Abstract
Glycosylation is the most abundant and diverse posttranslational modification of proteins. While several types of glycosylation can be predicted by the protein sequence context, and substantial knowledge of these glycoproteomes is available, our knowledge of the GalNAc-type O-glycosylation is highly limited. This type of glycosylation is unique in being regulated by 20 polypeptide GalNAc-transferases attaching the initiating GalNAc monosaccharides to Ser and Thr (and likely some Tyr) residues. We have developed a genetic engineering approach using human cell lines to simplify O-glycosylation (SimpleCells) that enables proteome-wide discovery of O-glycan sites using ‘bottom-up’ ETD-based mass spectrometric analysis. We implemented this on 12 human cell lines from different organs, and present a first map of the human O-glycoproteome with almost 3000 glycosites in over 600 O-glycoproteins as well as an improved NetOGlyc4.0 model for prediction of O-glycosylation. The finding of unique subsets of O-glycoproteins in each cell line provides evidence that the O-glycoproteome is differentially regulated and dynamic. The greatly expanded view of the O-glycoproteome should facilitate the exploration of how site-specific O-glycosylation regulates protein function.
Keywords: GALNT, GPCRs, LDLR, TNFα, Zinc finger nucleases
Introduction
Posttranslational modifications expand the diversity of the proteome enormously, with protein glycosylation being the most abundant and diverse form of modification serving different functions for protein folding, trafficking, processing, stability, and biological activity. A number of different types of human protein glycosylation exist, including N-linked to Asn, several types of O-linked to Ser, Thr, hydroxylysine, and Tyr residues, and C-mannosylation to Trp, and the glycans attached to proteins exhibit tremendous structural variation (Rana and Haltiwanger, 2011; Stanley, 2011). By far, the most complex-regulated type of protein glycosylation is the GalNAc-type, which is initiated by up to 20 distinct polypeptide GalNAc-transferase isoenzymes (GalNAc-Ts) with different but partially overlapping peptide specificities (Gill et al, 2011; Bennett et al, 2012; Tran and Ten Hagen, 2013). This unique scenario allows for an incomparable level of cell- and protein-specific regulation of where O-glycans are attached to proteins and ultimately the nature of the O-glycoproteome (Bennett et al, 2012). Other types of protein O-glycosylation are initiated by one or two isoenzymes and N-linked glycosylation by a single oligosaccharyltransferase complex, which leaves limited room for differential regulation of sites of glycosylation in cells.
Traditionally GalNAc-type O-glycosylation (hereafter simply O-glycosylation) has been considered a form of protein glycosylation occurring in dense clusters on mucin proteins and in mucin-like domains of proteins, hence its designation mucin-type O-glycosylation (Jentoft, 1990). However, it is becoming increasingly clear that this type of O-glycosylation is more widely distributed in isolated sites or regions of many different proteins not exhibiting mucin-like features (Halim et al, 2011; Steentoft et al, 2011; Halim et al, 2012; Schjoldager et al, 2012). Our knowledge of the O-glycoproteome is limited and biased towards abundant secreted proteins, which have mainly been analysed individually after isolation from a single source or expressed recombinantly. Substantial heterogeneity in O-glycan structures, lack of enzymes that can release all types of O-glycans, and constraints in analytical techniques have long hampered progress in mapping the O-glycoproteome, although some limited progress has been made in this respect (Nilsson et al, 2009; Darula et al, 2012). Furthermore, there are no straightforward consensus sequence motifs to provide a basis for prediction and identification of O-glycoproteins, and the current NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/) prediction algorithm performs poorly on isolated O-glycosylation sites (Julenius et al, 2005; Steentoft et al, 2011).
Site-specific O-glycosylation directed by distinct GalNAc-T isoforms is emerging as an important regulator of protein function, and further insight into the O-glycoproteome and specific sites of O-glycan attachments are paramount for progress (Schjoldager and Clausen, 2012). The true importance of site-specific O-glycosylation was first fully appreciated when Topaz et al (2004) discovered that familial tumoral calcinosis was associated with deficiency in a single GalNAc-T isoform, GALNT3. Loss of GALNT3 was subsequently shown to result in lack of site-specific O-glycosylation of the growth factor, fibroblast growth factor 23 (FGF23) at Thr178 in a proprotein convertase (PC)-processing site RHTR179 (Kato et al, 2006). O-glycosylation of Thr178 blocked inactivating furin processing of FGF23 and rescued secretion of active FGF23 in CHO cells. PC processing is a fundamental step in protein maturation where limited proteolysis activates or inactivates proteins, and more than 3500 proteins are predicted to undergo PC processing (Seidah, 2011). We have recently found another example of site-specific O-glycosylation adjacent to a PC-processing site that may underlie a disease condition associated with a dysfunctional GalNAc-T isoform, GALNT2 (Schjoldager et al, 2010). Furthermore, we predict that over 700 proteins are potentially affected by interplay between site-specific O-glycosylation and furin PC processing (Gram Schjoldager et al, 2011). It is thus clear that there is a huge potential for important undiscovered biological functions of O-glycosylation of proteins, and our knowledge of these is limited by lack of insight into the O-glycoproteome and where precisely O-glycans are attached to proteins.
In order to explore the O-glycoproteome, we have therefore developed a universal and stable genetic engineering strategy that simplifies O-glycosylation in cells, such that elongation of O-glycans is blocked and only simple truncated and homogeneous O-glycans are produced enabling efficient lectin enrichment followed by nLC-HCD/ETD-MS2 analysis (Steentoft et al, 2011). The strategy involves zinc finger nuclease (ZFN) targeting of COSMC, a private chaperone for the enzyme, C1GalT1, that controls O-glycan elongation (Ju and Cummings, 2002). We previously applied this strategy to a few human cell lines and demonstrated efficient identification of O-glycoproteins and sites of O-glycosylation. We showed that the O-glycoproteome identified in K562 SimpleCells (SC) was essentially identical to that found in wild-type K562 cells using a similar strategy of analysis (Steentoft et al, 2011). We also applied the SimpleCell strategy to pinpoint non-redundant O-glycosylation performed by a single polypeptide GalNAc-T using differential analysis of O-glycoproteomes produced in an isogenic cell model with and without knockout of one GalNAc-T isoform (Schjoldager et al, 2012). Here, we have systematically applied the SimpleCell strategy to 12 human cancer cell lines derived from diverse organ origin to obtain a first-generation view of the human O-glycoproteome. The size of the resulting data set yields an unprecedented opportunity to improve the performance of the NetOGlyc prediction algorithm, as well as to provide a first global view of acceptor substrate peptide specificities of a group of GalNAc-T isoforms.
Results
Precision mapping of O-glycosylation sites using human SC
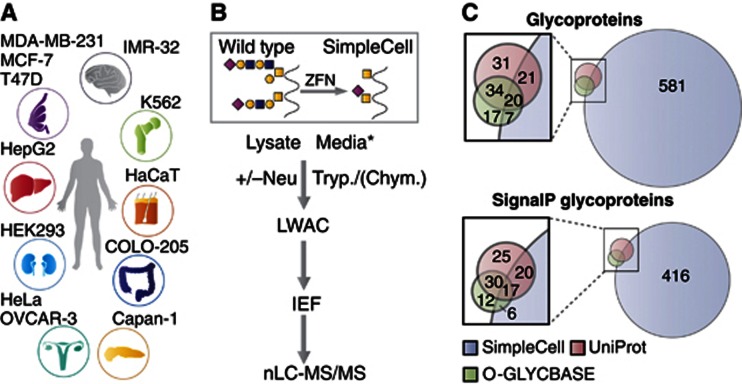
The SimpleCell strategy is depicted in Figure 1. The key feature is stable gene inactivation of the first step in the common elongation process of O-glycosylation to simplify O-glycans to immature Tn (GalNAcα1-O-Ser/Thr) and/or STn (NeuAcα2-6GalNAcα1-O-Ser/Thr) structures. We target the private chaperone, COSMC, for the core 1 β3galactosyltransferase (C1GalT1) elongation enzyme based on previous studies demonstrating that cells without a functional COSMC chaperone lack the C1GalT1 synthase activity and O-glycan elongation (Ju et al, 2008). We generated 12 human SimpleCell lines from diverse organs to be able to probe cell and organ variation in the O-glycoproteome Figure 1A. Four of the cell lines were described previously and limited analysis performed (Steentoft et al, 2011; Schjoldager et al, 2012). All the SimpleCell lines were selected by glycophenotype (Supplementary Figure S1) and confirmed by DNA sequencing (Supplementary Table S1).
