (CD90) on acute myeloid leukemia cells with long-term proliferative ability in vitro and in vivo. Blood. 1997;89(9):3104–3112. [PubMed] [Google Scholar]
- 45.Blair A, Sutherland HJ. Primitive acute myeloid leukemia cells with long-term proliferative ability in vitro and in vivo lack surface expression of c-kit (CD117). Exp.Hematol. 2000;28(6):660–671. doi: 10.1016/s0301-472x(00)00155-7. [DOI] [PubMed] [Google Scholar]
- 46.Goardon N, Marchi E, Atzberger A, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19(1):138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Wunderlich M, Chou FS, Link KA, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24(10):1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feuring-Buske M, Gerhard B, Cashman J, et al. Improved engraftment of human acute myeloid leukemia progenitor cells in beta 2-microglobulin-deficient NOD/SCID mice and in NOD/SCID mice transgenic for human growth factors. Leukemia. 2003;17(4):760–763. doi: 10.1038/sj.leu.2402882. [DOI] [PubMed] [Google Scholar]
- 49.Agliano A, Martin-Padura I, Mancuso P, et al. Human acute leukemia cells injected in NOD/LtSz-scid/IL-2Rgamma null mice generate a faster and more efficient disease compared to other NOD/scid-related strains. Int.J.Cancer. 2008;123(9):2222–2227. doi: 10.1002/ijc.23772. [DOI] [PubMed] [Google Scholar]
- 50.Sarry JE, Murphy K, Perry R, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rgammac-deficient mice. J.Clin.Invest. 2011;121(1):384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–5012. [PubMed] [Google Scholar]
- 52.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 53.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 54.Al Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc.Natl.Acad.Sci.U.S.A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quintana E, Shackleton M, Sabel MS, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson A, Murphy MJ, Oskarsson T, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18(22):2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr.Biol. 2001;11(8):558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 59.Hu Z, Gu X, Baraoidan K, et al. RUNX1 regulates corepressor interactions of PU.1. Blood. 2011 doi: 10.1182/blood-2010-10-312512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negrotto S, Ng KP, Jankowska AM, et al. CpG methylation patterns and decitabine treatment response in acute myeloid leukemia cells and normal hematopoietic precursors. Leukemia. 2011 doi: 10.1038/leu.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rocquain J, Carbuccia N, Trouplin V, et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC.Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boultwood J, Perry J, Pellagatti A, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24(5):1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- 63.Makishima H, Jankowska AM, Tiu RV, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24(10):1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 64.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin-Perez D, Piris MA, Sanchez-Beato M. Polycomb proteins in hematologic malignancies. Blood. 2010;116(25):5465–5475. doi: 10.1182/blood-2010-05-267096. [DOI] [PubMed] [Google Scholar]
- 66.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat.Rev.Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc.Natl.Acad.Sci.U.S.A. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum.Mol.Genet. 2007;16:R28–R49. doi: 10.1093/hmg/ddm021. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 69.Tuma RS. Targeted epigenetic therapies: the next frontier? J.Natl.Cancer Inst. 2010;102(24):1824–1825. doi: 10.1093/jnci/djq520. [DOI] [PubMed] [Google Scholar]
