Abstract
Stem cell differentiation depends on transcriptional activation driven by lineage-specific regulators as well as changes in chromatin organization. However, the coordination of these events is poorly understood. Here, we show that T-box proteins team up with chromatin modifying enzymes to drive the expression of the key lineage regulator, Eomes during endodermal differentiation of embryonic stem (ES) cells. The Eomes locus is maintained in a transcriptionally poised configuration in ES cells. During early differentiation steps, the ES cell factor Tbx3 associates with the histone demethylase Jmjd3 at the enhancer element of the Eomes locus to allow enhancer–promoter interactions. This spatial reorganization of the chromatin primes the cells to respond to Activin signalling, which promotes the binding of Jmjd3 and Eomes to its own bivalent promoter region to further stimulate Eomes expression in a positive feedback loop. In addition, Eomes activates a transcriptional network of core regulators of endodermal differentiation. Our results demonstrate that Jmjd3 sequentially associates with two T-box factors, Tbx3 and Eomes to drive stem cell differentiation towards the definitive endoderm lineage.
Keywords: definitive endoderm, Jmjd3, promoter-enhancer looping, stem cell differentiation, Tbx3
Introduction
ES cells maintain the ability to self-renew while retaining the ability to differentiate into any mature cell lineages. Differentiation of ES cells involves switching molecular circuits that maintain self-renewal and pluripotency to those that are associated with activation of lineage specification and, ultimately maintenance of cell type-specific gene expression. The switch in molecular circuits is enabled by dynamic genomic reorganization to alter chromatin structure and regulate transcriptional activation of lineage-specific genes. Chromatin reorganization controls accessibility for the transcriptional machinery around key lineage-specific loci to prepare ES cells to respond to signals to differentiate along a particular lineage. Genome-wide studies have shown that the promoters of lineage regulators in ES cells are enriched for histone modifications associated both with gene activation such as trimethylation of histone3 lysine4 (H3K4me3) and with polycomb-mediated repression such as trimethylation of histone3 lysine27 (H3K27me3) (Bernstein et al, 2006; Mikkelsen et al, 2007; Pan et al, 2007; Zhao et al, 2007; Ku et al, 2008). This bivalent chromatin profile keeps lineage regulators poised for rapid activation. In response to differentiation cues, the bivalent domains are resolved with the removal of H3K27me3 and activation of lineage-specific developmental genes. The Jumonji domain-containing proteins, Utx (Kdm6a) and Jmjd3 (Kdm6b), function as H3K27me3 demethylases (Agger et al, 2007; De Santa et al, 2007; Hong et al, 2007; Issaeva et al, 2007; Lan et al, 2007; Lee et al, 2007). Some studies have linked them to the resolution of bivalent domains during ES cell differentiation (Mikkelsen et al, 2007; Burgold et al, 2008; Kim et al, 2011). How the histone demethylases act in concert with lineage specification genes to reorganize the chromatin and direct differentiation along a particular lineage is poorly understood. Deciphering the detailed molecular mechanisms that control these epigenetic events is essential to understand how gene expression patterns are established and conveyed during ES cell differentiation and, ultimately, to manipulate ES cells for tissue engineering and regenerative medicine.
Differentiation of definitive endoderm is of great interest as its derivatives including lung, liver, and pancreas are of interest for regenerative medicine, but efforts to generate these cell types from ES cells remains a significant challenge. During development, epiblast cells ingress through the anterior segment of the primitive streak where they undergo an epithelial-to-mesenchymal transition to give rise to mesoderm and definitive endoderm (Grapin-Botton and Melton, 2000). Nascent definitive endoderm is marked by expression of transcription factors, Sox17 and Foxa2 (Ang and Rossant, 1994; Weinstein et al, 1994; Kanai-Azuma et al, 2002). Genetic studies in mice that delete these factors demonstrate their requirement for definitive endoderm formation (Ang and Rossant, 1994; Weinstein et al, 1994; Kanai-Azuma et al, 2002). Deletion of T-box transcription factor Eomesodermin (Eomes or Tbr2) in epiblast cells results in embryos that entirely lack of definitive endoderm. Chimera analysis show that Eomes-null ES cells can efficiently contribute to the mesodermal lineage but are excluded from definitive endoderm (Arnold et al, 2008). As epiblast cells express Nodal, the involvement of TGF-β signalling in definitive endoderm formation was demonstrated by chimera analysis showing that Smad2-null ES cells were unable to contribute to the definitive endoderm lineage (Tremblay et al, 2000). Consistent with this observation, Nodal signalling was shown to induce and maintain the transcriptional network present in definitive endoderm in the embryo (Vincent et al, 2003).
These genetic studies provided a dynamic picture of endoderm formation in the embryos and have guided studies of directed differentiation of ES cells. Application of Activin A or Nodal to mouse or human ES cells plays an important role in directing the formation of definitive endoderm (Kubo et al, 2004; D'Amour et al, 2005; Yasunaga et al, 2005; Borowiak et al, 2009). In human ES cells, it has been suggested that Activin A via Nanog-Smad2/3 complex (Vallier et al, 2009) induces the expression of Eomes, which in turn activates the transcriptional network in the formation of definitive endoderm (Costello et al, 2011; Teo et al, 2011). Both in vitro and in vivo data indicate that Eomes plays an essential role in definitive endoderm differentiation, although the early steps that lead to Eomes activation from pluripotent state remain elusive. It is also not clear how transcriptional activation of Eomes is coordinated with the reconfiguration of the chromatin associated with ES cell differentiation towards definitive endoderm lineage.
In this study, we show Eomes is maintained in a transcriptionally poised configuration in ES cells. During early steps of differentiation, the T-box protein Tbx3 and the demethylase Jmjd3 bound to the Eomes enhancer promote spatial reorganization to allow the enhancer region to engage in a direct physical interaction with the promoter proximal region. The promoter proximal region is then depleted of ubiquitination of histone2A (H2Aub) and phosphorylation of RNA polymerase II at Serine2 (RNAP-Ser2P), resulted in release of Eomes from the poised configuration. Following Activin A signalling, Eomes interacts with Smad2 to act on the bivalent domain within the Eomes promoter, transactivating its own expression in a positive feedback loop. Eomes in turn cooperates with Jmjd3 and Smad2 and acts on bivalent domains within the promoters of core endodermal regulators to activate a transcriptional network leading to definitive endoderm specification. Our results show conserved mechanisms in mouse and human during endoderm differentiation whereby the two crucial T-box transcription factors; Tbx3 and Eomes sequentially team up with an epigenetic modifier, Jmjd3 to drive stem cell differentiation towards definitive endoderm lineage.
Results
Release of poised RNAP leads to transcriptional activation of Eomes
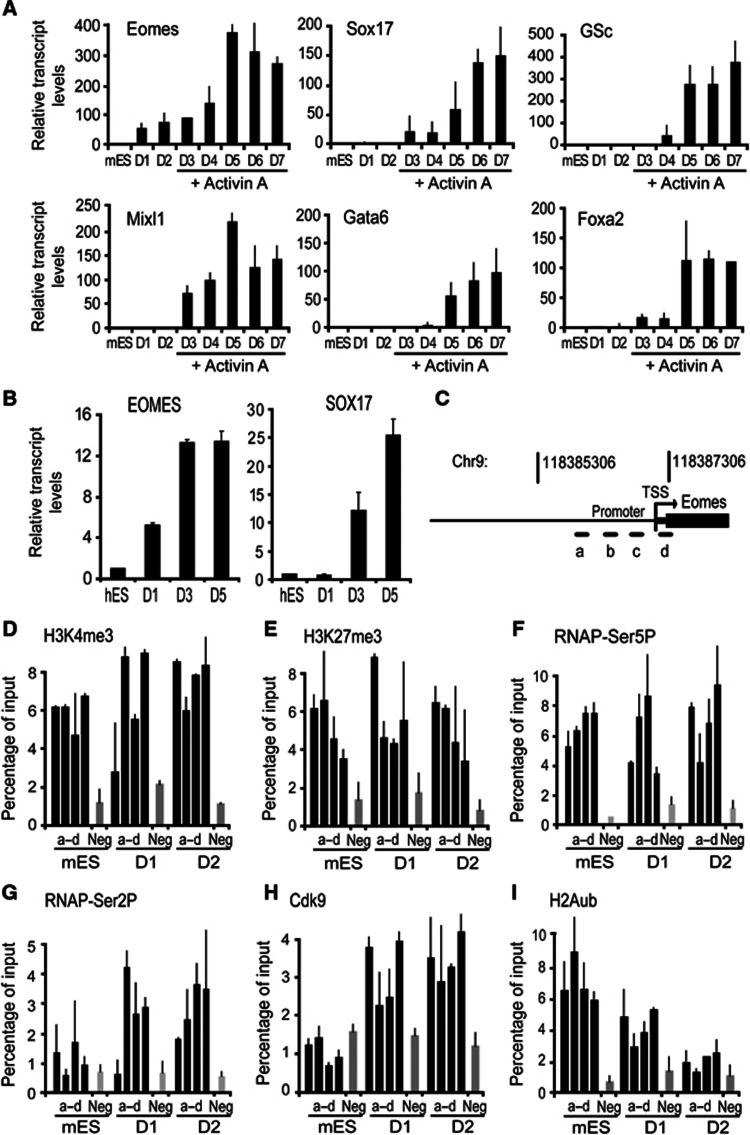
To study the temporal sequence of transcriptional activation of endodermal genes during directed differentiation of definitive endoderm, we used a step-wise differentiation protocol to direct mES cells towards definitive endodermal fate (Kubo et al, 2004; Morrison et al, 2008). Leukaemia inhibitory factor (LIF) was removed from culture medium to allow ES cells to aggregate and differentiate to form embryoid bodies (EBs). After 2 days, EBs were subsequently cultured for 5 days with medium supplemented with Activin A (see Materials and methods). Detailed expression analysis for transcription factors known involved in endodermal specification revealed that Eomes was induced in the early steps of differentiation and did not require Activin A for its induction (Figure 1A). Activin A treatment did however increase Eomes expression levels by eight-fold over the levels observed during EB stage (Figure 1A). The induction of other endodermal-specific transcription factors tested including Sox17, Gsc, Mixl1, Gata6 and Foxa2 was not observed during early stages and Activin A treatment was required for transcriptional activation these genes (Figure 1A). The rapid induction of Eomes during the earliest steps of ES cell differentiation suggested that Eomes is held in a transcriptionally poised state in ES cells. Induction of endoderm in hES cell line, HSF1, and induced pluripotent stem cells line, hiPS2 using previously established protocols (D'Amour et al, 2005; Borowiak et al, 2009; Patterson et al, 2011) showed that induction of EOMES preceded SOX17 expression (Figure 1B). Thus, the temporal sequence of transcriptional activation of endodermal genes was similar in mouse and human ES cell differentiation.
