Abstract
Extensive research on antiviral small molecules starting in the early 1970s has led to the identification of 10-carboxymethyl-9-acridanone (CMA) as a potent type I interferon (IFN) inducer. Up to date, the mode of action of this antiviral molecule has remained elusive. Here we demonstrate that CMA mediates a cell-intrinsic type I IFN response, depending on the ER-resident protein STING. CMA directly binds to STING and triggers a strong antiviral response through the TBK1/IRF3 route. Interestingly, while CMA displays extraordinary activity in phosphorylating IRF3 in the murine system, CMA fails to activate human cells that are otherwise responsive to STING ligands. This failure to activate human STING can be ascribed to its inability to bind to the C-terminal ligand-binding domain of human STING. Crystallographic studies show that two CMA molecules bind to the central Cyclic diguanylate (c-diGMP)-binding pocket of the STING dimer and fold the lid region in a fashion similar, but partially distinct, to c-diGMP. Altogether, these results provide novel insight into ligand-sensing properties of STING and, furthermore, unravel unexpected species-specific differences of this innate sensor.
Keywords: antiviral activity, innate immunity, STING, type I interferon
Introduction
The innate immune system operates to sense microbial infection. To this effect, it expresses a variety of so-called pattern-recognition receptors (PRRs) that are able to sense certain highly conserved microbial patterns, known as microbe-associated molecular patterns (MAMPs). Upon MAMP sensing, cells of the innate immune system elicit certain effector functions that are geared at eliminating the invading pathogen. As such, cytokines play an important role in orchestrating subsequent cellular immune responses or by inducing antimicrobial effector functions in non-immune cells. In this regard, cytokines of the type I interferon (IFN) family play a pivotal role in eliciting antiviral, but also antibacterial, effector functions. Soon after the discovery of type I IFNs by Isaacs and Lindenmann (1957), many research groups tried to delineate the mechanisms of its induction. Early on, it was already noted that nucleic acid preparations derived from viruses or enzymatic preparations were able to trigger potent type I IFN responses. One of the most potent triggers that emerged from these studies was the double-stranded RNA polynucleotide mimic poly(I:C) (Isaacs et al, 1963; Rotem et al, 1963), but other polynucleotide preparations, including double-stranded DNA (dsDNA), were also reported to initiate antiviral immunity by eliciting type I IFN responses. Moreover, in the early 1970s, several groups tried to develop small-molecule compounds with oral bioavailability that were able to trigger type I IFN responses, thereby blocking viral replication. In the course of these studies, the first small-molecule compound reported was the tricyclic compound tilorone (2,7-bis(2-diethylaminoethoxy)fluoren-9-one), which exhibited broad antiviral activities against many viruses (Krueger and Mayer, 1970; Mayer and Krueger, 1970). Subsequently, additional heterocyclic compounds were reported to induce type I IFNs, including various quinoline, anthraquinone and acridine derivatives. Another structurally related molecule that even surpassed most of these compounds in antiviral activity was 10-carboxymethyl-9-acridanone (CMA), discovered by Grunberg and colleagues, 1976. CMA was shown to harbour potent antiviral activity and this could be mainly ascribed to its ability to induce type I IFN production (Taylor et al, 1980b; Kramer et al, 1981; Storch and Kirchner, 1982; Brehm et al, 1986; Storch et al, 1986).
However, while most of these small-molecule compounds showed excellent type I IFN induction and antiviral activities in rodents (Kramer et al, 1976; Taylor et al, 1980a, 1980b), these promising results failed to translate into the human system. Tilorone, for example, showed no type I IFN induction in the human system, both upon systemic or topic administration (Kaufman et al, 1971), and at the same time, research on CMA was abandoned by most groups. Nevertheless, CMA is currently widely distributed and applied in Russia for antiviral therapy, including hepatitis B virus, hepatitis C virus, HIV or herpes simplex virus infection (Silin et al, 2009).
Three independent research approaches have led to the discovery of STING as a protein that strongly induced type I IFN production upon overexpression (Ishikawa and Barber, 2008; Zhong et al, 2008; Sun et al, 2009). Through its N-terminal four-pass transmembrane region STING is tethered to the ER, whereas its C-terminal region faces the cytoplasmic lumen. STING-deficient cells show a profound defect in sensing DNA viruses, and also synthetic DNA ligands are strongly blunted in their capacity to induce pro-inflammatory gene expression in STING-deficient cells (Ishikawa et al, 2009). At the same time, bacteria that replicate in the cytoplasm, such as Listeria monocytogenes, trigger type I IFN production in a STING-dependent manner (Ishikawa et al, 2009). This finding was initially explained by bacterial DNA being sensed in the cytoplasm in a STING-dependent fashion. However, it turned out that type I IFN production by L. monocytogenes could be mainly ascribed to the cytoplasmic presence of the bacterial quorum-sensing molecule cyclic diadenylate (c-diAMPs; Woodward et al, 2010). Indeed, bacteria-derived c-diAMP or cyclic diguanylate (c-diGMP) had already been described as potent triggers of innate immune responses (Karaolis et al, 2007a, 2007b; McWhirter et al, 2009). Both c-diAMP and c-diGMP function as quorum-sensing molecules in bacteria, regulating cell motility, biofilm formation and bacterial growth. Surprisingly, STING turned out to be the direct sensor for c-diGMP and c-diAMP, with its C-terminal part harbouring the binding domain (Burdette et al, 2011). In addition, most recently, it was discovered that cytosolic DNA sensing triggers the formation of a novel second messenger, cyclic GMP–AMP, which in turn binds to and activates STING (Sun et al, 2013; Wu et al, 2013). This finding reconciles the puzzling concept of STING serving as a sensor for microbial cyclic dinucleotides and DNA at the same time.