- 70.Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2008 doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 72.Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am.J.Pathol. 2009;174(5):1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Advani AS, Gibson SE, Douglas E, et al. Histone H4 acetylation by immunohistochemistry and prognosis in newly diagnosed adult acute lymphoblastic leukemia (ALL) patients. BMC.Cancer. 2010;10:387. doi: 10.1186/1471-2407-10-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J.Clin.Oncol. 2007;25(28):4358–4364. doi: 10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- 75.Manuyakorn A, Paulus R, Farrell J, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J.Clin.Oncol. 2010;28(8):1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elsheikh SE, Green AR, Rakha EA, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69(9):3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 77.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum.Mol.Genet. 2007;16:R88–R95. doi: 10.1093/hmg/ddm051. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 78.Li J. Quiescence Regulators for Hematopoietic Stem Cell. Exp.Hematol. 2011 doi: 10.1016/j.exphem.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 79.Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113(6):1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Postovit LM, Margaryan NV, Seftor EA, et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc.Natl.Acad.Sci.U.S.A. 2008;105(11):4329–4334. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309(5739):1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 82.Allegrucci C, Rushton MD, Dixon JE, et al. Epigenetic reprogramming of breast cancer cells with oocyte extracts. Mol.Cancer. 2011;10(1):7. doi: 10.1186/1476-4598-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gozzini A, Rovida E, Dello SP, Galimberti S, Santini V. Butyrates, as a single drug, induce histone acetylation and granulocytic maturation: possible selectivity on core binding factor-acute myeloid leukemia blasts. Cancer Res. 2003;63(24):8955–8961. [PubMed] [Google Scholar]
- 84.Kosugi H, Towatari M, Hatano S, et al. Histone deacetylase inhibitors are the potent inducer/enhancer of differentiation in acute myeloid leukemia: a new approach to anti-leukemia therapy. Leukemia. 1999;13(9):1316–1324. doi: 10.1038/sj.leu.2401508. [DOI] [PubMed] [Google Scholar]
- 85.Nowak D, Stewart D, Koeffler HP. Differentiation therapy of leukemia: 3 decades of development. Blood. 2009;113(16):3655–3665. doi: 10.1182/blood-2009-01-198911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spira AI, Carducci MA. Differentiation therapy. Curr.Opin.Pharmacol. 2003;3(4):338–343. doi: 10.1016/s1471-4892(03)00081-x. [DOI] [PubMed] [Google Scholar]
- 87.Gore SD, Samid D, Weng LJ. Impact of the putative differentiating agents sodium phenylbutyrate and sodium phenylacetate on proliferation, differentiation, and apoptosis of primary neoplastic myeloid cells. Clin.Cancer Res. 1997;3(10):1755–1762. [PubMed] [Google Scholar]
- 88.Wang J, Saunthararajah Y, Redner RL, Liu JM. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1-ETO leukemia cells. Cancer Res. 1999;59(12):2766–2769. [PubMed] [Google Scholar]
- 89.Moldenhauer A, Frank RC, Pinilla-Ibarz J, et al. Histone deacetylase inhibition improves dendritic cell differentiation of leukemic blasts with AML1-containing fusion proteins. J.Leukoc.Biol. 2004;76(3):623–633. doi: 10.1189/jlb.1103581. [DOI] [PubMed] [Google Scholar]
- 90.Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol.Ther. 1995;65(1):19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]
- 91.Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol.Cell Biol. 2005;25(11):4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 93.Pinto A, Attadia V, Fusco A, et al. 5-Aza-2′-deoxycytidine induces terminal differentiation of leukemic blasts from patients with acute myeloid leukemias. Blood. 1984;64(4):922–929. [PubMed] [Google Scholar]
- 94.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J.Biol.Chem. 1982;257(4):2041–2048. [PubMed] [Google Scholar]