Figure 1.
Release of poised RNAP leads to transcriptional activation of Eomes during differentiation. (A) Induction of Eomes occurred early during definitive endoderm differentiation, preceded the expression of the core transcription factors of definitive endoderm (D, days in culture). Transcript levels were measured using quantitative RT–PCR. (B) EOMES was upregulated (D1) prior to the induction of SOX17 expression (D3–D5) in hES cell differentiation. Transcript levels were measured using quantitative RT–PCR. (C–F) Transcriptional activation of Eomes is not accompanied by resolution of the bivalent domain. (C) The Eomes promoter proximal region analysed by ChIP using the four primer sets (a–d). Enrichment of H3K4me3 (D), H3K27me3 (E), and RNAP-Ser5P (F) is shown at the Eomes proximal-promoter regions (a–d) and negative region (Neg) in differentiated mES cells, D1–D2. (G–I) Transcriptional activation of Eomes involves release of poised RNAP into productive elongation upon differentiation. (G) RNAP is phosphorylated at Serine2 in differentiated mES cells. (H) Cdk9 occupancy accompanies increased phosphorylation of RNAP at Serine2. (I) H2Aub enrichment is diminished at the Eomes promoter proximal region in differentiated mES cells, D1–D2. Enrichment of RNAP-Ser2P, Cdk9, and H2Aub was measured using ChIP at the Eomes proximal-promoter regions (a–d) and negative control region (Neg) in differentiated mES cells, D1–D2. All values are mean±s.e.m. (n=2–3).
To assess whether Eomes is maintained in a transcriptionally poised state in ES cells, we analysed the histone modification profile and assembly of RNA polymerase II (RNAP) throughout the Eomes promoter proximal region during the early steps of ES cell differentiation (Figure 1C). Chromatin immunoprecipitation (ChIP) analysis revealed that like many other lineage regulators in ES cells, the Eomes promoter is simultaneously enriched for H3K4me3 (Figure 1D) and H3K27me3 (Figure 1E) and bound by RNAP phosphorylated at Serine5 (RNAP-Ser5P) (Figure 1F). The absence of binding of antibody 8WG16, which recognizes non-phosphorylated Serine2 residue of RNAP, to the Eomes promoter proximal region and the gene body reinforced the idea that Eomes is held in a poised configuration in ES cells (Supplementary Figure 1A and B). Surprisingly, the bivalent histone enrichment profile at the Eomes promoter did not change with transcriptional activation of Eomes in the first 48 h of differentiation (Figure 1D and E). The levels of RNAP-Ser5P at the Eomes promoter remained high (Figure 1F). The levels of RNAP-Ser2P, a form of RNAP associated with elongation, were initially low in ES cells, but increased markedly within 24 h of LIF withdrawal in both promoter and the gene body (Figure 1G; Supplementary Figure 1C). This suggested that RNAP is restrained at the promoter proximal region of Eomes in ES cells and released into productive elongation upon differentiation.
The onset of productive elongation requires Cdk9 kinase activity of the P-TEFβ complex, which phosphorylates RNAP at Serine2 (Marshall et al, 1996; Stock et al, 2007). ChIP analysis of Cdk9 on the Eomes promoter indeed showed that increased Cdk9 binding during EB formation correlated with increased RNAP-Ser2P levels (Figure 1H). The release of poised RNAP is enforced by a marked decrease in Polycomb-mediated H2Aub (Figure 1I). Decreased H2Aub levels were accompanied by reduced Ring1 binding to the Eomes promoter (Supplementary Figure 1D). No changes in Ezh2 binding were observed (Supplementary Figure 1E). Taken together, these analyses indicate that in ES cells, H2Aub enrichment and RNAP-Ser5P maintains Eomes in a poised configuration. H2Aub depletion and RNAP-Ser2P enrichment early in differentiation induces release of Eomes from the poised configuration.
Tbx3 recruits Jmjd3 to the enhancer region of Eomes locus
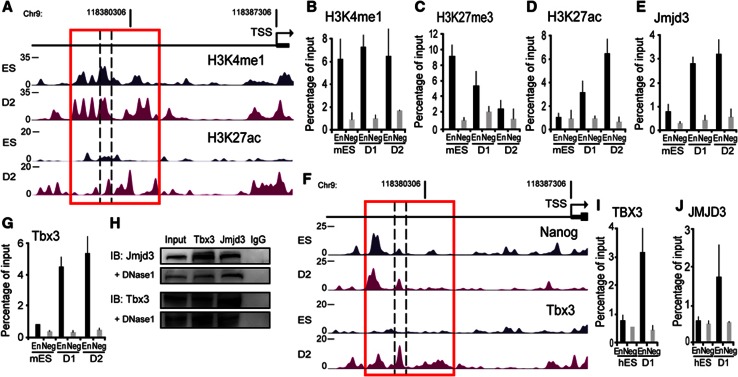
To investigate whether regulatory elements could be involved in the release of poised RNAP from the Eomes promoter during differentiation, we carried out epigenomic profiling of monomethylation of histone3 lysine 4 (H3K4me1), and acetylation of histone3 lysine 27 (H3K27ac). We performed ChIPs followed by high-throughput sequencing (ChIP-Seq) to identify enrichment of these marks. We identified a region 7 kb upstream of the transcription start site that was conserved in vertebrates (Supplementary Figure 2A) and marked by both H3K27me3 and H3K4me1 (Figure 2A–C). This chromatin signature is typically present in developmental regulators that are inactive in ES cells but poised for activation during lineage specification (Creyghton et al, 2010; Rada-Iglesias et al, 2010; Zentner et al, 2011). The marked change in H3K27ac on the Eomes enhancer during differentiation is followed by a significant decrease in the H3K27me3 mark (Figure 2A, C) consistent with activation of this enhancer element (Rada-Iglesias et al, 2010). We next investigated whether the enzymes responsible for removal of H3K27 methylation were recruited to the Eomes enhancer during differentiation. ChIP analysis showed increased Jmjd3 binding at the Eomes enhancer in differentiated cells compared to ES cells (Figure 2E; Supplementary Figure 2B and C). Jmjd3 binding was specific to the Eomes enhancer and binding was not detected at the Eomes proximal-promoter region (Supplementary Figure 2C). These results indicated that increased occupancy of Jmjd3 corresponded with decrease in methylation of H3K27 at the Eomes enhancer.
Figure 2.
Tbx3 and Jmjd3 are physically associated and cooccupy the enhancer region of Eomes locus. (A) An active enhancer region enriched in H3K4me1 and H3K27ac is identified ∼7 kb upstream region of the TSS of Eomes (red box). Genome browser representations of H3K4me1 and H3K27ac enrichment profiles at the Eomes locus in ES cells and differentiated cells. (B–D) Enrichment of histone modifications at the enhancer region of Eomes is altered to reflect a poised to active state of enhancer. Enrichment of H3K4me1 was unchanged (B), H3K27me3 was decreased (C) H3K27ac was increased (D) at the Eomes enhancer in differentiated mES cells. ChIP experiments were performed in ES cells and D1–D2-differentiated cells using a primer pair within the enhancer region of Eomes (En), and the negative control region (Neg). (E) Jmjd3 promotes Eomes enhancer activation. ChIP analyses show increased Jmjd3 binding to the Eomes enhancer in D1–D2-differentiated cells. (F, G) The binding of Tbx3 and not Nanog to the Eomes enhancer region (red box) significantly increased in differentiated cells compared to ES cells. (F) Genome browser representations Nanog and Tbx3 enrichment at the Eomes locus in ES cells and differentiated cells analysed using ChIP-seq. (G) ChIP analyses show increased Tbx3 binding at the Eomes enhancer during D1–D2 differentiation. (H) Tbx3 and Jmjd3 are physically associated during early differentiation. D2-differentiated cells were coimmunoprecipitated with Tbx3 antibody and subsequently subjected to western blot with Jmjd3 antibody. Reverse and self-coimmunoprecipitations are also shown. DNAse1 treatment was added to test if the interaction is independent of DNA. IgG served as negative controls. (I, J) Similar to mouse cells, TBX3 and the activating JMJD3 bound to the EOMES enhancer in D1-differentiated hES cells. Enrichment of TBX3 and JMJD3 was measured using ChIP analyses. ChIP data are mean±s.e.m. (n=2–3).
Source data for this figure is available on the online supplementary information page.