Figure 1.
The SimpleCell O-GalNAc glycoproteomics strategy. (A) SimpleCell lines originating from different organs as illustrated were generated by ZFN-mediated knockout of COSMC. (B) SC express homogeneous truncated O-glycans (Tn/STn), and glycopeptides from cell lysates as well as conditioned media can be isolated by LWAC after protease digestion with trypsin or chymotrypsin. If necessary, sialic acids are removed by neuraminidase treatment. GalNAc-glycopeptides are isolated on LWAC and separated by IEF prior to nLC-MS/MS analysis. *Glycoproteins from conditioned media were pre-concentrated by passing the media over a short VVA column (see Extended Experimental Procedures). (C) The SimpleCell strategy identified 629 glycoproteins (not including possible GlcNAc cases), with only 48 in common with glycoproteins previously reported in UniProt (106 total) and O-GLYCBASE (78 total). Of the 771 glycoproteins in these three data sets, only 526 are predicted to have a signal peptide by the SignalP predictor.
Cells producing homogeneous truncated Tn and/or STn O-glycan structures simplify all analytical steps, from lectin-based capture or enrichment to nLC-MS/MS protocols, spectral interpretation, and data processing for identification and sequencing of O-glycosites. Isolation of GalNAc glycopeptides from protease digests by lectin weak affinity chromatography (LWAC) with Vicia villosa agglutinin (VVA) efficiently enriches O-glycopeptides for mass spectrometry and overcomes difficulties with suppression of signals from unglycosylated peptides (Steentoft et al, 2011). We further optimized the process by including analysis of both cell lysates and secretomes, and orthogonal fractionation by isoelectric focusing (IEF) prior to nLC-MS analysis (Figure 1B) (Vakhrushev et al, 2013). IEF fractions were analysed by nLC-MS with data-dependent acquisition protocols, including HCD-MS2 and ETD-MS2 from the same precursors. Identified glycoproteins, glycopeptides, and glycosites are listed in Supplementary Table S2A–E. The data set includes over 2100 unambiguously identified O-glycosites, having sufficient fragments defining sequence and HexNAc position, and another 700 in O-glycopeptides where the site could not be unambiguously determined, where ETD spectra were either missing or insufficient to confirm HexNAc position (Extended Experimental Procedures). The results present a first-generation view of the GalNAc O-glycoproteome, and we expect that analysis of additional cell lines, use of different proteases to enhance coverage, prior release of, for example, N-glycans to identify glycopeptides with both N- and O-glycosites, and use of instrumentation with enhanced sensitivity, such as the OrbiTrap Velos Pro or Elite, will lead to substantial increases in number of detected O-glycoproteins and glycosites. Furthermore, it is important to note that the SimpleCell strategy may suffer from several shortcomings: (i) elimination of the O-glycan elongation pathway in cells may facilitate enhanced density of O-GalNAc glycosylation due to lack of competition with the lectin-mediated functions of GalNAc-Ts (Bennett et al, 2012), although we have found no evidence of such, and a comparative analysis of the O-glycoproteomes of wild type and SC of K562 showed they were similar (Steentoft et al, 2011); (ii) densely O-glycosylated regions may be less susceptible to proteolysis, and hence poorly detected by the mass spectrometric approach; however, this should be less pronounced with short GalNAc O-glycans, as evidenced by many identified O-glycopeptides with proteolytic cleavage in close proximity or adjacent to glycosites; and (iii) mass spectrometry can easily identify HexNAc modifications, but cannot distinguish between the isomeric/isobaric O-GalNAc and O-GlcNAc residues; however, O-GlcNAc is primarily found on cytosolic proteins without signal sequences (Hart and Akimoto, 2009) or, as recently demonstrated, in some EGF-like repeats on Notch and a few other glycoproteins (Sakaidani et al, 2011; Alfaro et al, 2012).
It is important to note that in correlating O-glycosites with the O-glycoproteins of origin, overall peptide sequence coverage is not a useful parameter for establishing confidence of identification since the major harvest consists of low numbers of O-glycosites distributed over a very limited set of O-glycopeptides representing each protein (Supplementary Figure S2). We employed Orbitrap FT-MS detection for all three stages of acquisition, MS1, HCD-MS2, and ETD-MS2; in this, we have chosen to trade possible increased depth of coverage for the positive benefit of more stringent m/z tolerance constraints that could be applied to fragment ion matching in database searches, increasing the confidence of O-glycoprotein and O-glycosite identifications. With respect to peptide confidence, a false discovery rate (FDR) of 5% was applied, but this was a less critical parameter than scoring rank due to the process of individual validation, which serves to further increase confidence in the data set. A small set of sequences shared by more than one protein was identified and excluded from statistical analysis (Supplementary Table S2F).
Since GalNAc and GlcNAc are exact isobars, it is also important to minimize the potential for inclusion of O-GlcNAc sites found on cytosolic proteins without signal peptides (Hart and Akimoto, 2009) as well as distinct O-GlcNAc glycosylation of Notch EGF repeats (Sakaidani et al, 2011). Glycoproteins that are not predicted to have a signal peptide (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al, 2011) and by manual inspection have no other annotations to the secretory pathway, are marked in Supplementary Table S2 and excluded from statistical analysis.