- 95.Niitsu N, Hayashi Y, Sugita K, Honma Y. Sensitization by 5-aza-2′-deoxycytidine of leukaemia cells with MLL abnormalities to induction of differentiation by all-trans retinoic acid and 1alpha,25-dihydroxyvitamin D3. Br.J.Haematol. 2001;112(2):315–326. doi: 10.1046/j.1365-2141.2001.02523.x. [DOI] [PubMed] [Google Scholar]
- 96.Attardi LD, Donehower LA. Probing p53 biological functions through the use of genetically engineered mouse models. Mutat.Res. 2005;576(1-2):4–21. doi: 10.1016/j.mrfmmm.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 97.Serrano M, Lee H, Chin L, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 98.Tuma RS. Epigenetic therapies move into new territory, but how exactly do they work? J.Natl.Cancer Inst. 2009;101(19):1300–1301. doi: 10.1093/jnci/djp342. [DOI] [PubMed] [Google Scholar]
- 99.Covey JM, D'Incalci M, Tilchen EJ, Zaharko DS, Kohn KW. Differences in DNA damage produced by incorporation of 5-aza-2′-deoxycytidine or 5,6-dihydro-5-azacytidine into DNA of mammalian cells. Cancer Res. 1986;46(11):5511–5517. [PubMed] [Google Scholar]
- 100.Schermelleh L, Haemmer A, Spada F, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35(13):4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Momparler RL, Goodman J. In vitro cytotoxic and biochemical effects of 5-aza-2′-deoxycytidine. Cancer Res. 1977;37(6):1636–1639. [PubMed] [Google Scholar]
- 102.Halaban R, Krauthammer M, Pelizzola M, et al. Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS.ONE. 2009;4(2):e4563. doi: 10.1371/journal.pone.0004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saunthararajah Y, Hillery CA, Lavelle D, et al. Effects of 5-aza-2′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood. 2003;102(12):3865–3870. doi: 10.1182/blood-2003-05-1738. [DOI] [PubMed] [Google Scholar]
- 104.Fero ML, Rivkin M, Tasch M, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85(5):733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 105.Kiyokawa H, Kineman RD, Manova-Todorova KO, et al. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell. 1996;85(5):721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 106.Nakayama K, Ishida N, Shirane M, et al. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85(5):707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 107.Furukawa Y, Kikuchi J, Nakamura M, et al. Lineage-specific regulation of cell cycle control gene expression during haematopoietic cell differentiation. Br.J.Haematol. 2000;110(3):663–673. doi: 10.1046/j.1365-2141.2000.02253.x. [DOI] [PubMed] [Google Scholar]
- 108.Ng KP, Negrotto S, Hu ZB, et al. Non-p53 Dependent, Leukemia Initiating-Cell Selective, Therapy. Blood. 2009;114(22):820. [Google Scholar]
- 109.Hu Z, Negrotto S, Gu X, et al. Decitabine maintains hematopoietic precursor self-renewal by preventing repression of stem cell genes by a differentiation-inducing stimulus. Mol.Cancer Ther. 2010;9(6):1536–1543. doi: 10.1158/1535-7163.MCT-10-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Chromatin challenges during DNA replication: a systems representation. Mol.Biol.Cell. 2008;19(1):1–7. doi: 10.1091/mbc.E07-06-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Milhem M, Mahmud N, Lavelle D, et al. Modification of hematopoietic stem cell fate by 5aza 2'deoxycytidine and trichostatin A. Blood. 2004;103(11):4102–4110. doi: 10.1182/blood-2003-07-2431. [DOI] [PubMed] [Google Scholar]
- 112.De Felice L, Tatarelli C, Mascolo MG, et al. Histone deacetylase inhibitor valproic acid enhances the cytokine-induced expansion of human hematopoietic stem cells. Cancer Res. 2005;65(4):1505–1513. doi: 10.1158/0008-5472.CAN-04-3063. [DOI] [PubMed] [Google Scholar]
- 113.Bug G, Gul H, Schwarz K, et al. Valproic acid stimulates proliferation and self-renewal of hematopoietic stem cells. Cancer Res. 2005;65(7):2537–2541. doi: 10.1158/0008-5472.CAN-04-3011. [DOI] [PubMed] [Google Scholar]
- 114.Young JC, Wu S, Hansteen G, et al. Inhibitors of histone deacetylases promote hematopoietic stem cell self-renewal. Cytotherapy. 2004;6(4):328–336. doi: 10.1080/14653240410004899. [DOI] [PubMed] [Google Scholar]