There is growing recognition that genes that regulate ES cell pluripotency are also involved in germ layer lineage commitment (Loh and Lim, 2011; Thomson et al, 2011; Wang et al, 2012). We hypothesized that ES cell factors could bind to the Eomes enhancer elements and carried out epigenomic profiling of a number of ES cells factors (Figure 2F). Previous studies showed that another pluripotency factor Nanog was bound to the Eomes enhancer (Teo et al, 2011). Our analysis confirmed that Nanog was bound to the Eomes enhancer in ES cells. We also identified Tbx3, the predominant T-box family member expressed in ES cells, that bound to the Eomes enhancer during differentiation (Figure 2F and G; Supplementary Figure 2D and E). ChIP analysis confirmed that Tbx3 was bound to the Eomes enhancer region in the differentiated cells (Figure 2G). Our analysis also revealed that unlike Jmjd3 and Tbx3, Nanog was already bound to the Eomes enhancer in ES cells and binding of Nanog decreased during EB differentiation (Supplementary Figure 2F). The recruitment of Jmjd3 and Tbx3 during the EB differentiation suggested the possibility that these factors were physically associated at the Eomes enhancer region. Coimmunoprecipitation of EB using Tbx3 antibody subjected to western blotting with Jmjd3 antibody showed that Tbx3 co-precipitated with Jmjd3 (Figure 2H). Reverse coimmunoprecipitation with Jmjd3 antibody followed by western blotting with Tbx3 antibody confirmed the interaction between the two proteins (Figure 2H). Furthermore, the interaction remained when the cell lysate was treated with DNase1 (Figure 2H) that completely eliminated the presence of DNA (Supplementary Figure 2G). These results indicate that Tbx3–Jmjd3 interaction is independent of DNA. We next investigated whether the recruitment of TBX3 and JMJD3 to the EOMES enhancer also occurred during hES differentiation. Using ChIP analyses, the EOMES enhancer displayed increased occupancy of both TBX3 and JMJD3 early in differentiation (Figure 2I and J). These analyses suggested that Jmjd3 and Tbx3 were physically associated and co-occupied the Eomes enhancer during the early steps of differentiation.
Tbx3 and Jmjd3 co-occupy the enhancer region of Eomes Locus
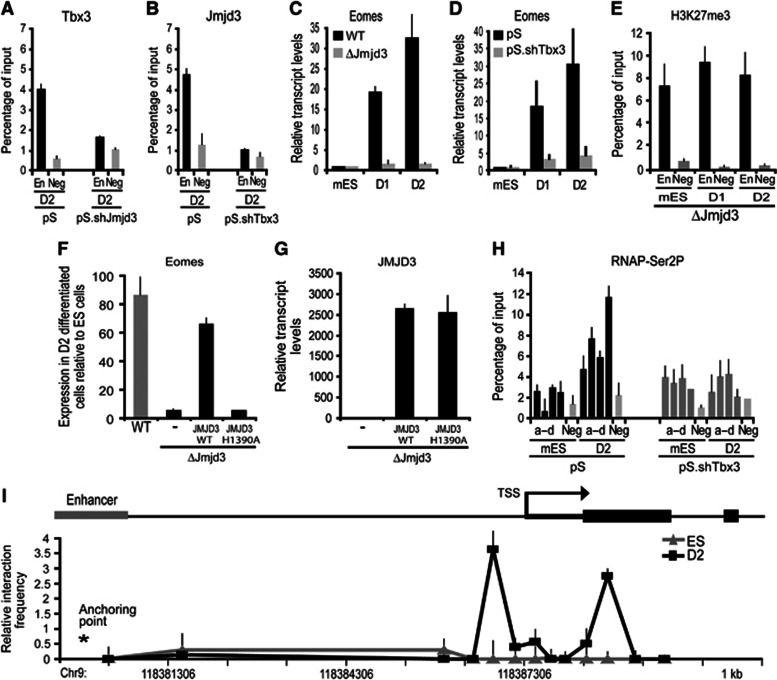
To further investigate whether Jmjd3 and Tbx3 were mutually required for Eomes activation, we employed short-hairpin RNA (shRNA)-mediated knockdown of Jmjd3 and Tbx3 (Supplementary Figure 3A and B). ChIP analysis on EBs formed from Jmjd3-knockdown ES cells showed that Tbx3 recruitment to the Eomes enhancer is prevented (Figure 3A). Conversely, Jmjd3 was not bound to the Eomes enhancer in EBs formed from Tbx3-knockdown ES cells (Figure 3B), reinforcing the idea that Tbx3 and Jmjd3 were associated. Unlike wild-type ES cells, Jmjd3-null ES cells failed to activate Eomes expression during differentiation (Figure 3C; Supplementary Figure 3C). Similarly, Tbx3 knockdown using either shRNA (Figure 3D) or SMARTPool siRNA (Supplementary Figure 3D and E) prevented Eomes activation during differentiation. These results suggest that both Jmjd3 and Tbx3 binding to the Eomes enhancer were required for Eomes transcriptional activation. We also noted that enrichment of H3K27me3 at the Eomes enhancer was unaffected when Jmjd3-null cells were used in the differentiation (Figure 3E). This indicated that the enzymatic activity of Jmjd3 was critical for the transcriptional activation of Eomes. To address this, we overexpressed either JMJD3-wild type or JMJD3-H1350A—a point mutation that renders deficiency in demethylase activity (Sen et al, 2008) in Jmjd3 null cells. Expression of the WT-Jmjd3 was sufficient to rescue transcriptional activation of Eomes. In contrast, expression of JMJD3-H1350A failed to activate Eomes transcription (Figure 3F and G). These experiments indicate that the demethylase activity of Jmjd3 was essential for transcriptional activation of Eomes during differentiation. We next investigated whether binding of the Jmjd3–Tbx3 complex to the Eomes enhancer affected the release of poised RNAP from the Eomes promoter. ChIP analysis revealed that enrichment of RNAP-Ser2P at the Eomes promoter was prevented in the absence of Tbx3 (Figure 3H). These results suggest that the Tbx3–Jmjd3 complex modifies the Eomes enhancer to promote long-range effects on the release of poised RNAP from the Eomes promoter.
Figure 3.
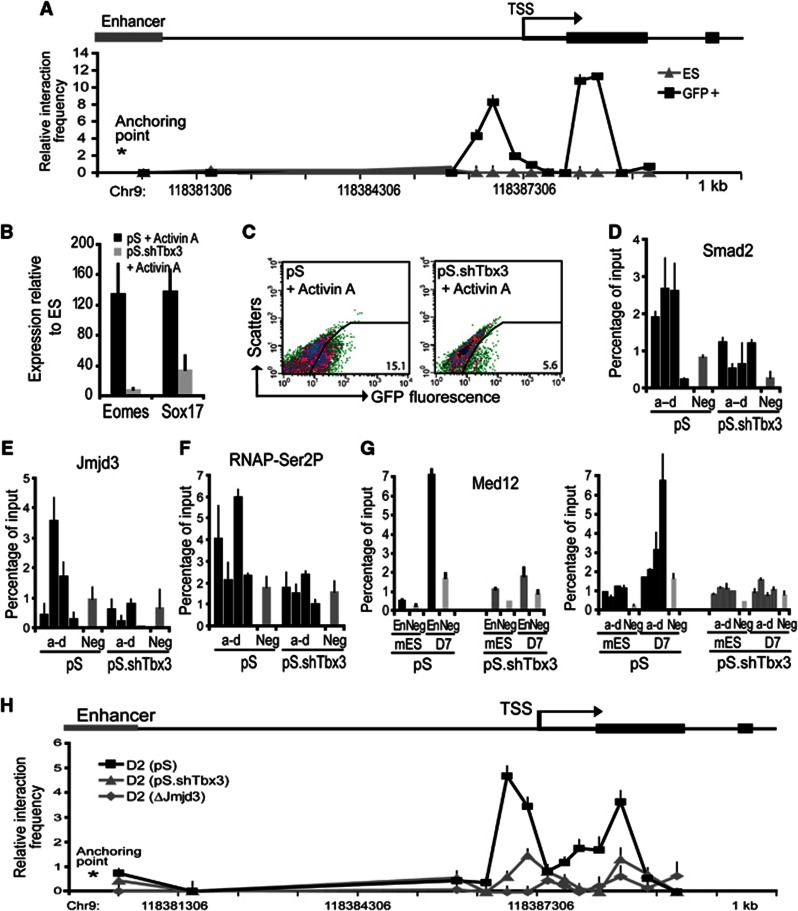
Eomes gene activation is dependent on Jmjd3 demethylase activity and involving enhancer–promoter DNA looping. (A, B) Tbx3 and Jmjd3 are mutually required for their binding to the Eomes enhancer. (A) ChIP analyses were carried out at the Eomes enhancer. Recruitment of Tbx3 was reduced in D2-differentiated Jmjd3-knockdown (pS.shJmjd3) compared to control (pS) cells. (B) Jmjd3 binding to the Eomes enhancer was decreased in D2-differentiated Tbx3-knockdown (pS.shTbx3) compared to the control (pS) cells. (C, D) Tbx3 and Jmjd3 are required for Eomes transcriptional activation. (C) Eomes expression was reduced in the Jmjd3-null (ΔJmjd3) compared to the wild-type (WT) cells during D1–D2 differentiation. (D) Similarly, Eomes expression measured by quantitative RT–PCR in the Tbx3 knockdown was repressed during differentiation. (E) H3K27me3 binding to the enhancer region of Eomes was performed using chromatin of undifferentiated Jmjd3-null mES cells and D1–D2-differentiated cells. Levels of H3K27me3 on the enhancer of Eomes in Jmjd3-null cells did not change following differentiation. (F, G) Transcriptional activation of Eomes is rescued in D2-differentiated Jmjd3-null cells (ΔJmjd3) by overexpressing JMJD3-WT (JMJD3 wild-type protein). No rescue was observed using overexpression of JMJD3 with a demethylase-inactive point mutation H1390A. (H) Phosphorylation of RNAP at Serine2 is dependent on Tbx3 binding at the enhancer. ChIP analyses was performed at the Eomes proximal promoter (a–d) in the Tbx3-knockdown cells during D1–D2 differentiation. No enrichment of RNAP-Ser2P was observed. (I) Long-range enhancer–promoter interaction accompanies Eomes transcriptional activation. Quantitative 3C analyses were performed on cross-linked chromatin extracted from ES cells and D2-differentiated cells. Chromatin was digested with Msp1, relegated and subjected to qPCR using an anchor primer at the enhancer region and selected primers facing the Msp1 digest sites. Taqman probe for detection of the amplicons was located close to the anchor primer. Association frequency between the anchor and selected primers is depicted on y axis. The chromosomal coordinate (mm9) of the 5′ Eomes locus is shown on x axis. All values are mean±s.e.m. (n=2–3).