Several groups have recently been able to solve the crystal structure of c-diGMP binding to the C-terminal domain of STING (Huang et al, 2012; Ouyang et al, 2012; Shang et al, 2012; Shu et al, 2012; Yin et al, 2012). These studies have shown that the ligand-binding domain (LBD) of STING is present as a preformed dimer that forms a V-shaped structure harbouring a binding pocket for one c-diGMP molecule. While ligand-binding does not induce a major conformational change of the LBD of STING, a plausible model of activation suggests that the C-terminal tail (CTT) of STING is displaced from the pocket upon binding, so that it can interact with TBK1 (Yin et al, 2012). TBK1 subsequently leads to the phosphorylation of IRF3 and thereby induces transcription of antiviral genes.
In this study, we elucidate the molecular mechanism of CMA, a long-known small-molecule inducer of antiviral responses. In the murine system, CMA was found to be a potent activator of type I IFN production, yet in the human system it failed to elicit detectable antiviral responses. CMA activity depends on STING, and non-responsive human cells can be conferred responsive by overexpressing murine STING or a chimeric version of human STING that contains the LBD of murine STING. Differential scanning fluorimetry (DSF) studies using the LBDs of murine and human STING furthermore indicate that unresponsiveness of human STING to CMA is due to a lack of ligand binding. The crystal structure of CMA bound to murine STING shows that two CMA molecules bind the central c-diGMP-binding cavity in a fashion representing the inherent c-diGMP molecule symmetry, whereas differences are found in the folding of the lid region. While the structural studies cannot explain the species specificity in CMA detection, we provide additional data that suggest a differential involvement of the STING ‘lid region’ in CMA versus c-diGMP recognition. Altogether, these data reveal important insight into the species-specific recognition of a novel class of STING ligands.
Results
CMA is a potent trigger of the type I IFN response in murine macrophages
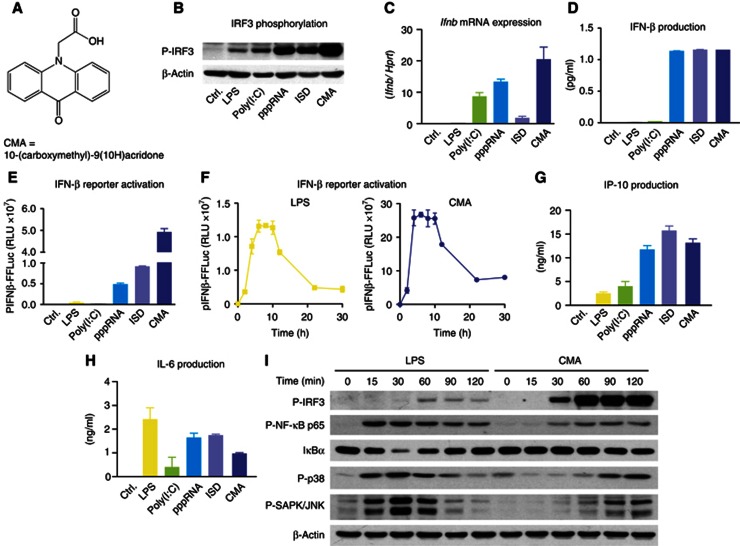
Intrigued by earlier publications on the antiviral activity of CMA (Figure 1A), we were interested in the molecular mechanisms of its activation, especially its putative receptor and associated signalling routes. We first assessed its ability to induce type I IFN production in macrophages in comparison to defined ligands for various PRR systems. To this effect, LPS (TLR4), transfected poly(I:C) (TLR3 and MDA5), transfected 5′triphosphate RNA (pppRNA/RIG-I) and transfected 45mer dsDNA (ISD/STING) were used. All ligands were tested at their previously determined optimal concentrations, and as readouts we assessed phosphorylation of IRF3, transcription of Ifnb mRNA, IFNβ protein levels and transactivation of the IFNβ promoter, using pIFNβ-Luc reporter mouse macrophages (Lienenklaus et al, 2009). Indeed, CMA induced robust IRF3 phosphorylation that was followed by strong Ifnb mRNA induction and translation (Figure 1B–E). CMA-mediated IFNβ production reached peak levels already 4 h after stimulation, even exceeding LPS in its readiness to trigger IFNβ synthesis (Figure 1F). Of note, for the various ligands studied, IRF3 phosphorylation, Ifnb mRNA induction and its translation did not go in parallel, which is attributable to the fact that additional ligand properties can play important roles in regulating transcription and translation of this key cytokine. Assessing other IRF3-dependent targets, such as IP-10, corroborated the notion of CMA being a potent trigger of antiviral immunity, yet NF-kB target cytokines (e.g., IL-6) were induced to a lesser degree in CMA-stimulated cells (Figure 1G,H). In line with this observation, assessing IRF3, MAPK and NF-kB activation following CMA stimulation showed that CMA led to synchronous and rapid activation of all these signalling cascades, with a predominant IRF3 signature. LPS, on the other hand, showed stronger NF-kB and MAPK activation, with a slight delay in IRF3 phosphorylation (Figure 1I). At the functional level, CMA blocked viral gene expression of VSV-based replicon particles in a dose-dependent manner (Supplementary Figure S1; Berger Rentsch and Zimmer, 2011). Altogether, these studies indicated that CMA is a rapid and potent inducer of antiviral immune responses in murine macrophages.