A first-generation human GalNAc-type O-glycoproteome
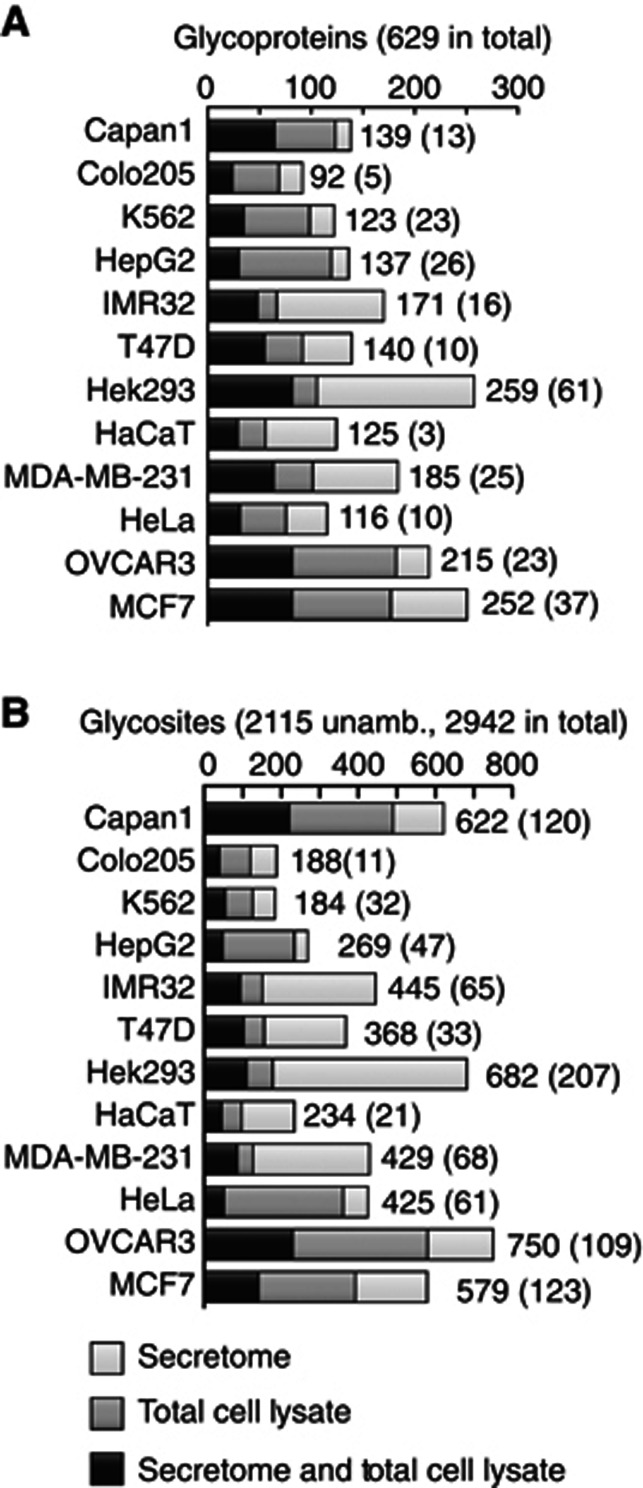
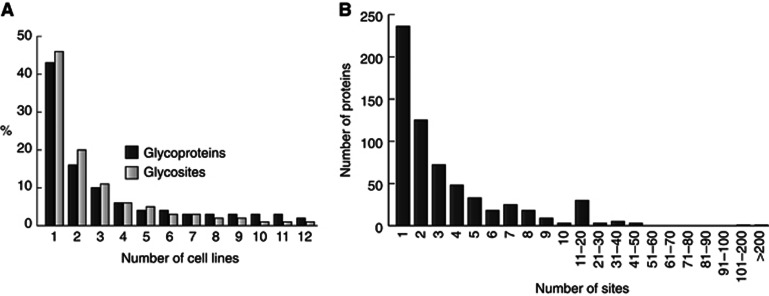
The analysis of 12 SimpleCell lines resulted in identification of 629 O-glycoproteins, of which only 48 were previously annotated (Figure 1C, Supplementary Table S2), although the number of annotated glycoproteins is an underestimate of previously identified O-glycoproteins. The distribution of glycoprotein and glycosite identifications in different cell lines varied substantially (Figure 2). Interestingly, almost 50% of the identified glycoproteins and glycosites were found only in one cell line, and each cell line contributed a number of unique glycoproteins (Figure 3A). Only 14 proteins were identified in all 12 cell lines. Thus in all cell lines, we found a new subset of O-glycoproteins (and O-glycosites) not found in the other cell lines. This is in contrast to general proteomic strategies where a major fraction of proteins can be identified in many cell lines (Geiger et al, 2012). These proteomic strategies can, however, rely on identification of multiple peptides from each protein, increasing chances of identification. The unique feature of O-glycosylation is that the repertoire of GalNAc-T isoforms in cells can determine whether a glycosite is occupied or not. The result therefore provides strong evidence for the existence of differential O-glycoproteomes in cells, consistent with the complex regulation of O-glycan initiation by the 20 GalNAc-Ts. Almost 50% of the identified glycoproteins showed only a single glycosite (Figure 3B) and we expect that many of these single sites may be selectively glycosylated by specific GalNAc-T isoforms, which further suggests that a large number of proteins may only become O-glycoproteins when expressed in certain cell types with the appropriate GalNAc-T repertoire. We tested the expression of 10 GalNAc-Ts in the 12 SC lines by ICC using our panel of MAbs and demonstrated differential expression of these isoforms (Supplementary Table S3); however, it is currently premature to try to correlate expression of individual GalNAc-T isoforms with the O-glycoproteome data.
Figure 2.
Number of O-glycoproteins and O-glycosites identified in individual SimpleCell lines. The distribution of glycoproteins (A) and unambiguous O-glycosites (B) identified in secretome, in TCLs, and in both are shown. Total numbers are given for each cell line as well as in parenthesis unique identifications in the particular cell line. Chymotrypsin data (Capan-1 and HEK293) omitted.
Figure 3.
Cellular distribution of O-glycoproteins and O-glycosites. (A) Distribution of identified glycoproteins and sites by number of cell lines. Chymotrypsin data (Capan-1 and HEK293) omitted and only unambigously assigned sites included. (B) Number of O-glycosites identified per number of proteins (all sites included).
Our strategy identified only 48 (37%) of the human O-glycoproteins annotated in UniProt (106 total) and/or O-GLYCBASE (78 total) (Figure 1). Possible explanations are that not all proteins are expressed in sufficient amounts to be detectable in the cells tested, or they may require specific GalNAc-T isoforms for O-glycosylation not co-expressed. The major part of our data set is based on trypsin digestion only, and other proteases may be required to obtain suitable peptide sizes for identification. In support of this, we included analysis of chymotrypsin digests of cell lysates from two cell lines and found additional glycoprotein identifications compared with trypsin (Supplementary Table S2). Blatantly missing from our data set are the tandem repeat regions of mucins with dense O-glycosylation. This may be in large part ascribed to difficulties in digesting these regions because of their dense glycosylation and dominance of S, T, and P residues. While the majority of identified glycosites did occur in sequences containing P as well as multiple S and T residues, a total of 74 glycosites were observed that had no S, T, or P within five amino acids N or C terminal from the site (Supplementary Figure S3).
One recent surprise in the field has been the identification of GalNAc O-glycosylation of Tyr residues. The first site was identified in an intriguing position in the amyloid P protein close to the β′-processing site of BACE-1 (Halim et al, 2011), and we identified sites in several other proteins (Steentoft et al, 2011; Vakhrushev et al, 2013). Here, we identified another 17 sites in diverse proteins, to bring the total to 23 identified Tyr O-glycosites (Supplementary Table S2G). It is unclear if these sites are glycosylated by GalNAc-Ts, but in preliminary studies we have observed incorporation of GalNAc on Tyr in peptide substrates by in-vitro enzyme assays (not shown).