- 115.Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38(1):32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 116.Araki H, Mahmud N, Milhem M, et al. Expansion of human umbilical cord blood SCID-repopulating cells using chromatin-modifying agents. Exp.Hematol. 2006;34(2):140–149. doi: 10.1016/j.exphem.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 117.Suzuki M, Harashima A, Okochi A, et al. 5-Azacytidine supports the long-term repopulating activity of cord blood CD34(+) cells. Am.J.Hematol. 2004;77(3):313–315. doi: 10.1002/ajh.20178. [DOI] [PubMed] [Google Scholar]
- 118.Chung YS, Kim HJ, Kim TM, et al. Undifferentiated hematopoietic cells are characterized by a genome-wide undermethylation dip around the transcription start site and a hierarchical epigenetic plasticity. Blood. 2009;114(24):4968–4978. doi: 10.1182/blood-2009-01-197780. [DOI] [PubMed] [Google Scholar]
- 119.PIERCE GB, Jr., VERNEY EL. An in vitro and in vivo study of differentiation in teratocarcinomas. Cancer. 1961;14:1017–1029. doi: 10.1002/1097-0142(196109/10)14:5<1017::aid-cncr2820140516>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 120.Michalewicz R, Lotem J, Sachs L. Cell differentiation and therapeutic effect of low doses of cytosine arabinoside in human myeloid leukemia. Leuk.Res. 1984;8(5):783–790. doi: 10.1016/0145-2126(84)90099-7. [DOI] [PubMed] [Google Scholar]
- 121.SEILERN-ASPANG F, KRATOCHWIL K. Induction and differentiation of an epithelial tumour in the newt (Triturus cristatus). J.Embryol.Exp.Morphol. 1962;10:337–356. [PubMed] [Google Scholar]
- 122.Huang ME, Ye YC, Chen SR, et al. All-trans retinoic acid with or without low dose cytosine arabinoside in acute promyelocytic leukemia. Report of 6 cases. Chin Med.J.(Engl.) 1987;100(12):949–953. [PubMed] [Google Scholar]
- 123.Bernt KM, Zhu N, Sinha AU, et al. MLL-Rearranged Leukemia Is Dependent on Aberrant H3K79 Methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Scuto A, Kirschbaum M, Kowolik C, et al. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph- acute lymphoblastic leukemia cells. Blood. 2008;111(10):5093–5100. doi: 10.1182/blood-2007-10-117762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc.Natl.Acad.Sci.U.S.A. 2010;107(33):14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Conti C, Leo E, Eichler GS, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70(11):4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sorm F, Vesely J. Effect of 5-aza-2′-deoxycytidine against leukemic and hemopoietic tissues in AKR mice. Neoplasma. 1968;15(4):339–343. [PubMed] [Google Scholar]
- 128.Rivard GE, Momparler RL, Demers J, et al. Phase I study on 5-aza-2′-deoxycytidine in children with acute leukemia. Leuk.Res. 1981;5(6):453–462. doi: 10.1016/0145-2126(81)90116-8. [DOI] [PubMed] [Google Scholar]
- 129.Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 130.Steensma DP, Baer MR, Slack JL, et al. Multicenter study of decitabine administered daily for 5 days every 4 weeks to adults with myelodysplastic syndromes: the alternative dosing for outpatient treatment (ADOPT) trial. J.Clin.Oncol. 2009;27(23):3842–3848. doi: 10.1200/JCO.2008.19.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kitano H. Cancer robustness: tumour tactics. Nature. 2003;426(6963):125. doi: 10.1038/426125a. [DOI] [PubMed] [Google Scholar]
- 132.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127(5):905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 133.Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106(8):1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 134.Kantarjian HM, O'Brien S, Huang X, et al. Survival advantage with decitabine versus intensive chemotherapy in patients with higher risk myelodysplastic syndrome: comparison with historical experience. Cancer. 2007;109(6):1133–1137. doi: 10.1002/cncr.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kantarjian HM, O'Brien S, Shan J, et al. Update of the decitabine experience in higher risk myelodysplastic syndrome and analysis of prognostic factors associated with outcome. Cancer. 2007;109(2):265–273. doi: 10.1002/cncr.22376. [DOI] [PubMed] [Google Scholar]