Figure 1.
CMA strongly induces type I IFN in primary mouse macrophages. (A) The chemical structure of CMA is depicted. (B–E and G–H) Bone marrow-derived macrophages were transfected with poly(I:C), pppRNA and ISD, or stimulated with LPS or CMA (500 μg/ml). (B) After 2 h, cells were collected and subjected to SDS–PAGE, and western blotting for phospho-IRF3 (P-IRF3) was performed. (C) Four hours after stimulation, transcription of the IFNβ gene (Ifnb) was assessed by quantitative RT–PCR, with normalization to HPRT1. (D) Eighteen hours after stimulation, IFNβ was measured in the supernatants by enzyme-linked immunosorbent assay (ELISA). (E) pIFNβ-firefly-luciferase (pIFNβ-FFLuc) macrophages were stimulated as indicated. After 18 h, cells were lysed with passive lysis buffer, and FFLuc activity was measured in the lysates. (F) pIFNβ-FFLuc macrophages were stimulated with LPS or CMA (500 μg/ml). Luciferase activity was assessed at the indicated time points (in hours). (G,H) Eighteen hours after stimulation, IP-10 (G) and IL-6 (H) were measured in the supernatants by ELISA. (I) Bone marrow-derived macrophages were stimulated with LPS or CMA (500 μg/ml). Total protein was collected at indicated time points (in minutes) after stimulation and was assessed for P-IRF3, phospho-NF-κB-p65 (P-NF-κB p65), IkBα, phospho-p38 (P-p38) or phospho-SAPK/JNK (P-SAPK/JNK). Representative results out of three independent experiments are depicted.
Source data for this figure is available on the online supplementary information page.
CMA-dependent type I IFN production requires STING
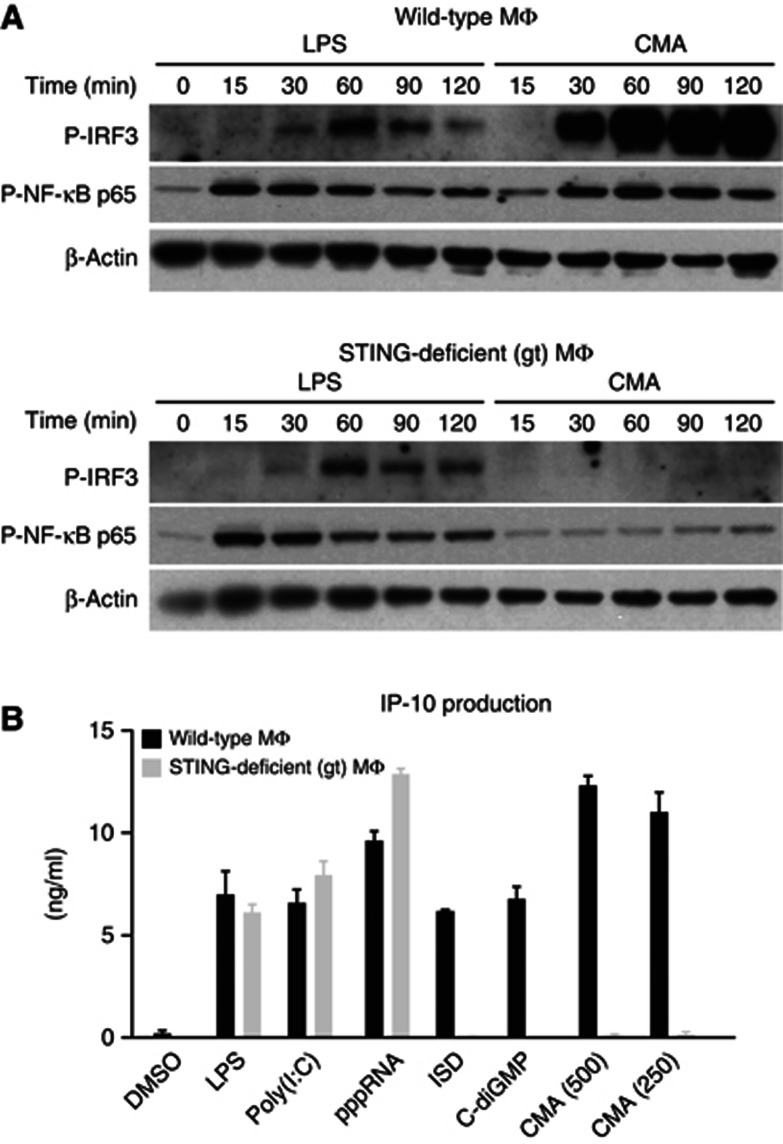
While most immortalized cell lines did not respond to CMA (e.g., immortalized murine embryonic fibroblasts (MEFs), HEK 293T cells, HeLa cells), early-passage MEFs could be stimulated with CMA. This allowed us to use MEFs from TBK1/IKKε-deficient mice, to determine the role of these canonical kinases in IRF3 phosphorylation. As expected, IRF3 phosphorylation was completed blunted in MEFs deficient for TBK1 and IKKε (Supplementary Figure S2A). We next assessed the role of TLR signalling pathways using macrophages from mice deficient in MyD88 or TRIF. Neither MyD88 nor TRIF were required for CMA sensing, which ruled out an involvement of TLRs (Supplementary Figure S2B–D). At the same time, absence of MAVS, the shared signalling adapter of RIG-I and MDA5, had no impact on CMA-mediated antiviral immunity (Supplementary Figure S2E,F). We next went on to study the role of STING in CMA-triggered type I IFN production. To this effect, we used macrophages from a mutant mouse strain, Goldenticket, that harbours a missense mutation (I199N) in STING (Sauer et al, 2011). Indeed, macrophages from these mice showed no detectable activation of IRF3 or NF-kB (Figure 2A) and also a complete absence of cytokine production (Figure 2B). In addition, the antiviral activity elicited by CMA treatment was completely abolished when STING-deficient macrophages were stimulated (Supplementary Figure S3). In line with the critical requirement of STING in CMA sensing, a human HEK 293T cell line engineered to stably express murine STING responded to CMA, with a strong increase in pIFNβ or pELAM reporter activity (Supplementary Figure S4). As expected, this cell line also showed a strong gain-of-function signal with regards to c-diGMP sensing, which is usually non-functional in unmodified HEK 293T cells. Altogether, these results indicated that STING was required and also sufficient for CMA recognition.