We thus hypothesized that the chromatin at the Eomes locus underwent spatial reorganization during EB formation to allow the enhancer region to engage in a direct physical interaction with the proximal-promoter region. This promoter–enhancer interaction could in turn facilitate release of poised RNAP and transcriptional activation of Eomes. We investigated whether higher order packaging of the chromatin at the Eomes locus changed during EB formation using the Chromosome Conformation Capture (3C) assay. This assay determines formation of chromatin loops between regulatory elements (Dekker et al, 2002). Intact nuclei from ES cells and EBs were subjected to paraformaldehyde cross-linking to fix segments of genomic DNA that are in close physical proximity to each other. Cross-linked genomic DNA was digested with MspI followed by ligation. We designed a series of primers positioned at all unique MspI restriction digestion sites across the Eomes locus. Using an anchor primer located in the Eomes enhancer, we performed qPCR assays to determine the proximity of different regions of the locus to the Eomes enhancer. The presence of a given PCR product is an indication of the relative proximity of the two restriction sites to one another captured at any given time. Comparison of cross-linking frequencies between ES cells and EBs showed that the Eomes locus underwent a dramatic three-dimensional change during EB formation (Figure 3I). The primers near the restriction sites located at the promoter showed increased cross-linking frequencies in EBs compared to ES cells indicating enhancer–promoter interactions (Figure 3I). The 3C results are consistent with a model in which Tbx3–Jmjd3 binding during EB formation promotes Eomes gene activation through enhancer–promoter DNA looping.
Activin A signalling potentiates Eomes expression and differentiation to definitive endoderm
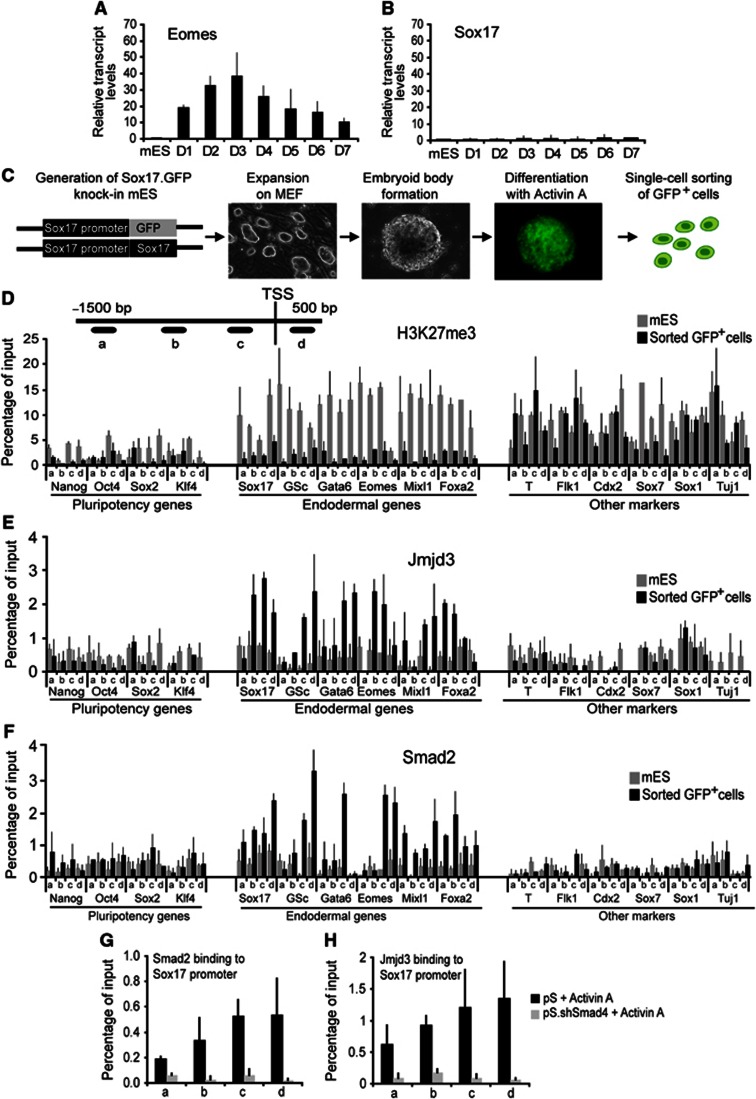
The expression of Eomes was not sustained during EB differentiation and core definitive endodermal regulators such as Sox17 were not induced without supplementation of Activin A in the culture medium (Figures 1B, 4A), suggesting that the low levels of Eomes induced during EB differentiation did not correlate with formation of definitive endoderm. To address the molecular mechanism that regulates definitive endoderm formation, we used a mouse ES cell line containing a loxed cassette acceptor allele to insert a Cre-GFP (Green fluorescent protein) fusion protein to replace the Sox17 coding sequences by recombinase-mediated cassette exchange (RMCE) (Long et al, 2004). We verified that GFP expression accurately reflected endogenous Sox17 expression by comparing the expression of GFP in embryos from Sox17.Cre-GFP mice with localization of the Sox17 endogenous protein. This ES cell line enabled us to directly visualize Sox17 expression in the differentiation protocol and to isolate Sox17-expressing cells.
Figure 4.
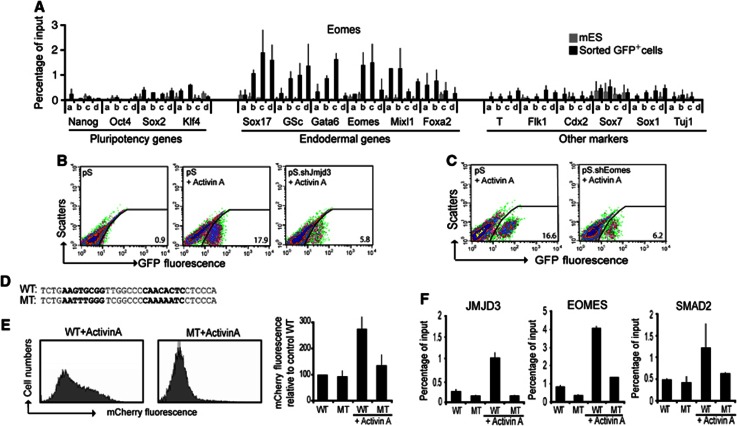
Jmjd3 and Smad2 bind and resolve bivalent domain within the promoters of core definitive endoderm regulators. (A, B) Activin A is required for maintenance of Eomes expression and induction of Sox17 expression. Quantitative RT–PCR shows expression of Eomes (A) and Sox17 (B) in ES cells and differentiated embryoid bodies cultured without Activin A for 7 days. (C) Schematic of a step-wise definitive endoderm differentiation protocol using Sox17.GFP knock-in ES cells for isolation of Sox17.GFP+ cells. (D–F) Removal of H3K27me3 repressive mark from the promoters of core definitive endoderm regulators in Sox17.GFP+ cells (D) coincides with Jmjd3 (E) and Smad2 (F) recruitment to these promoters. ChIP experiments were performed on ES and Sox17.GFP+ cells with the indicated antibodies. Four pairs of primers at the proximal-promoter regions (a–d) were used on select promoters from three groups; core pluripotency regulators, core definitive endoderm, and key transcriptional regulators of other germ layers. (G, H) Jmjd3 and Smad2 are mutually required for their recruitment to the Sox17 promoter. ChIP analyses of Smad2 (G) and Jmjd3 (H) at the Sox17 promoter regions (a–d) show reduced binding in the Activin A-differentiated Smad4-knockdown (pS.shSmad4) cells compared to the control (pS). All values are mean±s.e.m. (n=2–4).
The Sox17.Cre-GFP ES cells had no detectable levels of GFP expression. Following the two-step differentiation protocol, intense GFP expression was observed (Figure 4C) and GFP+ cells were isolated by flow cytometry and subjected to a microarray expression analysis. The levels of pluripotent transcriptional regulators such as Nanog, Oct4, Sox2, and Klf4 were downregulated in the isolated GFP+ cells compared to ES cells (Supplementary Figure 4A). The core regulators of definitive endoderm such as Foxa2, Sox17, Mix1, and Gata6 were highly expressed in the GFP+ cells (Supplementary Figure 4A). Transcriptional regulators that are selectively activated in other lineages, such as Sox1 for neuroectoderm, Sox7 for extraembryonic endoderm, Brachyury for mesoderm, and Cdx2 for trophectoderm were not upregulated (Supplementary Figure 4A). This suggests that directed differentiation preferentially to the definitive endoderm germ layer is accomplished with our protocol.