Cellular and functional classification of O-glycoproteins
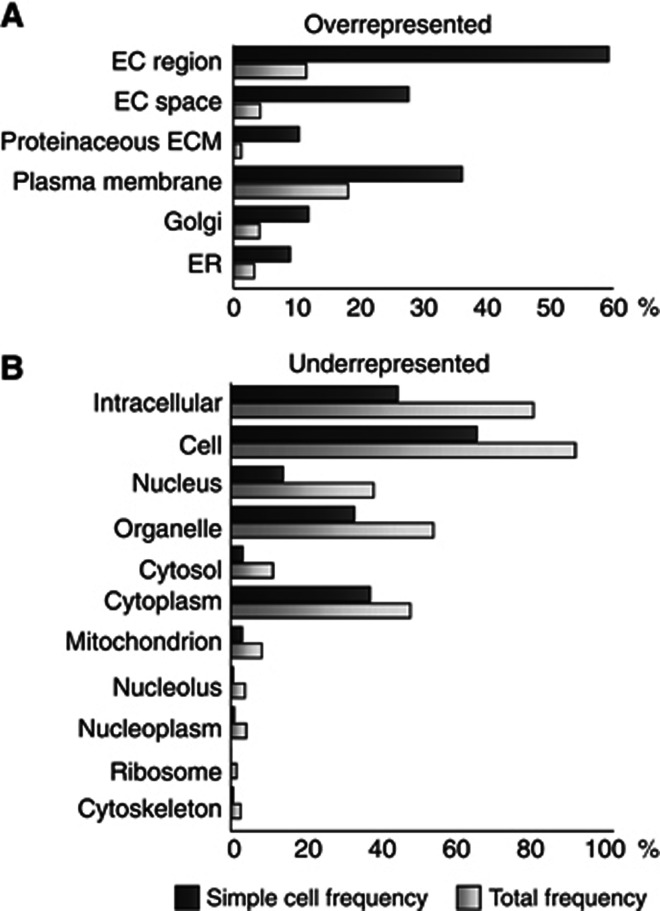
Cellular component gene ontology analysis (GO) (Maere et al, 2005) of the identified O-glycoproteins in SC showed a clear overrepresentation in the extracellular region and plasma membrane, and underrepresentation in nuclear and cytosolic regions as expected (Figure 4), even though, intriguingly, nuclear proteins represented 14%. However, in the majority of cases the GO assignments to these proteins were nonexclusive and differed from the UniProt assignments. While O-glycosylation of ER- and Golgi-located proteins have not been widely found before, our SimpleCell approach has revealed extensive O-glycosylation of resident ER and Golgi proteins. In agreement with this, we found strong overrepresentation in these organelles similar to what has been observed for N-glycosylation (Zielinska et al, 2010).
Figure 4.
Cellular component GO analysis. Cellular components that are significantly over (A) or under (B) represented in the SimpleCell data set compared to the entire human proteome using BinGO plugin for Cytoscape (www.cytoscape.org). The individual annotations for each protein have not been validated manually as the analysis is merely for relative representation purposes. EC, extracellular; ECM, extracellular matrix.
Domain and structure preference of O-glycosylation
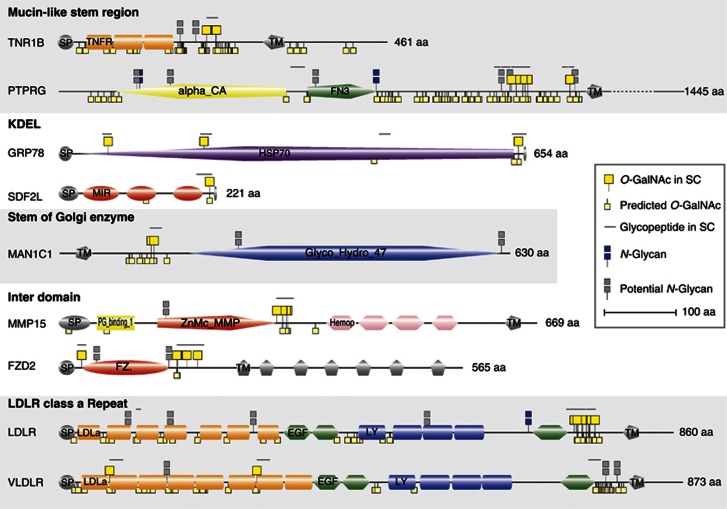
In order to enable more detailed studies of positions of O-glycosites in proteins and predictions of potential functions of site-specific O-glycosylation and relationship between protein structure and glycosylation, we have developed a graphic tool (GlycoDomainViewer) that incorporates curated protein domain annotations, as well as both identified and predicted sites of N- and O-glycosylation (N-glycosylation sites based on UniProt data) (http://cbs.dtu.dk/biotools/glycodomainviewer).
Using the GlycoDomainViewer, we determined that over 80% of the sites identified by our SimpleCell strategy were located outside the curated set of domains. In contrast, analysing the same protein subset for UniProt-annotated N-glycosylation sites (only sites with experimental references included), we found that ∼78% of the N-glycan sites were located within annotated protein domains. A number of interesting general tendencies emerge for substructures where O-glycosylation occurs on proteins, as exemplified in Figure 5: (i) juxtamembrane stalk regions of transmembrane proteins, which is in agreement with a role in protrusion of functional domains by inducing extended conformation of the peptide backbone as well as providing protection from proteolysis (Jentoft, 1990); (ii) in close proximity to C-terminal KDEL ER-retention signals, which could suggest a role in co-regulation of retrieval and resident time of many ER proteins; (iii) juxtamembrane stalk regions of Golgi and ER-resident membrane proteins; (iv) linker regions between functional domains, which could also serve to extend and protect these; and (v) O-glycosites in the short linker regions of class A repeats in lipoprotein receptors.
Figure 5.
Graphic depiction of glycosites in proteins by a novel GlycoDomainViewer. Representative examples of selected O-glycoproteins depicted based on the GlycoDomainViewer. O-GalNAc sites identified in SC are listed in the upper side of each protein while sites predicted by NetOGlyc4.0 are located on the lower half. N-glycan sites, experimentally verified as well as predicted, are obtained from UniProt. Designations are as follows: SP, signal peptide; TNFR, tumour necrosis factor receptor; TM, transmembrane; alpha_CA, carbonic anhydrase alpha; FN3, fibronectin type 3; HSP70, heat shock protein 70; MIR, domain in ryanodine and inositol trisphosphate receptors and protein O-mannosyltransferases; Glyco_Hydro_47, Glycosyl hydrolase family 47; PG_binding_1, putative peptidoglycan-binding domain; ZnMc_MMP, zinc-dependent metalloprotease; Hemop, hemopexin; FZ.; frizzled; LDLa, low-density lipoprotein receptor domain class A; EGF, calcium-binding EGF domain; LY, low-density lipoprotein-receptor YWTD domain. The listed proteins are TNF1B (tumour necrosis factor receptor superfamily member 1B, P20333), PTPRG (receptor-type tyrosine-protein phosphatase gamma, P23470), GRP78 (78 kDa glucose-regulated protein, P11021), SDF2L (stromal cell-derived factor 2-like protein 1, Q9HCN8), MAN1C1 (Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC, Q9NR34), MMP15 (matrix metalloproteinase-15, P51511), FZD2 (frizzled-2, Q14332), LDLR (low-density lipoprotein receptor, P01130), and VLDLR (very low-density lipoprotein receptor, P98155).