Figure 2.
Loss of STING leads to complete abrogation of CMA-induced cytokine production. (A) Bone marrow-derived wild-type and STING-deficient macrophages were stimulated with LPS and CMA (500 μg/ml). Phospho-IRF3 (P-IRF3) and phospho-NF-κB-p65 (P-NF-κB p65) were assessed at indicated time points (in minutes) after stimulation. (B) Bone marrow-derived wild-type and STING-deficient macrophages were transfected with poly(I:C), pppRNA, ISD and c-diGMP, or stimulated with LPS and CMA (500 or 250 μg/ml) in duplicates. Supernatants were collected after 18 h, and IP-10 levels were measured by enzyme-linked immunosorbent assay. Representative results out of two independent experiments are depicted, whereas data are presented as mean values+s.e.m.
Source data for this figure is available on the online supplementary information page.
CMA fails to activate human STING
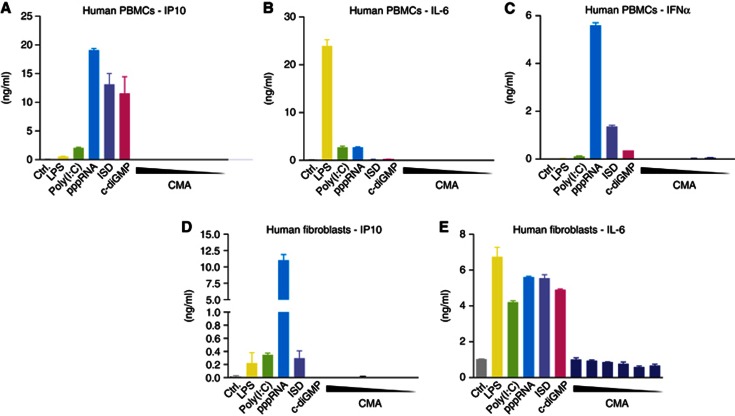
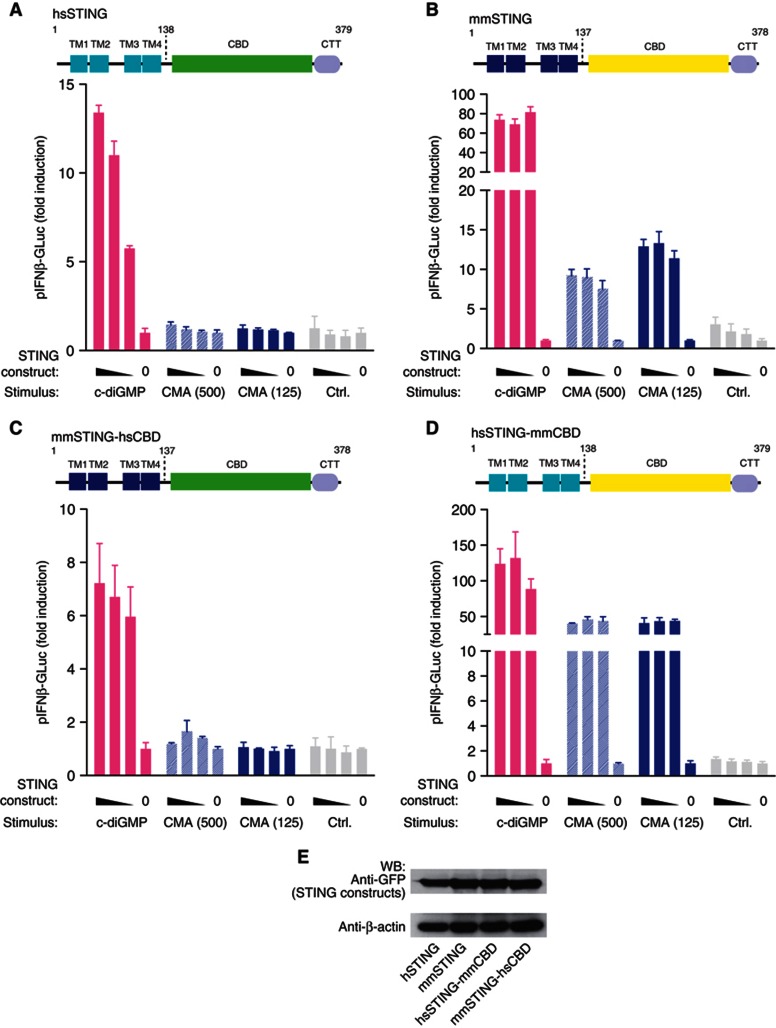
Despite the fact that CMA had been reported to induce type I IFN responses in human cells (Silin et al, 2009), we were unable to elicit type I IFN production in various cell types of the human system. Human PBMCs readily responded to 5′pppRNA, poly(I:C), DNA, c-diGMP and LPS stimulation, yet CMA failed to induce detectable cytokine responses, even at high doses (Figure 3A–C). A similar picture was seen when primary human fibroblasts were used (Figure 3 D). In line with this finding, when we transiently transfected HEK 293T cells with a human STING construct, no CMA response could be detected, despite the fact that overexpression of human STING rendered 293T cells sensitive to c-diGMP (Figure 4A). At the same time, transient overexpression of murine STING made HEK 293T cells responsive towards both c-diGMP and CMA (Figure 4B). The fact that human STING displayed c-diGMP-dependent signalling capacity in HEK 293T cells suggested that the C-terminal LBD of STING could be held responsible for the insensitivity towards CMA. To address this question, we constructed chimeric STING constructs, in which the N- and C-terminal domains of the murine or human STING were exchanged: mmSTING(1-137)-hSTING(139-379) or hSTING(1-138)-mmSTING(138-378) (Figure 4C,D). Testing these constructs in HEK 293T cells revealed that as long as the murine LBD was present (hSTING(1-138)-mmSTING(138-378); Figure 4D), CMA was able to trigger type I IFN production. Altogether, these results indicated that unresponsiveness of human cells towards CMA could be explained by species-dependent differences in the LBD of STING.