Jmjd3 and Smad2 resolve bivalent domain within promoters of core definitive endoderm regulators
We next investigated whether selective activation of core definitive endodermal regulators was accompanied by changes in the histone modifications. ChIP analysis of bivalent histone modifications revealed that the promoters of core definitive endoderm regulators were depleted of H3K27me3 in the isolated GFP+ cells (Figure 4D). In contrast, promoters of lineages markers from other germ layers were enriched for this repressive mark (Figure 4D). Notably, unlike the EB stage of differentiation, the Eomes proximal-promoter region was depleted of H3K27me3 at the definitive endoderm stage (Figure 4D). The depletion of H3K27me3 was accompanied by binding of Jmjd3 at the promoters of definitive endodermal markers (Figure 4E). H3K4me3, on the other hand, was maintained at the promoters of definitive endoderm markers but depleted from promoters of the pluripotency and other lineage markers (Supplementary Figure 4B). We also observed that the histone methyltransferase Set7/9 was bound to the promoters of definitive endoderm markers (Supplementary Figure 4C). These results indicate that enzymes that can resolve bivalent domains occupy promoters of core definitive endoderm regulators during differentiation.
As Activin A signalling was required to activate core endodermal regulators and potentiate Eomes expression, we assessed whether Smad2, the intracellular mediator of Activin A signalling was recruited to the promoters of definitive endoderm markers. Smad2 ChIP analysis showed binding of Smad2 to the promoters of core definitive endoderm regulators in the GFP+ cells (Figure 4F). However, Smad2 did not occupy the promoters of pluripotency regulators and regulators of other lineages (Figure 4F). Knockdown of Smad4 an essential partner of Smad2 in the nucleus resulted in diminished binding of Smad2 to the Sox17 promoter (Figure 4G). Interestingly, Jmjd3 binding to the Sox17 promoter was also diminished suggesting co-dependency of Smad2 and Jmjd3 binding to the Sox17 promoter (Figure 4H). These analyses indicate that Jmjd3 and Smad2 are functionally associated at the promoters of core definitive endoderm regulators.
Eomes participates in a positive feedback loop to potentiate its own expression and target the promoters of core definitive endoderm regulators
To elucidate how the Jmjd3 and Smad2 are targeted to the promoters of core definitive endoderm regulators during differentiation, we sought to identify a transcription factor present in EBs that could interact with Jmjd3. We reasoned that Eomes, the definitive endoderm regulator that has been expressed in EBs directs Jmjd3 and Smad2 to definitive endoderm promoters. ChIP analysis indeed showed that Eomes selectively bound to its own promoter and the promoters of other definitive endoderm regulators (Figure 5A). The co-occupancy of Eomes, Jmjd3, and Smad2 at the promoters of definitive endoderm regulators indicates that these factors function together to control the expression of core definitive endoderm regulators. Utilizing sequential ChIP analyses as well as ChIP analyses on the differentiated Jmjd3, Eomes, and Smad4 knockdown cells, we confirm the co-occupancy of Eomes, Jmjd3, and Smad2 at the Eomes promoter (Supplementary Figure 5A–E). We would also expect knockdown of these factors would have inhibiting effects on definitive endoderm differentiation. Indeed, knockdown of Jmjd3, Eomes, and Smad4 using shRNAs resulted in reduced expression of all definitive endoderm markers and moreover flow cytometry revealed that the proportion of GFP+ cells was markedly diminished (Figure 5B and C; Supplementary Figure 5F and G). Similarly, knockdown of Smad4 using SMARTpool siRNA inhibited Eomes gene activation (Supplementary Figure 5H).
Figure 5.
Eomes recruits Jmjd3 and Smad2 to the promoter regions of core definitive endoderm genes. (A) Eomes selectively occupies the promoters of definitive endoderm genes in sorted Sox17.GFP+ cells. ChIP experiments were using four pairs of primers at the proximal-promoter regions (a–d) of select promoters from three groups; core pluripotency regulators, core definitive endoderm, and key transcriptional regulators of other germ layers. (B, C) Definitive endoderm specification by Activin A requires both Eomes and Jmjd3. (B) The number of GFP+ cells formed was analysed by flow cytometry in control (pS) cells differentiated without Activin A (left panel), with Activin A (middle panel), and in Jmjd3-knockdown (pS.shJmjd3) cells differentiated with Activin A (right panel). (C) Flow cytometry was performed in control (pS) cells (left panel) and Eomes-knockdown cells (pS.shEomes), both differentiated with Activin A (right panel). (D–F) Eomes-Smad2-Jmjd3 binding requires the T-box binding motif on the promoter of Sox17. (D) Sequences of the wild-type and the mutated Eomes binding motif on Sox17 promoter, containing two tandem core motifs (in bold). (E) mCherry fluorescence in Hek293T transfected with the Sox17 promoter mcherry reporter constructs containing the wild-type Eomes binding sequence motif (left panel) was higher than the mutant one (middle panel) following stimulation with Activin A. The graph shows mean±s.e.m. of the relative mcherry fluorescence from two independent experiments. (F) ChIP analyses of JMJD3, EOMES, and SMAD2 show increased binding to the wild-type but not to the mutated Sox17 promoter constructs in Hek293T following Activin A treatment. Data values are mean±s.e.m. (n=2–3).
To further investigate how Eomes can specifically target the promoters of definitive endoderm regulators, we developed an exhaustive motif search algorithm by combining T-box binding motif specificity (Conlon et al, 2001) with our ChIP data on Eomes binding. We identified putative T-box binding sites that consisted of two tandem core motifs separated by four to eight nucleotides within the promoter regions of many definitive endodermal regulators (Supplementary Figure 5I and J). We then generated reporter constructs by cloning the 750-bp promoter region of Sox17 into a promoterless vector containing the fluorescence reporter gene mCherry. We also generated a mutant Sox17 promoter reporter construct by introducing nucleotide substitutions to the putative Eomes binding site (Figure 5D). Transfection of the Sox17 promoter construct into human embryonic kidney cell line 293T (HEK293T) did not yield appreciable levels of fluorescence. However, when these transfected cells were treated with Activin A, robust mCherry fluorescence was observed (Figure 5E). In contrast, Activin A treatment of HEK293T cells transfected with the mutant Sox17 reporter construct did not yield appreciable levels of fluorescence (Figure 5E). ChIP analysis also showed binding of Eomes, Smad2, and Jmjd3 to the wild-type Sox17 promoter region following Activin A treatment. However, no binding of Eomes, Smad2, and Jmjd3 was observed in Activin A-treated HEK293T cells transfected with the mutant Sox17 reporter construct (Figure 5F). Taken together, these results confirm that in response to Activin A signalling, Eomes directs Smad2 and Jmjd3 to promoters of definitive endoderm regulators to potentiate its own expression as well as activate expression of other definitive endoderm regulators.
Sequential two-step activation of Eomes locus leads to definitive endoderm differentiation
Our results suggest that formation of definitive endoderm occurs in two steps: (1) enhancer–promoter DNA loop formation leading to induction of Eomes and (2) Activin A-mediated potentiation of Eomes expression and transcriptional activation of definitive endodermal regulators. We hypothesized that transcriptional activation of Eomes involving loop formation rendered cells competent to respond to Activin A signalling and form definitive endoderm. We first examined whether the enhancer–promoter DNA loop of the Eomes locus was maintained in definitive endoderm. We carried out 3C analysis on the Eomes locus on ES cells and GFP+ sorted cells using the two-step differentiation protocol. We observed an increased interaction frequency between the promoter proximal region and the enhancer of the Eomes locus in the GFP+ cells compared to ES cells (Figure 6A). Interaction frequency profile of the Eomes locus in definitive endoderm and EB cells was similar (Figures 3F and 6A) suggesting that the enhancer–promoter loop of the Eomes locus formed during EB differentiation is maintained in definitive endoderm. To investigate whether induction of Eomes primes cells to respond to Activin A and form definitive endoderm, we knocked down Tbx3, which is required for initial induction of Eomes but not involved in the Activin A-mediated potentiation of Eomes expression and transcriptional activation of definitive endodermal regulators (Supplementary Figure 6A). Gene expression analysis of Tbx3-knockdown ES cells subjected to the endoderm differentiation protocol revealed that Eomes and Sox17 expression was severely diminished (Figure 6B). Correspondingly, flow cytometry analysis showed marked reduction in GFP+ cells (Figure 6C). Moreover, in Tbx3 knockdown cells, ChIP analysis showed that Activin A treatment did not promote the binding of Smad2 and Jmjd3 to the Eomes promoter or resulted in the release of poised RNAP (Figure 6D–F; Supplementary Figure 6B). We further analysed binding of the Mediator complex, a transcriptional coactivator that has been shown to bridge enhancer-bound transcription factors and promoter-bound RNAP (Heintzman et al, 2009; Kagey et al, 2010). ChIP analyses of Med12, a subunit of the Mediator complex, revealed that Mediator was not bound to the Eomes locus in ES cells but bound to both the enhancer and promoter proximal region in definitive endoderm-differentiated cells (Figure 6G). The binding of Med12 at both enhancer and promoter proximal region is consistent with DNA loop formation of the Eomes locus. In contrast, Med12 binding to the Eomes locus is not observed with knockdown of Tbx3 (Figure 6G). Using 3C analyses, we confirmed that the enhancer–promoter looping formation was blocked in both Tbx3-knockdown and Jmjd3-null cells during endoderm differentiation (Figure 6H; Supplementary Figure 6C). These results are consistent with a model that the initial DNA loop formation leads to the induction of Eomes and is required for Activin A-mediated definitive endoderm differentiation.
Figure 6.