Towards sequence recognition motifs for O-glycosylation
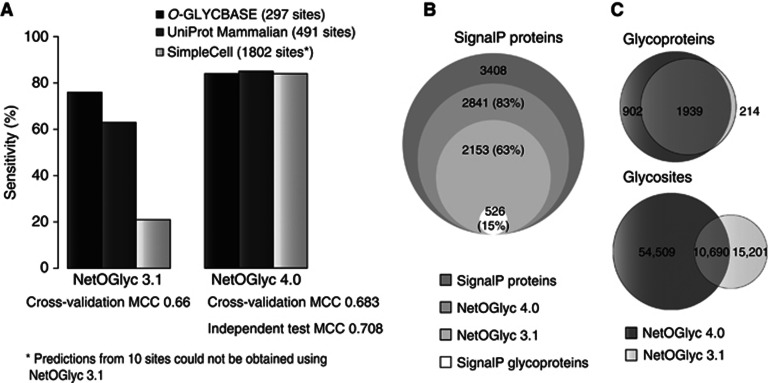
It is clear that a simple consensus sequence motif like the one for N-glycosylation does not exist and this is at least partly because of the complex regulation of O-glycosylation sites by up to 20 GalNAc-T isoforms with different, albeit partly overlapping, substrate recognition (Gerken et al, 2011; Bennett et al, 2012). The neural network-trained prediction algorithm (NetOGlyc 3.1) has been widely used in the past; however, we found that it only predicted 21% of the O-glycosites identified in SC (Figure 6A). NetOGlyc 3.1 was trained on a set of known O-glycoproteins biased towards abundant serum O-glycoproteins (Julenius et al, 2005). Here, we used the first proteome-wide generated data set to develop a new support vector machine-based predictor, NetOGlyc4.0 (http://www.cbs.dtu.dk/services/NetOGlyc-4.0/). The new predictor showed high general sensitivity (over 84%) when tested on different sets of glycosites (Figure 6A). To better characterize its performance, a four-fold cross-validation (CV) analysis was performed (Supplementary Figure S4), taking care to maintain independence between data folds. A commonly used metric to measure predictor performance is the Matthews Correlation Coefficient (MCC) (Matthews, 1975). Existing predictors have reported MCC values from ∼0.3 to 0.8, although the numbers are not directly comparable. We obtained an average MCC of 0.683, as well as an average MCC of 0.708 testing each CV iteration on an independent data set (primarily UniProt-annotated O-GalNAc sites). This is an improvement over NetOGlyc 3.1, which exhibited a CV MCC of 0.66 with overall lower sensitivities. To achieve this performance, the predictor now incorporates secondary structure predictions from Netsurfp and Disembl, and transmembrane predictions from TMHMM, as well as sourcing mass spectrometric data for information on non-glycosylated sites. Applying the NetOGlyc4.0 predictor to the SignalP proteome, more than 83% of the proteome is predicted to be O-glycosylated, compared to over 63% with NetOGlyc 3.1 (Figure 6B). Perhaps, more importantly, the overlap with v3.1 predictions of O-glycoproteins is high (90%), but extremely low with respect to glycosites (41%), where the majority of predictions are different (Figure 6C).
Figure 6.
NetOGlyc4.0 predictor performance. (A) A comparison of the performance of NetOGlyc 3.1 and the novel v4.0 predictors on three different data sets. CV MCC values indicated. The v3.1 shows poor sensitivity with the SimpleCell data set. By comparison, v4.0 exhibits a marked general improvement in sensitivity when testing with the O-GLYCBASE subset used to train v3.1, annotated experimental O-GalNAc from the curated subset of UniProt, as well as the SimpleCell data set. (B) Comparative analysis of predictors on the SignalP proteome (human curated UniProt). (C) Comparative analysis of O-glycoprotein and O-glycosite predictions.
Probing GalNAc-T isoform functions by in-vitro enzyme analysis
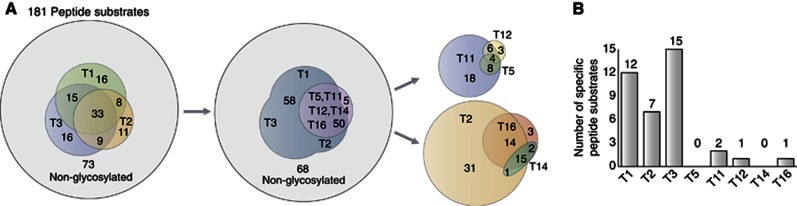
Our knowledge of substrate specificities and functions of individual GalNAc-Ts relies to a large extent on in-vitro enzyme assays using short synthetic peptides as acceptor substrates (Ten Hagen et al, 2003; Gerken et al, 2011; Bennett et al, 2012). Several studies have confirmed the validity of this approach (Bennett et al, 1996; Nehrke et al, 1997; DeFrees et al, 2006; Kato et al, 2006). To date, the peptide substrates tested have been limited to a small selected set from a few proteins (Schwientek et al, 2002). In order to get a first snapshot of global functions of GalNAc-Ts, we tested 181 peptide substrates derived from randomly selected O-glycoproteins with eight GalNAc-T isoforms (Figure 7 and Supplementary Table S4). For simplicity, all enzyme reactions were performed with equal amounts of purified recombinant enzymes in a standard reaction mixture.
Figure 7.
In-vitro analysis of GalNAc-T isoform substrate specifities. (A) One hundred and eighty-one peptide substrates from the SimpleCell data set were tested by in-vitro enzyme assays with recombinant GalNAc-Ts. Seventy-three peptides were not glycosylated by either GalNAc-T1, T2, or T3. When testing additional five enzymes (GalNAc-T5, T11, T12, T14, and T16), only five more peptides were glycosylated. (B) Number of unique peptide substrates for each GalNAc-T isoform.
The three most widely expressed and best-characterized GalNAc-Ts (T1, T2, and T3) glycosylated ∼60% of the peptides, and exhibited partly overlapping substrate specificities (Figure 7A), as well as apparent unique substrate specificities (Figure 7B). Interestingly, except for five peptides, the 40% of the peptide substrates not glycosylated by these isoforms were not significantly glycosylated by the five other isoforms tested either. We recently proposed a classification of the 20 GalNAc-Ts into evolutionary conserved subfamilies (Bennett et al, 2012), and the isoforms tested here represent members of different subfamilies as well as three members of subfamily Ib (GalNAc-T2, T14, and T16). We expected that distinct subfamilies would contribute substantially to glycosylation of peptide substrates not glycosylated by GalNAc-T1-3, but isoforms representative of these subfamilies generally glycosylated an overlapping subset, and only one or two unique substrates were identified for GalNAc-T5, T11, and T12 (Figure 7B). Interestingly, the three members of the closely homologous subfamily Ib (GalNAc-T2, T14, and T16) showed very limited overlap. This is in contrast to our previous experience with, for example, subfamily Ic consisting of GalNAc-T3 and T6, as well as Ia consisting of GalNAc-T1 and T13 (not shown).
We analysed the 181 peptide substrates tested by in-vitro glycosylation (Figure 7 and Supplementary Table S4) with Gerkens IsoGlyP predictor (Gerken et al, 2011). The positively in-vitro glycosylated peptides were predicted quite correctly in 84, 96, 82, 91, 100, and 88% of the cases for GalNAc-T1, T2, T3, T5, T12, and T16 isoforms, respectively. However, the predictor predicted glycosylation of 43–55% of the peptides that were not glycosylated in vitro by the respective enzymes. Thus, there is relatively poor agreement and either the in-vitro analysis underpredicts or the IsoGlyP overpredicts glycosylation.
Discussion
The SimpleCell strategy for mapping the O-glycoproteome
The GalNAc-type O-glycoproteome has long been an elusive target. Introduction of the SimpleCell strategy (Figure 1) has for the first time enabled a proteome-wide discovery of the O-glycoproteome and determination of sites of O-glycan attachments (Steentoft et al, 2011). The key feature of the strategy is simplification of the O-glycan structures to a homogeneous glycoform that is amenable for selective lectin enrichment, which is essential for sensitive detection of O-glycopeptides by mass spectrometry. The SimpleCell strategy is limited so far to use with immortalized genetically engineered cell lines, but mapping of the glycoproteome in these cell lines can provide a view of the finite aggregate O-glycoproteome, which opens the field for targeted analysis of particular O-glycoproteins in other biological settings, as well as providing a basis for future-targeted glycoproteome-wide strategies, ultimately without the need for simplification of O-glycan structures. Applying the strategy to 12 human cancer cell lines provided a first global view of the human O-glycoproteome, with more than 600 O-glycoproteins and almost 3000 glycosites. This represents an almost 10-fold expansion of known O-glycosites, reaching the level of experimentally validated N-glycosites in mammalian cells and tissues. Furthermore, the results provide the first evidence that the O-glycoproteome is differentially regulated in cells.