Figure 3.
Human cells do not respond to CMA. (A–E) Primary human PBMCs and fibroblasts were transfected with poly(I:C), pppRNA, ISD and c-diGMP, or stimulated with LPS and decreasing concentrations of CMA (4000—125 μg/ml in twofold dilutions). IP-10, IL-6 and IFNα levels in the supernatants of stimulated PBMCs (A–C) and fibroblasts (D,E) were determined by enzyme-linked immunosorbent assay. Representative results out of three independent experiments are depicted.
Figure 4.
Species-specificity CMA activity is determined by the C-terminal LBD of STING. (A–D) 293T cells were transiently transfected with the indicated STING constructs (25, 12.5, 6.25 and 0 ng), whereas 12.5 ng of pIFNα-GLuc reporter plasmid were included. For titrations, an empty pCI vector served as a stuffer to obtain 200 ng total plasmid DNA. After 24 h, cells were transfected with c-diGMP or stimulated with CMA (500 and 125 μg/ml). Luciferase activity was measured after an additional period of 24 h in the supernatant, and data were normalized to the condition without STING overexpression. Plasmids coding for full-length hSTING (A), mmSTING (B), mmSTING(1-137)-hSTING(139-379) (C) and hSTING(1-138)-mmSTING(138-378) (D) were tested. (E) Expression of the above described constructs was studied in 293T cells 24 h after transfection (200 ng per 96-well plate) using western blot, whereas β-actin served as a loading control. Representative results out of three independent experiments are depicted.
Source data for this figure is available on the online supplementary information page.
Using the murine STING construct, we next wanted to address the role of functionally relevant point mutations for the recognition of CMA. The previously described null-mutant I199N most likely perturbs the structure of STING and thereby abolishes c-diGMP binding and signalling (Burdette et al, 2011). In line with this notion, overexpression of mmSTING-I199N showed no pIFNβ-Luc transactivation upon CMA- or c-diGMP-mediated stimulation (Supplementary Figure S5A,B). Interestingly, mmSTING-R231A, a mutant that has previously been described to completely blunt c-diGMP-mediated STING activation despite binding, showed normal activity upon CMA stimulation (Supplementary Figure S5C). These data demonstrated that c-diGMP sensing could be dissociated from CMA recognition at the receptor level.