Tbx3-mediated induction of Eomes is required for definitive endoderm differentiation. (A) Interaction of the Eomes enhancer with its promoter is maintained in definitive endoderm. Quantitative 3C analyses of ES and the sorted Sox17.GFP+ cells were performed using an anchor primer at the enhancer region and selected primers facing the Msp1 digest sites. Association frequency between the anchor and selected primers is depicted on y axis. The chromosomal coordinate (mm9) of the 5′ Eomes locus is shown on x axis. (B, C) Loss of Tbx3 prevents definitive endoderm differentiation. (B) Expression of Eomes and Sox17 and (C) flow cytometry analyses of GFP+ cells from the Activin A-differentiated Tbx3-knockdown and control cells. (D, E) ChIP analyses on Tbx3-knockdown cells show that Tbx3 is required for Jmjd3 (D) and Smad2 (E) binding to the Eomes proximal-promoter region following Activin A-induced differentiation. (F) ChIP analyses on Tbx3-knockdown cells show that Tbx3 is required for the enrichment of RNAP-Ser2P elongation complex on the Eomes proximal-promoter region following Activin A-induced differentiation. (G) ChIP analyses on Tbx3-knockdown cells show no binding of Med12 to both promoter and enhancer regions of Eomes. All ChIP experiments were performed using cross-linked chromatin with the indicated antibodies and primers at the Eomes proximal-promoter regions (a–d), the Eomes enhancer region (En) and the negative control region (Neg), as indicated. (H) Tbx3 and Jmjd3 are required for initial chromosomal interaction between enhancer and promoter of the Eomes locus. The enhancer–promoter interactions in the Eomes locus were monitored using the quantitative 3C analysis. Cross-linked chromatin was extracted from the cells. Association frequency between the anchor point and the regions of the selected primers is depicted on y axis. The chromosomal coordinate (mm9) of the 5′ Eomes locus is shown on x axis. DNA looping between enhancer and promoter of Eomes was observed in the differentiated wild-type cells (pS), but diminished in Tbx3-knockdown (pS.shTbx3) and Jmjd3-null cells (ΔJmjd3). Values are mean±s.e.m. (n=2–3).
Mechanisms for endoderm differentiation are conserved in mouse and human ES cells
We used previously established protocols to induce endoderm differentiation in hES cell line, HSF1, and hiPS cell line, hiPS2 (D'Amour et al, 2005; Borowiak et al, 2009; Patterson et al, 2011). After 5 days of Activin A treatment, ∼ 95% of cells in culture expressed SOX17 many of which coexpressed FOXA2 protein (Supplementary Figure 7A). Similar to mouse ES cells, gene expression analysis showed upregulation of definitive endoderm markers such as GSC, MIXL1, GATA6, and FOXA2, while transcriptional regulators that are selectively activated in other lineages, such as SOX1 for neuroectoderm, SOX7 for extraembryonic endoderm, BRACHYURY for mesoderm and CDX2 for trophectoderm were not changed (Supplementary Figure 7B and C).
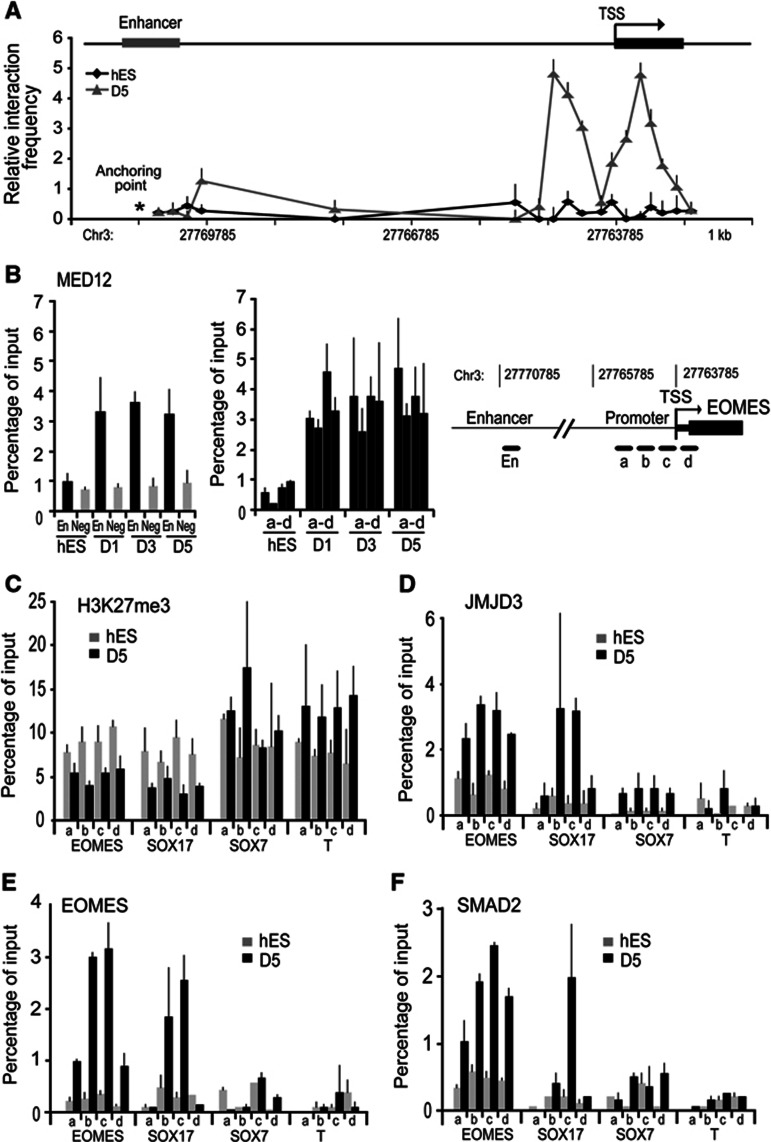
The temporal sequence of transcriptional activation of endodermal genes was similar in mouse and human ES cell differentiation. Notably, induction of EOMES occurred early in differentiation and preceded SOX17 expression (Figure 1B). The enhancer region of EOMES displayed increased occupancy of JMJD3 and TBX3 early in differentiation that was accompanied by reduction in H3K27me3 levels within the enhancer region (Figure 2I and J). The 3C analysis shows that the enhancer–promoter DNA looping accompanies accompanied by EOMES gene activation during hES cells differentiation to endoderm (Figure 7A). Analysis of Mediator binding showed that MED12 subunit was not bound to the EOMES locus in hES cells but bound to both the enhancer and promoter proximal region in differentiated endoderm cells, consistent with DNA loop formation (Figure 7B). We also observed decreased levels of H3K27me3 and increased recruitment of JMJD3, EOMES, and SMAD2 at the promoter region of EOMES later in differentiation (Figure 7C–F). These results are consistent with a sequential two-step activation of EOMES locus during hES cell differentiation. In differentiated endoderm, the SOX17 promoter was occupied by SMAD2, JMJD3, and EOMES while promoters of other lineage regulators, SOX7 and BRACHYURY did not display any binding of these factors (Figure 7C–F; Supplementary Figure 7D). We also observed that SMAD2, JMJD3, and EOMES occupied the SOX17 promoter when hiPS was used to generate definitive endoderm (Supplementary Figure 7E and F). These results indicate that the epigenetic mechanisms during definitive endoderm differentiation are conserved in mouse and human ES cells.
Figure 7.
The sequential two-step activation of EOMES locus is conserved between human and mouse. (A) The quantitative 3C analyses on Eomes locus was carried out in hES cells and definitive endoderm derived from hES cells (D5). Similar to mES cells, enhancer–promoter looping is absent in undifferentiated hES cells but accompanies EOMES gene activation in differentiated cells. (B) Using ChIP analyses, MED12 bound to both enhancer (En) and promoter (a–d) of the EOMES locus but not to the negative control region (Neg), confirming looping formation in Activin A-differentiated hES cells. (C–F) EOMES activates selectively definitive endoderm loci in Activin A-differentiated hES. ChIP analyses on definitive endoderm derived from hES cells (D5) that H3K27me3 (C) is reduced and JMJD3 (D), EOMES (E), and SMAD2 (F) are enriched at the promoter regions (a–d) of EOMES and SOX17, but not SOX7 and BRA. Values are mean±s.e.m. (n=3–4).
Discussion
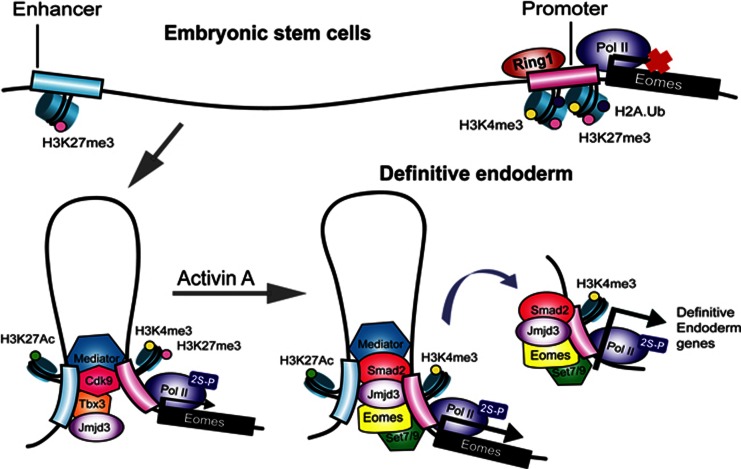
Embryonic stem cells differentiate into a number of cell types, which makes them attractive for cell replacement strategies. How chromatin dynamics regulate cell fate decisions is often difficult to study in the embryo and ES cells are a good model system to mimic early differentiation in the embryo. In this study, we elucidate the molecular mechanisms that progressively restrict this differentiation potential and direct stem cells to differentiate towards definitive endoderm lineage (Figure 8). Our results lead us to suggest a factor relay model whereby an ES cell factor, Tbx3 acts on the enhancer elements of Eomes to bring it in close proximity to the transcription apparatus at the core promoter. This DNA loop formation of the Eomes locus establishes a permissive chromatin states that restricts differentiation potential as well as primes the cells to respond to Activin A differentiation signal. Activin A signalling allows Eomes to act on bivalent domains within the Eomes promoter to transactivate its own expression in a positive feedback loop. The positive feedback loop contributes to the formation of a stable switch to maintain definitive endoderm fate. Eomes also cooperates with Smad2 to act on bivalent domains within the promoters of core endodermal regulators to activate a transcriptional network leading to definitive endoderm specification. While we have focused on definitive endoderm specification in this study, other germ layer specification could utilize the factor relay model. For instance, the Brachyury locus could be regulated in a similar fashion as Eomes during mesodermal specification. Analysis of the factor relay model in mesoderm specification could shed light on whether mesoderm and endoderm share a permissive chromatin state or whether segregation of these germ layers is driven by ES cell factors at the earliest steps of differentiation.