- 136.Olivieri NF, Saunthararajah Y, Thayalasuthan V, et al. A pilot study of subcutaneous decitabine in {beta}-thalassemia intermedia. Blood. 2011 doi: 10.1182/blood-2011-03-341909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu Z, Marcucci G, Byrd JC, et al. Characterization of decomposition products and preclinical and low dose clinical pharmacokinetics of decitabine (5-aza-2′-deoxycytidine) by a new liquid chromatography/tandem mass spectrometry quantification method. Rapid Commun.Mass Spectrom. 2006;20(7):1117–1126. doi: 10.1002/rcm.2423. [DOI] [PubMed] [Google Scholar]
- 138.Steuart CD, Burke PJ. Cytidine deaminase and the development of resistance to arabinosyl cytosine. Nat.New Biol. 1971;233(38):109–110. doi: 10.1038/newbio233109a0. [DOI] [PubMed] [Google Scholar]
- 139.Ohta T, Hori H, Ogawa M, et al. Impact of cytidine deaminase activity on intrinsic resistance to cytarabine in carcinoma cells. Oncol.Rep. 2004;12(5):1115–1120. [PubMed] [Google Scholar]
- 140.Momparler RL, Laliberte J. Induction of cytidine deaminase in HL-60 myeloid leukemic cells by 5-aza-2′-deoxycytidine. Leuk.Res. 1990;14(9):751–754. doi: 10.1016/0145-2126(90)90067-j. [DOI] [PubMed] [Google Scholar]
- 141.Yin B, Tsai ML, Hasz DE, et al. A microarray study of altered gene expression after cytarabine resistance in acute myeloid leukemia. Leukemia. 2007;21(5):1093–1097. doi: 10.1038/sj.leu.2404595. [DOI] [PubMed] [Google Scholar]
- 142.Ge Y, Jensen TL, Stout ML, et al. The role of cytidine deaminase and GATA1 mutations in the increased cytosine arabinoside sensitivity of Down syndrome myeloblasts and leukemia cell lines. Cancer Res. 2004;64(2):728–735. doi: 10.1158/0008-5472.can-03-2456. [DOI] [PubMed] [Google Scholar]
- 143.Braess J, Wegendt C, Feuring-Buske M, et al. Leukaemic blasts differ from normal bone marrow mononuclear cells and CD34+ haemopoietic stem cells in their metabolism of cytosine arabinoside. Br.J.Haematol. 1999;105(2):388–393. [PubMed] [Google Scholar]
- 144.Qin T, Jelinek J, Si J, Shu J, Issa JP. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood. 2009;113(3):659–667. doi: 10.1182/blood-2008-02-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kreis W, Lesser M, Budman DR, et al. Phenotypic analysis of 1-B-D-arabinofuranosylcytosine deamination in patients treated with high doses and correlation with response. Cancer Chemother.Pharmacol. 1992;30(2):126–130. doi: 10.1007/BF00686404. [DOI] [PubMed] [Google Scholar]
- 146.Beumer JH, Eiseman JL, Parise RA, et al. Modulation of gemcitabine (2′,2′-difluoro-2′-deoxycytidine) pharmacokinetics, metabolism, and bioavailability in mice by 3,4,5,6-tetrahydrouridine. Clin.Cancer Res. 2008;14(11):3529–3535. doi: 10.1158/1078-0432.CCR-07-4885. [DOI] [PubMed] [Google Scholar]
- 147.Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc.Natl.Acad.Sci.U.S.A. 2011 doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stirewalt DL, Meshinchi S, Kopecky KJ, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes.Cancer. 2008;47(1):8–20. doi: 10.1002/gcc.20500. [DOI] [PubMed] [Google Scholar]
- 149.Ferrari F, Bortoluzzi S, Coppe A, et al. Genomic expression during human myelopoiesis. BMC.Genomics. 2007;8:264. doi: 10.1186/1471-2164-8-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Majeti R, Becker MW, Tian Q, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc.Natl.Acad.Sci.U.S.A. 2009;106(9):3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cheung AM, Chow HC, Liang R, Leung AY. A comparative study of bone marrow and peripheral blood CD34+ myeloblasts in acute myeloid leukaemia. Br.J.Haematol. 2009;144(4):484–491. doi: 10.1111/j.1365-2141.2008.07431.x. [DOI] [PubMed] [Google Scholar]
- 152.Pellagatti A, Cazzola M, Giagounidis AA, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108(1):337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]