CMA does not bind the human STING LBD
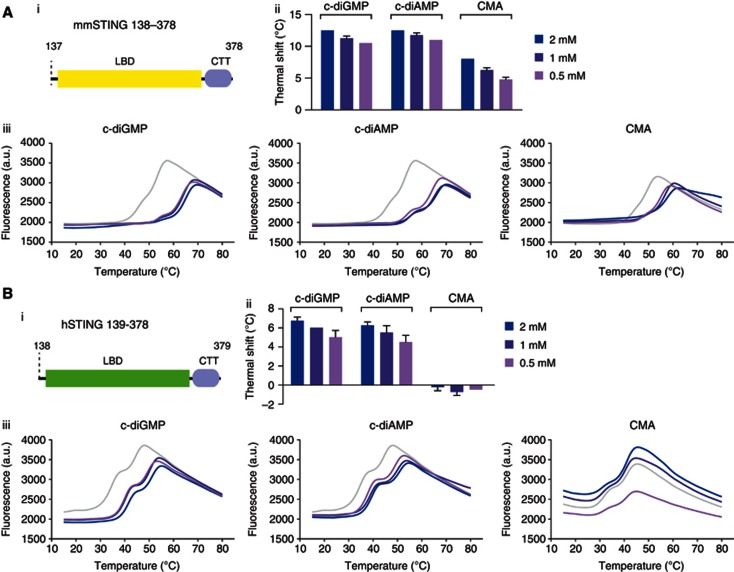
The failure of CMA to activate human STING could be explained by several scenarios. Foremost, we wanted to rule out the possibility that human STING does not bind to CMA. To address this question, we expressed and purified the LBD of murine and human STING in E. coli and tested its ability to associate with various ligands, using DSF. DSF indirectly assesses the association of a low molecular weight compound to a purified protein by measuring the stability of a protein–compound complex as a function of temperature (Niesen et al, 2007). As indicated by a robust thermal shift, the LBD of murine STING associated with its known ligands c-diGMP and c-diAMP, and also binding to CMA, could be observed (Figure 5A). At the same time, also the LBD of human STING displayed association with c-diGMP and c-diAMP, as indicated by a dose-dependent shift in thermal stability. However, addition of CMA had no detectable impact on the thermal stability of human STING, indicating that CMA does not bind human STING (Figure 5B). Despite high homology, murine and human STING show least conservation in their CTT region. As such, we additionally wanted to rule out that this part of STING was required for CMA binding. To this effect, we generated truncated versions of the murine STING LBD lacking the CTT. However, testing this variant showed similar binding properties as the full-length LBD, indicating that the CTT is not required for ligand binding by STING (Supplementary Figure S6).
Figure 5.
DSF implicates direct binding of CMA to murine, but not to human STING. (A,B) The interaction of STING with c-diGMP, c-diAMP and CMA were analysed by thermal shift assay. Purified murine STING (A) and human STING (B) were tested with different concentrations of c-diGMP/c-diAMP/CMA; (i) Schematic views of the protein domains used for binding studies are shown; (ii) thermal shifts of (iii) fluorescence intensity versus temperature are shown. Representative results out of two independent experiments are depicted for the temperature curves (ii), whereas mean values+s.e.m. out of two independent experiments are depicted for the thermal shift graphs (iii).
The crystal structure shows a c-diGMP-like STING interaction for CMA
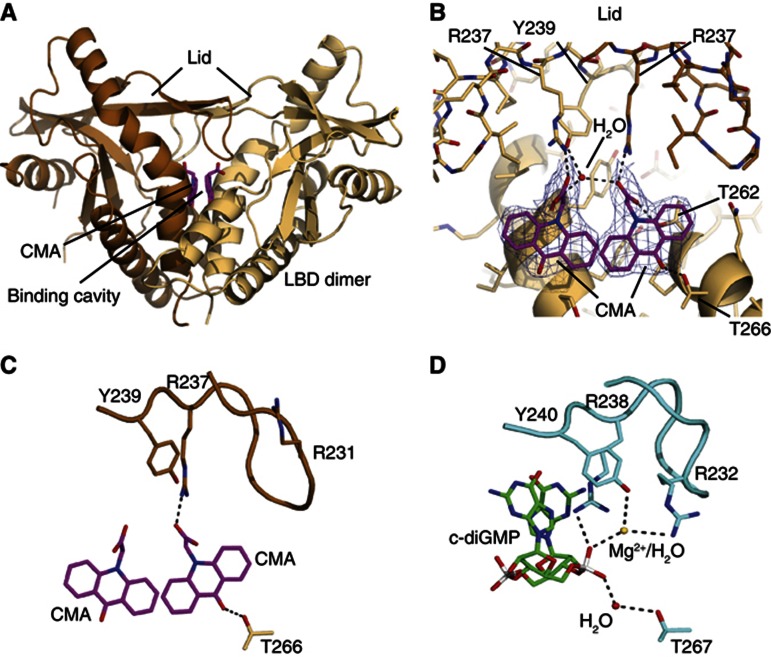
Recently, five independent groups determined the crystal structure of the LBD of human STING bound to c-diGMP (Huang et al, 2012; Ouyang et al, 2012; Shang et al, 2012; Shu et al, 2012; Yin et al, 2012). In these studies it was shown that the LBD of STING constitutes a preformed dimer that is stabilized through homotypic interaction at a hydrophobic interface. This STING dimer forms a V-shaped pocket, in which one c-diGMP molecule is buried at its bottom. To see how CMA binds murine STING, we crystallized murine STING LBD in the presence of CMA and determined the crystal structure to 2.75 Å resolution (Supplementary Table S1). The final model comprises two STING LBDs along with two well-resolved CMA molecules (Figure 6A,B). Murine STING LBD forms a dimer with high overall structural similarity to the previously determined human STING LBDs (Supplementary Figure S7). Two CMA molecules are located in the deep central c-diGMP-binding pocket at the dimer interface. The acridone ring moieties of both CMAs partially stack to each other (∼4 Å distance) in a parallel, laterally shifted orientation, resembling the twofold symmetry of c-diGMP. They are situated near the phosphate–ribose binding site for c-diGMP, but due to the flat shape can wedge deep into the helical bundle core of the LBD dimer. As a result, CMA binds directly to Thr226 at the bottom of the ligand-binding pocket, whereas c-diGMP binds via a water molecule (Figure 6C,D).
Figure 6.