Figure 8.
A model of the two-step activation of Eomes locus during specification of definitive endoderm from ES cells, conserved between human and mouse. Eomes is poised in embryonic stem cells, marked by H3K4me3 and H3K27me3. During initial differentiation, Tbx3 associates with histone demethylase Jmjd3 and activates the Eomes enhancer, promoting enhancer–promoter interaction. This spatial reorganization of the chromatin primes the cells to respond to Activin A signalling, by allowing Eomes to promote Jmjd3-mediated transcriptional activation of the Eomes itself as well as the core regulators of endoderm.
We show that the sequence-specific transcription factors and epigenetic modifications of the chromatin interact in order to implement control pathways that drive differentiation of stem cells towards the definitive endoderm lineage. Both Tbx3 and Eomes are T-box transcription factors that act in a spatial and temporally distinct manner during differentiation by recruiting the histone demethylase, Jmjd3 to influence the dynamics of chromatin structure. Recruitment of Jmjd3 by Tbx3 to the enhancer element of Eomes locus promotes enhancer–promoter interaction by DNA looping resulting in the release of the PRC1 complex from the proximal-promoter region. The release of PRC1 complex was not accompanied by changes in PRC2 binding and H3K27me3 enrichment in the proximal-promoter region. Genome-wide analysis indicates that binding of PRC1 and methylated H3K27 may not always overlap (Schoeftner et al, 2006; Schwartz et al, 2006; Vincenz and Kerppola, 2008); however, DNA looping may link H3K27 methylation in the enhancer region with PRC1 binding in the proximal-promoter region of the Eomes locus. Release of PRC1 and loss of H2Aub have been suggested to remove a physical barrier in the nucleosome that prevents FACT recruitment and release the proximal-promoter pausing of RNA elongation (Zhou et al, 2008). Our results suggest that the release of the proximal-promoter pausing of RNA elongation may be sufficient for low levels of Eomes transcription but stable levels of Eomes require the feedback loop involving Eomes itself and the recruitment of Jmjd3 to its proximal-promoter region. Jmjd3 is recruited to the proximal-promoter region of Eomes to resolve the bivalent domain although the exact role of bivalent domains in regulating DNA–histone contacts and movement of RNA polymerase in gene transcription is not clear. Our results highlight how a few crucial factors can link changes in chromatin states to differentiation providing a simplification of the complex transcriptional circuits of lineage specification.
Jmjd3 is induced in response to different stimuli and subsequently contributes to the transcriptional activation of target genes. For example, Jmjd3 is induced by the transcription factor NF-κB in response to microbial stimuli to the control of gene expression in LPS-activated macrophages (De Santa et al, 2009). Jmjd3 is essential in M2 macrophage development for anti-helmith responses (Satoh et al, 2010). Jmjd3 is also induced upon activation of the RAS–RAF signalling pathway to activate the INK4A–ARF locus in response to oncogene- and stress-induced senescence (Agger et al, 2009). Jmjd3 is also induced by retinoic acid signalling in neural stem cells differentiation and interestingly, SMRT (also known as NCoR2, nuclear receptor co-repressor-2) is critical to repress Jmjd3 expression in the absence of retinoic acid stimulus (Jepsen et al, 2007). We show that Jmjd3 is induced during early differentiation to endoderm and whether SMRT is involved in repression of Jmjd3 in ES cells could be informative. Jmjd3 has also been shown to have a role in neural differentiation and immune cell development (Jepsen et al, 2007; Lan et al, 2007; Burgold et al, 2008; Dahle et al, 2010; Dai et al, 2010; Miller et al, 2010; Canovas et al 2012; Chen et al, 2012). Mice with partial deletion of Jmjd3 died perinataly due to premature defect in lung development (Satoh et al, 2010); however, the role of other H3K27 demethylase, Utx and Uty in compensating for Jmjd3 in endoderm formation was not evaluated in this study (Kawaguchi et al, 2012).
Analysis of histone demethylases in ES cells link the core transcription factor network to the regulation of chromatin status in pluripotent stem cell maintenance. Several ES cell core regulators target Jarid2 to maintain high levels of its expression in ES cells (Zhou et al, 2007). Oct4 targets other histone demethylases, Jmjd1a and Jmjd2c, to positively regulate their expression (Loh et al, 2007). Jmjd1a demethylates K9H3me2 at the promoter regions of Tcl1, Tcfcp2l1, and Zfp57 to positively regulate the expression of these pluripotency-associated genes while Jmjd2c acts as a positive regulator for Nanog. Interestingly, Jmjd1a or Jmjd2c depletion leads to ES cell differentiation, which is accompanied by a reduction in the expression of ES cell-specific genes and an induction of lineage marker genes (Loh et al, 2007). These studies suggest that repression of Jmjd3 in ES cells could be integrated in the transcriptional factor regulatory network of ES cells and links core transcription factor network to the regulation of chromatin status during cellular differentiation. Overall, our study provides a general framework for how ES factors regulate chromatin changes that provide a context for signal interpretation to drive differentiation of pluripotent stem cells into specific tissue lineages. Establishing the detailed principles on which cell lineages are defined and maintained is required for developing safe and successful cell therapies for a full potential of stem cell therapy.
Materials and methods
Generation of Sox17.Cre-GFP knock-in ES cells
The Sox17.Cre-GFP knock-in mouse ES cells were generated using a two step procedure. First, ES cells containing a loxed cassette acceptor (LCA) allele were made by standard homologous recombination. Second, RMCE was then applied using the vectors described (Chen et al, 2011) and a staggered positive-negative selection strategy as previously described in Long et al (2004). The net result was the replacement of exons 4 and 5 of the Sox17 gene with a fusion protein consisting of Cre-recombinase linked to GFP. A more complete description of the vectors used and validation of the Sox17.Cre-GFP allele are in preparation for publication elsewhere.
Mouse and human ES cell culture, differentiation, and RNA interference
All cells were cultured at 37°C with 5% CO2. Primary mouse embryonic fibroblasts (MEFs) were derived from E13.5 CF1 mouse embryos and mitotically inactivated using γ-irradiation (UCLA). Sox17.GFP knock-in ES cells and Jmjd3-null ES cells (KO-2211 from Knock-out Mouse Project Repository (KOMP); this cell line carries deleted Kdm6b allele: Kdm6btm1(KOMP)Wtsi) were maintained on irradiated MEFs in mouse ES medium containing Knock-out Dulbecco’s modified Eagle’s medium (KO-DMEM; GIBCO), supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone), 0.055 mM β-mercaptoethanol (GIBCO), 2 mM L-glutamine (GIBCO), 0.1 mM non-essential amino acid (GIBCO), 5000 U/ml penicillin-streptomycin (GIBCO), 15 mM HEPES (GIBCO) and 1000 U/ml LIF (Millipore/Chemicon). Jmjd3-null ES cells were selected for homozygous colonies in aforementioned medium using a highly concentrated G418 antibiotic (2.5 mg/ml) as previously described in Mortensen et al (1992). RNA interference was done as previously described (Loh et al, 2007) using shRNA-expressing pSuperpuro (Oligoengine) constructs with puromycin (Sigma) selection at 1 μg/ml or siRNA SMARTpool (Dharmacon). All transfections were performed using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. shRNA sequences are provided in Supplementary Table 1. For differentiation, mES cells were initially cultured in non-adherent conditions at a density of 1 × 104 cells/ml for 2 days in supplemented KO-DMEM except in the absence of LIF. The formed EBs were then differentiated into definitive endoderm using 5-day treatment of 20 ng/ml Activin A (R&D) in 1:1 KO-DMEM/Neurobasal medium (GIBCO) supplemented with N2 (GIBCO), B27 (GIBCO), 0.055 mM β-mercaptoethanol, 2 mM L-glutamine, 0.1 mM non-essential amino acid, 5000 U/ml penicillin-streptomycin, 15 mM HEPES, and 20 ng/ml EGF (R&D) modified from (Kubo et al (2004) and Morrison et al (2008). The formation of GFP+ cells was monitored using Leica DMLRE2 microscope. Human ES cells (HSF1) and human induced pluripotency stem cells (hiPS2) were maintained in human ES medium containing DMEM/F12 (GIBCO) supplemented with 2 mM L-glutamine, 0.1 mM non-essential amino acids, 5000 U/ml penicillin-streptomycin, 15 mM HEPES, 20% knockout serum replacement (Invitrogen), and 10 ng/ml basic FGF (R&D Systems) as previously described (Lowry and Plath, 2008). Hek293T cells were maintained in DMEM (GIBCO) supplemented with 10% heat-inactivated FBS (GIBCO), 2 mM L-glutamine and 5000 U/ml penicillin/streptomycin. HSF1 and hiPS2 were differentiated into endoderm using 100 ng/ml Activin A for 5 days in DMEM/F12 with 2 mM L-glutamine, 15 mM HEPES, 5000 U/ml penicillin-streptomycin and supplemented with FBS (0% from Day 0 to 1, 1% from Day 1 to 2, 2% from Day 2 to 5) (D'Amour et al, 2005; Borowiak et al, 2009; Patterson et al, 2011).