When evaluating the status of O-glycoproteomics, it is perhaps valuable to compare the current state of N-glycoproteomics. The deepest analysis so far has been achieved with lectin capture of protease-digested N-glycopeptides via the common N-glycan mannose core structure followed by enzymatic release of the N-glycan by PNGase F (Zielinska et al, 2010). An important feature is that N-glycoproteins in general will be glycosylated if expressed in any cell, while this may not be the case for O-glycoproteins. A number of different strategies have been applied to identify O-glycosylation, but few have been applicable for proteome-wide screening of complex biological samples (for review see Zauner et al (2012)). The most promising alternative strategy currently is the selective capture of O-glycoproteins via sialic acid residues (Nilsson et al, 2009; Halim et al, 2012). This strategy can provide information on the structure as well as site of attachment of O-glycans capped by sialic acids, and applications to cerebrospinal fluid and urine have led to identification of almost 100 O-glycosites. We used 12 different human SimpleCell lines to probe the O-glycoproteome and found that almost 50% of the sites were identified in only one cell line (Figure 2). In contrast, in a similar analysis of the global proteome of 11 human cell lines, it was found that almost 90% of the identified proteins were found in all cell lines, albeit in different levels (Geiger et al, 2012). Analysis of the O-glycoproteome is, however, dependent not only on the expression of a particular protein but also the expression of the GalNAc-Ts. The study included three breast cancer cell lines, and it could be expected that these cell lines would express a common set of breast cancer-specific glycoproteins. We did not identify obvious subsets of the O-glycoproteins common to these three cell lines and this may illustrate the large variability of the O-glycoproteome.
Characterization of the O-glycoproteome
The O-glycoproteome is vast and clearly much larger than predicted so far. Our data suggest that we have not exhausted discovery, and our new NetOGlyc4.0 predicts that up to 83% of proteins entering the ER–Golgi secretory pathway are O-glycosylated (Figure 6). On the other hand, the data provide evidence that the O-glycoproteome is variable in cells, because analysis of each of the 12 cell lines incrementally provided identification of a substantial fraction of new O-glycoproteins (Figure 2). All classes of proteins appear to be represented, and O-glycosylation is found in clusters with and without mucin-like domains, as well as in isolated single sites (Figures 3 and 5). Our digestion strategy (trypsin and chymotrypsin) and use of cell lines may bias against classical mucins and their PTS-rich tandem repeat regions, but we did identify a few glycosites outside the tandem repeat of MUC1, MUC4, MUC5B, MUC17, and MUC20, as well as a larger number of glycosites in the large N-terminal PTS region of MUC16, which is not typical mucin-like with tandem repeats (Supplementary Table S2). It is noteworthy that this represents the first identification of O-glycosites in human mucins except for targeted analysis of endo-Arg-released MUC1 tandem repeats (Muller et al, 1999). We also identified several members of the GPCR family (GPR 37, 56, 64, 107, 108, and 116), many of which are known to have long N-terminal PTS-rich regions (Fredriksson et al, 2003). A large number of O-glycoproteins have clustered O-glycosites in juxtamembrane stem regions and in linker regions between known domains, some examples of which are shown in Figure 5. These include cell membrane receptors, for example, the TNR1B receptor, in which a long stretch of the stem region is densely O-glycosylated. This is particularly interesting, since the ectodomain, including the stem region, is part of the chimeric glycoprotein consisting of the TNFα receptor and the Fc region of human IgG1, which is a highly successful anti-inflammatory recombinant glycoprotein drug (Etanercept, Enbrel) (Peppel et al, 1991). They also include a large number of type II transmembrane glycosyltransferases, which are rarely found in general proteomic approaches, presumably due to their low abundance and difficulties in solubilization. To our knowledge, this class of proteins has only been identified in reasonable numbers in a proteomic analysis of fractionated ER–Golgi vesicles (Gilchrist et al, 2006). Our data also showed that O-glycosylation of Tyr residues is widely found in proteins with or without coexisting Ser and Thr O-glycosylation (Supplementary Table S2G). Tyr O-glycosylation may serve special functions and could, for example, compete with both phosphorylation and sulphation. We presume that the biosynthesis of Tyr O-glycosylation is controlled by GalNAc-Ts, but further studies are needed to evaluate this.
The lack of clear and searchable consensus sequence motifs for O-glycosylation comparable to the NXS/T motif for N-glycosylation is a strong barrier to developing a more global view of common protein features directing topology of O-glycans on proteins. The current O-glycoprotein data set provides clear evidence for location of O-glycans in disordered regions (>80% identified sites assigned), while analysis of identified N-glycans in the same subset of proteins showed that these are primarily located in folded domains (78% sites assigned). This is obviously consistent with the well-known role of N-glycosylation in protein folding and quality control (Parodi, 1977), and with initiation of GalNAc O-glycosylation in the Golgi after ER folding processes (Rottger et al, 1998). Interestingly, however, our SimpleCell approach has identified a large number of ER-located O-glycoproteins, including chaperones (e.g., HSPA5 (GRP78), HSPA13, and Calnexin), disulphide-isomerases (e.g., PDIA3/4), and glycosidases (e.g., PRKCSH). Many of these proteins are at least transiently exposed to the Golgi during ER retrieval, but recent evidence suggesting that the GalNAc-Ts can relocate to ER may also be part of the explanation (Gill et al, 2011). In order to promote the use of the O-glycosite data set, we have developed GlycoDomainViewer displaying graphic representations of O- and N-linked glycosites on individual proteins with sequence and domain topology (Figure 5). The GlycoDomainViewer allows for easy visualization of the location of individual sites in a given protein with the proviso that quality of domain representation is dependent on available predictors.
The O-glycoproteome points to functions of site-specific O-glycosylation
Dense O-glycosylation promotes extension of the protein backbone, and confers local resistance to proteolysis and overall stability to proteins (Jentoft, 1990). Apart from these general effects, we have previously demonstrated that site-specific O-glycosylation is a co-regulator of the important PC processing many proteins undergo for maturation (Kato et al, 2006; Schjoldager et al, 2010; Gram Schjoldager et al, 2011; Schjoldager and Clausen, 2012; Schjoldager et al, 2012). Here, we discovered a number of O-glycosites within +/−3 residues of potential furin-type PC-processing sites, which should be well within the distance from which O-glycosylation can modulate PC processing. This includes, for example, a furin-processing site in NGF that we previously demonstrated was modulated by O-glycosylation (Gram Schjoldager et al, 2011). The data thus clearly provide further support for a general co-regulatory function of site-specific O-glycosylation in PC processing, and presumably other regulated proteolytic events (Schjoldager and Clausen, 2012).
Perhaps, the most remarkable finding was the presence of O-glycosylation in the short linker regions in between the EGF-like class A repeats in most of the lipoprotein receptors (Figure 5). Classic studies have demonstrated the importance of O-glycosylation in the stem region of LDLR (Kingsley et al, 1986), and we did identify several of these O-glycosites. However, we also identified O-glycosites in a conserved Thr residue immediately before the first of six Cys residues in each ∼30–40 amino-acid repeat. The lipoprotein-binding regions of lipoprotein receptors are constituted by 7–11 class A repeats (reviewed in Jeon and Blacklow (2005)), and the presence of O-glycans in the short linker regions between these must have dramatic effects on the structure of the receptor and its function. In six of the LDLR family receptors (LDLR, VLDLR, LRP1, LRP1B, LRP2 (megalin), LRP8 (ApoER2), and SorLA), we find from 1 up to 4 O-glycosites per class A repeat cluster and in total more than 20 sites in between class A repeats. Although these O-glycans have been overlooked in the past, there are early reports that LDLR without the stem region appeared to have O-glycosylation (Davis et al, 1986; Seguchi et al, 1991). Further studies are clearly needed to address how and where O-glycosylation occurs as well as the specific roles the O-glycans play for these receptors, but this is an excellent example of how the inability to predict and identify O-glycosylation of proteins has obstructed discovery of potentially very important biological functions.