Structural basis for CMA recognition. (A) Ribbon model of the mouse STING dimer (light and dark brown) with highlighted secondary structure. The two bound CMA molecules are shown as magenta stick models. (B) Close-up view of the CMA-binding site with superimposed 2mFo-DFc electron density (blue; contoured at 1.4). One STING protomer is shown in light brown. For the other protomer (brown), only the lid is displayed. Folding of the lid via hydrogen bonds to Arg237 and Tyr239 suggests how CMA activates mouse STING. (C,D) Side-by-side comparison of CMA bound to mouse STING (C) and c-diGMP bound to human STING (D) showing selected interactions. CMA folds the lid differently from c-diGMP, due to steric clash with Arg231 (human Arg232), which binds c-diGMP via a magnesium ion/water molecule.
Significantly, we find two well-ordered lid domains in a four-stranded antiparallel β-sheet that closes the CMA/c-diGMP-binding pocket (Figure 6A,B). The lids are generally in a similar conformation than the folded lids in the human STING–c-diGMP complex reported by Huang et al (2012) (PDB-code: 4F5D), but there are also differences. The carboxymethyl groups of the CMA moieties face and stabilize the conformation of the lid by forming salt bridges to both Arg237 residues. The equivalent human Arg238 residues stack between the two c-diGMP guanine moieties, thereby stabilizing the lid. The space of the guanine moieties is unoccupied by CMA. As a result, murine Arg237 and Tyr167, plus Tyr240, directly stack instead of sandwiching guanine as observed in the human c-diGMP complex. The direct stacking induces or enables further closure of the V-shaped binding cleft compared to the c-diGMP-bound conformation, which may contribute to active signalling. However, the tips of the lids fold differently in the presence of CMA and c-diGMP. In particular, Arg231 (human Arg232), which binds the phosphates of c-diGMP via a magnesium ion or water molecule (Huang et al, 2012), is pushed to the surface due to steric hindrance from the acridone ring. In summary, CMA binds to the c-diGMP-binding pocket, with two stacked CMA molecules mimicking the symmetric c-diGMP. CMA induces a conformation in the LBD dimer that is similar, if not more pronounced than the proposed signalling conformation of human LBD, with folded lids in the presence of c-diGMP, but also results in a somewhat altered lid conformation. Altogether, the structure can explain why CMA activates murine STING.
Discussion
In line with early reports on the antiviral activity of CMA (Storch and Kirchner, 1982; Storch et al, 1986), our data confirm the extraordinary type I IFN-inducing capacity of CMA in the murine system. Further analysis of signalling cascades involved in IFNβ transactivation show a quick and strong phosphorylation of IRF3, whereas the activation of the NF-κB and MAPK pathways is slightly delayed and less prominent compared to TLR signalling. This observation is also reflected by the cytokine profile, as CMA induces high levels of IFNβ as opposed to IL-6. Studies using knockout cells revealed that CMA triggers antiviral immunity cells through the STING–TBK1–IRF3 route, which could be further corroborated by gain-of-function experiments in cells devoid of functional mmSTING.
Unexpectedly, human cells showed complete unresponsiveness towards CMA, although c-diGMP-dependent STING activity was observed. We could ascribe this to species-dependent differences within the C-terminal LBD of STING, as HEK 293T cells expressing a chimeric STING protein equipped with the murine LBD readily responded towards CMA. Further studies could pinpoint the failure of human STING to respond to CMA, to the inability of the human LBD to form a stable complex. Of note, these experiments do not formally proof that human STING cannot bind to CMA, yet in light of the functional data this appears to be the most likely scenario. The species-specific activation of murine STING by CMA is surprising, especially given the fact that several reports on antiviral activity in the human system both in vitro and in vivo exist (Vershinina et al, 2002; Zarubaev et al, 2003). One possibility is that these antiviral activities are due to direct inhibition of viral replication, independent of the innate immune response (e.g., inhibition of viral polymerases). At the same time, it is conceivable that higher local concentrations of CMA can be achieved during systemic application or that other cell types are activated, both leading to a STING-dependent type I IFN response. Nevertheless, our results clearly question a predominant role for type I IFN induction by CMA in the human system.
FAA (flavone-8-acetic acid) and DMXAA (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)-acetic acid) are two additional tricyclic small-molecule compounds that have been reported to trigger potent type I IFN responses in the murine system (Hornung et al, 1988; Futami et al, 1991; Perera et al, 1994; Roberts et al, 2007). Both compounds have been pursued for their potent antitumour activity in murine tumour models, and for both compounds it was shown that activation of the immune system plays a pivotal role in therapeutic activity (Ching and Baguley, 1987; Hornung et al, 1988; Pang et al, 1998). FAA, however, failed to induce type I IFN responses in human cells (Futami et al, 1991) and, moreover, it failed to display antitumour activity in clinical trials (Bibby and Double, 1993). In parallel, despite strong activity in the murine system, DMXAA by itself also showed limited immunostimulatory capacity in human cells (Patel et al, 1997; Philpott et al, 2001; Gobbi et al, 2006). At the same time, DMXAA in combination with a platinum-based chemotherapy did not show any efficacy in a large phase III clinical trial for the treatment of non-small cell lung cancer (Lara et al, 2011). Most recently, it has been reported that DMXAA triggers type I IFN responses in a STING-dependent fashion (Brunette et al, 2012; Prantner et al, 2012). In these reports, only murine cells were tested and direct binding of DMXAA to STING was not assessed. Nevertheless, given the similarity of CMA and DMXAA at the molecular level, we would expect that DMXAA also directly engages STING in a species-specific manner. As such, it appears likely that the failure of DMXAA in the human system can be attributed to its inability to trigger STING activation.