ChIP-qPCR and sequential ChIPs
ChIP was performed according to Dahl and Collas (2008) with minor modifications. Briefly, single cell suspension of 1 × 105 cells was cross-linked with 1% formaldehyde and incubated for 15 min at room temperature. Formaldehyde was deactivated by adding glycine to a final concentration of 125 mM. Cells were lysed using 100 μl lysis buffer consisted of 50 mM Tris–HCl pH 8.0, 10 mM EDTA, 1% SDS, supplemented with 1 × complete proteinase inhibitors (Calbiochem) and sonicated to yield DNA fragments with an average size of 500 bp using Bioruptor (Diagenode). A total of 1–2 μg of antibody was bound to 20 μl Protein-A/G Dynabeads (Invitrogen), depending on the antibody isotype, for 2 h at 4°C. One fourth fraction of the sonicated chromatin was incubated overnight at 4°C with the antibody-bead complexes in total volume of 200 μl in RIPA buffer containing 10 mM Tris–HCl pH 8.0, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS and 0.1% Na-deoxycholate supplemented with 1 × complete proteinase inhibitors. After 4 × washing with RIPA buffer and 1 × with TE buffer, chromatin was eluted, followed by reverse cross-linking at 68°C for 4 h with vigorous agitation in the presence of Proteinase K (Sigma). The DNA fragments were then purified using phenol-chloroform extraction and ethanol precipitation. For sequential ChIP experiments, washed protein–DNA complex from the first immunoprecipitation step was eluted for 30 min at 37°C using 75 μl TE buffer with 2% SDS, 15 mM DTT and 1 × complete proteinase inhibitors. This eluate was then diluted 20 times with RIPA buffer and subjected to the second ChIP assay. SYBR green-based qPCR was performed using ABI7900HT using 10–20 ng DNA per reaction. ChIP-qPCR signals were calculated as percentage of input. Data are representative of two to three independent experiments, and error bars indicate standard error of the replicates. The primer sequences and primary and secondary antibodies used are in Supplementary Tables 2 and 3, respectively.
Coimmunoprecipitation and western blotting
For each coimmunoprecipitation, 1 × 107 cells were lysed using 300 μl non-denaturing lysis buffer consisted of 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, supplemented with 1 × complete proteinase inhibitors for 20 min at 4°C. Coimmunoprecipitation was performed by incubation of the protein extract with 3–5 μg primary antibody at 4°C overnight, followed by incubation with 60 μl Protein-A/G Dynabeads for 5 h at 4°C. Each coimmunoprecipitation was washed three times, each for 5 min, with the lysis buffer. In some experiments, the cell lysate was treated with 100 U/ml DNAse1 (Roche) for 30 min before coimmunoprecipitation. Complete elimination of DNA in the treated lysate is monitored using Qubit HS DNA assay (Invitrogen) and tested using PCR of the Eomes enhancer region (850 bp) after DNA extraction (Qiagen). The captured proteins were reduced in sample buffer (Invitrogen) and denatured by boiling for 10 min. Proteins were run on Nupage gels (Invitrogen). Gels were blotted onto PVDF membrane (Amersham), then stained with primary antibody as indicated, followed by secondary conjugated to horseradish peroxidase (HRP). The antibodies used are listed in Supplementary Table 3. Membranes were developed using ECL western blotting detection system (Millipore) according to manufacturer's instructions.
ChIP-seq
ChIP-seq libraries were generated from the ChIP and input samples, using the Illumina ChIP-Seq DNA Sample Prep Kit according to manufacturer’s instructions. These libraries were then subjected to sequencing using Illumina HiSeq2000. Base-calling and QC statistics were generated using Illumina software. All unique sequences were mapped by Bowtie (Langmead et al, 2009) to the mouse mm9 genome. The number of unique reads was calculated in bins across the genome. Bins containing statistically significant Chip-seq enrichment were identified by comparison to a Poissonian background model. WIG files were generated for all ChIP-seq data sets. These files were subsequently used for visualization purposes and for obtaining average signal profiles.
Chromosome conformation capture assay
Chromosome conformation capture (3C) assays were performed as described in Gavrilov et al (2009) using MspI to digest cross-linked chromatin. 3C ligation products were measured by the Taqman-based qPCR technology. qPCR control for determination of primer efficiency of each primer combination was generated using BAC clone covering the Eomes genomic segment under study. The interaction frequencies were normalized with the log ratio of the interaction frequencies between fragments in the control region, to correct for the cross-linking and ligation efficiencies and the amount of the templates. Data are representative of two to three independent experiments, and error bars indicate standard error of the replicates. Probe and primer sequences are included in Supplementary Table 2. See Supplementary data for details.
Bioinformatic and statistical analyses
Mathematica7.0 was used to build an exhaustive motif search algorithm to map the Eomes binding motif on mouse promoters between −2500 and +500. The extracted consensus sequences were then graphically represented using Weblogo (UC Berkeley). Microarray raw data were subjected to RMA normalization after quality control scanning using the Affymetrix Suite Analysis software. Differential gene expression was analysed using Z-test false discovery rate (FDR)-adjusted P-values. Heatmap data were the log scale of the expression levels. Bound sites from the ChiP-seq data were analysed using Z-test FDR-adjusted P-values. A P-value cutoff of 10−6 was used to identify comparable numbers of bound sites. Other biological data are statistically analysed using Student’s t-test or Z-test, followed by Bonferroni correction for multiple comparisons. Genomic coordinates were based on build mm9 for mouse and hg19 for human. See Supplementary data for details.
Data Access
High-throughput data are available for download at GEO (GSE44764). Several raw data are included in Source data.
Supplementary Material
Acknowledgments
This work was supported by grants from NIDDK, Juvenile Diabetes Research Foundation and the Helmsley Trust to AB. We acknowledge the support from the UCLA Broad Stem Cell Research Center High-Throughput Sequencing Core.
Author contributions: AK and AS performed the experiments and analyses on mES cells. AK, JZ and MK performed the Chip-seq experiments and analyses. AK, DC and WL performed the experiments and analyses on hES cells. AG generated Sox17 GFP construct. MM generated Sox17 Cre-GFP mES cells. AB and AK conceived, planned the experiments and interpreted data. AB and AK wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K (2009) The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 23: 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang SL, Rossant J (1994) HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78: 561–574 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ (2008) Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA (2009) Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 4: 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G (2008) The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 3: e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas S, Cibelli JB, Ross PJ (2012) Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc Natl Acad Sci USA 109: 2400–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, Lv R, Li X, Villen J, Gygi SP, Liu XS, Shi Y (2012) The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev 26: 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Osipovich AB, Ustione A, Potter LA, Hipkens S, Gangula R, Yuan W, Piston DW, Magnuson MA (2011) Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech 4: 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC (2001) Determinants of T box protein specificity. Development 128: 3749–3758 [DOI] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ (2011) The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 13: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE (2005) Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541 [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P (2008) A rapid micro chromatin immunoprecipitation assay (microChIP). Nat Protoc 3: 1032–1045 [DOI] [PubMed] [Google Scholar]
- Dahle O, Kumar A, Kuehn MR (2010) Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 3: ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JP, Lu JY, Zhang Y, Shen YF (2010) Jmjd3 activates Mash1 gene in RA-induced neuronal differentiation of P19 cells. J Cell Biochem 110: 1457–1463 [DOI] [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, Testa G, Sung WK, Wei CL, Natoli G (2009) Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J 28: 3341–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, Vassetzky Y (2009) Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol Biol 567: 171–188 [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Melton DA (2000) Endoderm development: from patterning to organogenesis. Trends Genet 16: 124–130 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA 104: 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E (2007) Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG (2007) SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450: 415–419 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, Hayashi Y (2002) Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129: 2367–2379 [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Ochi H, Sudou N, Ogino H (2012) Comparative expression analysis of the H3K27 demethylases, JMJD3 and UTX, with the H3K27 methylase, EZH2, in Xenopus. Int J Dev Biol 56: 295–300 [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J (2011) Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357: 492–504 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G (2004) Development of definitive endoderm from embryonic stem cells in culture. Development 131: 1651–1662 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Loh KM, Lim B (2011) A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8: 363–369 [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21: 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Shelton KD, Lindner J, Jones JR, Magnuson MA (2004) Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive-negative selection. Genesis 39: 256–262 [DOI] [PubMed] [Google Scholar]
- Lowry WE, Plath K (2008) The many ways to make an iPS cell. Nat Biotechnol 26: 1246–1248 [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH (1996) Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS (2010) Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison GM, Oikonomopoulou I, Migueles RP, Soneji S, Livigni A, Enver T, Brickman JM (2008) Anterior definitive endoderm from ESCs reveals a role for FGF signaling. Cell Stem Cell 3: 402–415 [DOI] [PubMed] [Google Scholar]
- Mortensen RM, Conner DA, Chao S, Geisterfer-Lowrance AA, Seidman JG (1992) Production of homozygous mutant ES cells with a single targeting construct. Mol Cell Biol 12: 2391–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1: 299–312 [DOI] [PubMed] [Google Scholar]
- Patterson M, Chan DN, Ha I, Case D, Cui Y, Handel BV, Mikkola HK, Lowry WE (2011) Defining the nature of human pluripotent stem cell progeny. Cell Res 22: 178–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2010) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 11: 936–944 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA (2008) Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L (2011) Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S (2011) Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145: 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Hoodless PA, Bikoff EK, Robertson EJ (2000) Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development 127: 3079–3090 [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA (2009) Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ (2003) Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev 17: 1646–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz C, Kerppola TK (2008) Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci USA 105: 16572–16577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N (2012) Distinct Lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10: 440–454 [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE Jr (1994) The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78: 575–588 [DOI] [PubMed] [Google Scholar]
- Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T (2005) Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol 23: 1542–1550 [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1: 286–298 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH (2007) A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA 104: 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG (2008) Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.