A new prediction algorithm, NetOGlyc4.0
The new predictor trained on our large data set represents a major improvement compared with the 3.1 version (Figure 6). In particular, the general sensitivity across different data sets is improved. In this respect it is noteworthy that the 3.1 version, despite its lower sensitivity for our large data set, predicts the majority of its sites orthogonal to the new predictor (Figure 6C). Clearly, further refinement of the predictor is required to increase the sensitivity and specificity. To this end, there is a pressing need to better integrate disorder information with the prediction of glycosites. Further, there is a need to improve the negative (unglycosylated) data set. The current predictor is based on a unifying model for all GalNAc-Ts, but improved results may be obtained by collecting sufficient isoform-specific data to produce isoform-specific algorithms.
The O-glycoproteome is differentially regulated in cells
The repertoire of GalNAc-Ts in cells directs O-glycosylation, with several studies demonstrating that the expression of individual GalNAc-T isoforms differ in cells, tissues, during cell differentiation and also malignant progression (Sutherlin et al, 1997; Mandel et al, 1999; Young et al, 2003; Tian and Ten Hagen, 2006). Due to the limited number of known O-glycoproteins, studying the contribution of individual GalNAc-T isoforms has been difficult in the past, although it is clear that these isoforms have both overlapping and unique functions (Bennett et al, 2012). The availability of this new large data set allowed us to probe the functions of multiple isoforms on a large acceptor peptide panel, which confirmed that the three more commonly expressed isoforms (GalNAc-T1-3) play the major role in shaping the O-glycoproteome, while isoforms with more restricted expression appear to have more narrow specificities (Figure 7). These results complement studies using random peptide libraries to probe specificities (Gerken et al, 2011). Moreover, the results point to unique functions of individual enzyme isoforms that may produce phenotypes in individuals with dysfunctional enzymes. A number of studies point to individual GALNT genes as susceptibility genes for diseases, but uncovering causative roles is a very complicated exercise. However, with this broader information on substrate specificity, it is possible to select and validate unique candidate O-glycoproteins by targeted approaches (Schjoldager et al, 2012). Application of the SimpleCell and other strategies will help us understand the evolutionary need for the large GalNAc-transferase gene family.
Summary and outlook
We have presented a first-generation view of the O-glycoproteome using our SimpleCell-based mass spectrometry strategy and an improved model for predicting O-glycosylation. These tools are now available for the community to probe biological functions of the O-glycoproteome. This is a first view of the human O-glycoproteome, and we anticipate further expansion through use of additional cell lines, different protease digestion strategies, and more sensitive instrumentation. We have pointed to several interesting possible functions of site-specific O-glycosylation, but ultimately discovery of functions will have to be dealt with protein by protein in relevant cells or organisms, keeping in mind that the cellular capacity for O-glycosylation is variable, and perhaps dynamic. Our SimpleCell strategy, combined with targeted knockout of individual GALNT genes and analysis of differential glycoproteomes, is a promising approach for such studies as we have recently demonstrated (Schjoldager et al, 2012). We envision that the presented data set of O-glycosites can be used to develop targeted O-glycoproteomic strategies without the need for reducing natural O-glycan structural complexity so that information about site and glycan structure can be assessed simultaneously.
Materials and methods
Generation of O-GalNAc SC, sample preparation and lectin enrichment
Human SC were generated and processed as previously described (Steentoft et al, 2011; Schjoldager et al, 2012). Conditioned media obtained from 2 × T175 flasks (2 × 35 ml) cultured for 48–72 h were dialyzed, and glycoproteins were enriched by capture on a short (300 μl contained in 1 ml syringe) VVA agarose column. Unlike previous studies (Schjoldager et al, 2012), glycoproteins were eluted by heating the lectin (4 × 90 °C 10 min) with 0.05% RapiGest, eliminating a dialysis step (for details see Extended Experimental Procedures). Total cell lysates (TCL) were obtained by addition of 2 ml 0.05% RapiGest and sonication of cell pellets from 2 × cultured and scraped T175 flasks. Cell lysates and glycoprotein-enriched media were digested with trypsin or chymotrypsin, purified by C18 solid phase extraction, neuraminidase treated if necessary, and diluted in lectin-binding buffer. Lectin chromatography was performed as previously described (Steentoft et al, 2011, 2013). Briefly, protease-digested TCL and media were loaded on a 2.6-m-long VVA agarose column and eluted with 2 × 2 ml 0.2 M GalNAc and 1 × 2 ml 0.4 M GalNAc. Eluted glycopeptides were further fractionated into 12 samples by IEF (Vakhrushev et al, 2013), desalted by Stage Tips, and submitted for MS analysis.
Mass spectrometric analysis
Enriched glycopeptides were analysed on an EASY-nLC II interfaced via nanoSpray Flex ion source to an LTQ-Orbitrap XL ETD spectrometer (Thermo Fisher Scientific). Similar to previous (Steentoft et al, 2011; Vakhrushev et al, 2013), MS1, HCD-MS2, and ETD-MS2 spectra were all acquired in the Orbitrap sector; MS data analysis was performed using Proteome Discoverer 1.2 software, assisted by manual validation (see Extended Experimental Procedures for details on MS acquisition and data analysis). Annotated MS/MS spectra related to this paper can be downloaded from ProteomeCommons.org using the following tranche hash: eI8E81E3CPNnMiHn52O2Z7Wta/e9Ih8GE6b04v3Edq9aIGZfOj58pQ8E6+AmY6/ictzVOtCuc7Zaavr2VYIECMFgiKsAAAAAAAAKZw==.
NetOGlyc version 4.0
A support vector machine was used to produce a predictor of O-GalNAc glycosylation. Positive glycosylation site data were obtained from the SimpleCell data set, as well as mass spectrometric data supporting non-glycosylation from a number of cell lines. Features were generated to characterize various parameters surrounding each site, including transmembrane prediction (TMHMM), surface accessibility (NetSurfP), and protein disorder (DISEMBL). Individual features were evaluated by characterizing their learning curves. Coupled with data redundancy reduction to ensure data independence between training and testing sets, sensitivity as well as the more general MCC metrics were used to characterize the performance of the predictor (Supplementary Figure S4A–C) (Extended Experimental Procedures).
Functional analysis of GalNAc-T isoforms
From the O-glycoprotome, 181 peptides covering 1–3 identified or potential glycosites were synthesized (NeoBioSci) and glycosylated with recombinant GalNAc-Ts expressed as soluble secreted truncated proteins in insect cells (Extended Experimental Procedures).
Supplementary Material
Acknowledgments
This work was supported by Kirsten og Freddy Johansen Fonden, A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, The Carlsberg Foundation, The Novo Nordisk Foundation, The Danish Research Councils, a programme of excellence from the University of Copenhagen, the Sino-Danish Breast Cancer Centre, and The Danish National Research Foundation (DNRF107). We thank Frederic Bard for providing us with the MDA-MD-231 cell line as well as helpful discussion and comments on the manuscript. We thank Thomas Nordahl Petersen for helpful discussion on the predictor model and James C Paulson for comments on the manuscript.
Author contribution: CS, SYV, SBL, HJJ, and HC designed and performed experiments, analysed data, and wrote the paper. KT-BGS and HHW designed and performed experiments. MBV-C, YK, EPB, KL, SD, LM-S, and UM performed experiments. NBP, SB, and RG analysed data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG 2nd, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F, Smith RD (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci USA 109: 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Clausen H (1996) cDNA cloning and expression of a novel human UDP-N-acetyl-alpha-D-galactosamine polypeptide N-acetylgalactosaminyltransferase, GalNAc-T3. J Biol Chem 271: 17006–17012 [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22: 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darula Z, Sherman J, Medzihradszky KF (2012) How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Mol Cell Proteomics 11: O111.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CG, Elhammer A, Russell DW, Schneider WJ, Kornfeld S, Brown MS, Goldstein JL (1986) Deletion of clustered O-linked carbohydrates does not impair function of low density lipoprotein receptor in transfected fibroblasts. J Biol Chem 261: 2828–2838 [PubMed] [Google Scholar]
- DeFrees S, Wang ZG, Xing R, Scott AE, Wang J, Zopf D, Gouty DL, Sjoberg ER, Panneerselvam K, Brinkman-Van der Linden EC, Bayer RJ, Tarp MA, Clausen H (2006) GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology 16: 833–843 [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Gloriam DE, Hoglund PJ, Lagerstrom MC, Schioth HB (2003) There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem Biophys Res Commun 301: 725–734 [DOI] [PubMed] [Google Scholar]
- Geiger T, Wehner A, Schaab C, Cox J, Mann M (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics 11: M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA (2011) Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J Biol Chem 286: 14493–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127: 1265–1281 [DOI] [PubMed] [Google Scholar]
- Gill D, Clausen H, Bard F (2011) Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol 21: 149–158 [DOI] [PubMed] [Google Scholar]
- Gram Schjoldager KT, Vester-Christensen MB, Goth CK, Petersen TN, Brunak S, Bennett EP, Levery SB, Clausen H (2011) A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J Biol Chem 286: 40122–40132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A, Brinkmalm G, Ruetschi U, Westman-Brinkmalm A, Portelius E, Zetterberg H, Blennow K, Larson G, Nilsson J (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid β-peptides in human cerebrospinal fluid. Proc Natl Acad Sci USA 108: 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A, Nilsson J, Ruetschi U, Hesse C, Larson G (2012) Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol Cell Proteomics 11: M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Akimoto Y (2009) The O-GlcNAc modification. In Essentials of Glycobiology Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (eds) pp 263–279. Cold Spring Harbor: NY, [PubMed] [Google Scholar]
- Jentoft N (1990) Why are proteins O-glycosylated? Trends Biochem Sci 15: 291–294 [DOI] [PubMed] [Google Scholar]
- Jeon H, Blacklow SC (2005) Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem 74: 535–562 [DOI] [PubMed] [Google Scholar]
- Ju T, Aryal RP, Stowell CJ, Cummings RD (2008) Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J Cell Biol 182: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Cummings RD (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc Natl Acad Sci USA 99: 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S (2005) Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15: 153–164 [DOI] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem 281: 18370–18377 [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Kozarsky KF, Hobbie L, Krieger M (1986) Reversible defects in O-linked glycosylation and LDL receptor expression in a UDP-Gal/UDP-GalNAc 4-epimerase deficient mutant. Cell 44: 749–759 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mandel U, Hassan H, Therkildsen MH, Rygaard J, Jakobsen MH, Juhl BR, Dabelsteen E, Clausen H (1999) Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology 9: 43–52 [DOI] [PubMed] [Google Scholar]
- Matthews BW (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405: 442–451 [DOI] [PubMed] [Google Scholar]
- Muller S, Alving K, Peter-Katalinic J, Zachara N, Gooley AA, Hanisch FG (1999) High density O-glycosylation on tandem repeat peptide from secretory MUC1 of T47D breast cancer cells. J Biol Chem 274: 18165–18172 [DOI] [PubMed] [Google Scholar]
- Nehrke K, Ten Hagen KG, Hagen FK, Tabak LA (1997) Charge distribution of flanking amino acids inhibits O-glycosylation of several single-site acceptors in vivo. Glycobiology 7: 1053–1060 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Ruetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G (2009) Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods 6: 809–811 [DOI] [PubMed] [Google Scholar]
- Parodi AJ (1977) Synthesis of glycosyl-dolichol derivatives in bakers' yeast and their role in protein glycosylation. Eur J Biochem 75: 171–180 [DOI] [PubMed] [Google Scholar]
- Peppel K, Crawford D, Beutler B (1991) A tumor necrosis factor (TNF) receptor-IgG heavy chain chimeric protein as a bivalent antagonist of TNF activity. J Exp Med 174: 1483–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von HG, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Rana NA, Haltiwanger RS (2011) Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Cur Opin Struc Biol 21: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H, Nilsson T (1998) Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci 111(Pt 1): 45–60 [DOI] [PubMed] [Google Scholar]
- Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, Nadano D, Matsuda T, Furukawa K, Okajima T (2011) O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat Commun 2: 583. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Clausen H (2012) Site-specific protein O-glycosylation modulates proprotein processing - Deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta 1820: 2079–2094 [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Vakhrushev SY, Kong Y, Steentoft C, Nudelman AS, Pedersen NB, Wandall HH, Mandel U, Bennett EP, Levery SB, Clausen H (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc Natl Acad Sci USA 109: 9893–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Vester-Christensen MB, Bennett EP, Levery SB, Schwientek T, Yin W, Blixt O, Clausen H (2010) O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J Biol Chem 285: 36293–36303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, Reis CA, Behrens J, Mandel U, Keck B, Schafer MA, Haselmann K, Zubarev R, Roepstorff P, Burchell JM, Taylor-Papadimitriou J, Hollingsworth MA, Clausen H (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J Biol Chem 277: 22623–22638 [DOI] [PubMed] [Google Scholar]
- Seguchi T, Merkle RK, Ono M, Kuwano M, Cummings RD (1991) The dysfunctional LDL receptor in a monensin-resistant mutant of Chinese hamster ovary cells lacks selected O-linked oligosaccharides. Arch Biochem Biophys 284: 245–256 [DOI] [PubMed] [Google Scholar]
- Seidah NG (2011) The proprotein convertases, 20 years later. Methods Mol Biol 768: 23–57 [DOI] [PubMed] [Google Scholar]
- Stanley P (2011) Golgi Glycosylation. Cold Spring Harbor Perspect Biol 3: a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Bennett EP, Clausen H (2013) Glycoengineering of human cell lines using zinc finger nuclease gene targeting - SimpleCells with homogenous GalNAc O-glycosylation allow isolation of the O-glycoproteome by one-step lectin affinity chromatography. Methods Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Vester-Christensen MB, KTBG Schjoldager, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nature Methods 8: 977–982 [DOI] [PubMed] [Google Scholar]
- Sutherlin ME, Nishimori I, Caffrey T, Bennett EP, Hassan H, Mandel U, Mack D, Iwamura T, Clausen H, Hollingsworth MA (1997) Expression of three UDP-N-acetyl-alpha-D-galactosamine:polypeptide GalNAc N-acetylgalactosaminyltransferases in adenocarcinoma cell lines. Cancer Res 57: 4744–4748 [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA (2003) All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13: 1R–16R [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG (2006) Expression of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology 16: 83–95 [DOI] [PubMed] [Google Scholar]
- Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V, Zelikovic I, Raimer S, Metzker A, Richard G, Sprecher E (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet 36: 579–581 [DOI] [PubMed] [Google Scholar]
- Tran DT, Ten Hagen KG (2013) Mucin-type O-Glycosylation during Development. J Biol Chem 288: 6921–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrushev SY, Steentoft C, Vester-Christensen MB, Bennett EP, Clausen H, Levery SB (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol Cell Proteomics 12: 932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young WW Jr., Holcomb DR, Ten Hagen KG, Tabak LA (2003) Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isoforms in murine tissues determined by real-time PCR: a new view of a large family. Glycobiology 13: 549–557 [DOI] [PubMed] [Google Scholar]
- Zauner G, Kozak RP, Gardner RA, Fernandes DL, Deelder AM, Wuhrer M (2012) Protein O-glycosylation analysis. Biol Chem 393: 687–708 [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141: 897–907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.