At the structural level, the species-specific recognition of CMA is unlikely to be attributable to the binding pocket itself. This region, which is mainly formed by α-helices, α1 and α3, is more or less invariant between the human and the murine system, with all direct interactors of CMA being identical. It is possible that more subtle structural differences prevent binding of CMA to human STING. An alternative, probably more likely, explanation concerns loops β2 and β3 of the LBD, which are critically involved in the coordination of c-diGMP. While flexible in the apo form of human STING, this loop region undergoes a conformational rearrangement upon c-diGMP binding, thereby functioning as a ‘lid’ that keeps c-diGMP within the binding pocket (Huang et al, 2012). Two conserved residues within the tip of this loop (human: Arg238, Tyr240) have been shown to interact with c-diGMP, and point mutagenesis data support the critical role of the loop region in c-diGMP recognition. In support of a similar activation mechanism, CMA binds Arg237 of murine STING. Interestingly, the previously reported R231A mutation within this loop region impairs c-diGMP-mediated IFN induction even though DNA- and CMA-dependent IFN induction are not affected (Burdette et al, 2011). These data suggest that the loop region is differently involved in CMA sensing, as opposed to c-diGMP recognition. Consistent with the mutational data, Arg231 is not involved in recognition of CMA, and the tip of the lid, where this conserved residue is located, is folded differently compared to the human STING bound to c-diGMP (Figure 6C). As such, species-dependent differences that may stabilize or allow this altered conformation could account for the observed unresponsiveness of hSTING towards CMA. Yet, additional studies will be required to pinpoint this phenomenon to an exact structural determinant.
This is the first report of a direct STING ligand that is not a cyclic dinucleotide. Most intriguingly, CMA is not sensed as a monomeric ligand within the ligand-binding pocket of the STING dimer; yet, two molecules positioned in a rotational symmetry are required to activate STING. We assume that other reported tricyclic small-molecule antivirals (e.g., tilorone, DMXAA, etc) are also sensed as a dimeric ligand in a fashion similar to CMA. Moreover, it is tempting to speculate that beyond these synthetic compounds, natural ligands exist that also follow a CMA-like recognition mode. In this regard, several bioflavonoids reported to exert antiviral activity might be interesting candidates. At the same time, the novel class of STING ligands described here might open novel avenues to develop compounds that block STING activation, with favorable drug-like properties. Given the likely involvement of STING in sensing endogenous DNA in the course of sterile inflammatory conditions (Ahn et al, 2012; Gall et al, 2012), pharmacological targeting of STING might constitute a reasonable therapeutic venture.
Materials and methods
Reagents
Poly(I:C) and CMA were purchased from Sigma Aldrich. Ultra-pure LPS from E. coli was purchased from Invivogen. c-diGMP and c-diAMP were from BioLog. GeneJuice Transfection Reagent was from Novagen. Lipofectamine 2000 was from Life Technologies. Goat anti-rabbit-IgG-HRP, goat-anti-mouse-IgG-HRP and anti-β-actin-IgG-HRP were from Santa Cruz Biotechnology. Passive lysis buffer was purchased from Promega.
Plasmids
Expression plasmids coding for human or murine STING were cloned into pEFBOS coding for an N-terminal GFP or an N-terminal mCherry tag. Chimeric STING constructs, human STING (AA1–138)–murine STING (AA138–378) and murine STING (AA1–137)–human STING (AA139–379) were generated by ligation-independent cloning (Aslanidis and de Jong, 1990; Schmid-Burgk et al, 2012). The STING mutants (I199N, R231A in pEFBOS-mCherry) were generated by point mutagenesis PCR. pCI-empty was used as a stuffer. All primer sequences used for cloning are available upon request.
Cell culture
PBMCs were isolated from whole blood of healthy donors. After Ficoll density-gradient centrifugation (Biochrom), red blood cells were lysed using lysing buffer (BD Biosciences). PBMCs were seeded with a density of 4 × 106 per ml at 100 μl in a 96-well plate containing RPMI supplemented with 10% (v/v) FCS, sodium pyruvate, penicillin and streptomycin (all from Life Technologies). 293T and primary human fibroblasts were cultured in DMEM supplemented with 10% (v/v) FCS, sodium pyruvate (all from Life technologies) and Ciprofloxacin (Bayer Schering Pharma). Primary macrophages were generated from mouse bone marrow cells that were cultured for 7 days in DMEM with the same additives as described above and 30% (v/v) L929 supernatant.
Immunoblotting
Primary macrophages were lysed in 1 × Laemmli buffer and denatured at 95°C for 5 min. Probes were separated by 10% SDS–PAGE and transferred onto nitrocellulose membranes. Blots were incubated with anti-Phospho-IRF3 (number 4947), anti-Phospho-NF-kappaB-p65 (number 3033), anti-Phospho-p38 (number 4511), anti-IkappaB-alpha (number 4814) or anti-Phospho-SAPK/JNK (number 9255) from Cell Signaling Technology.
Transfection
For transfection experiments, primary macrophages were seeded with a density of 1 × 105 per ml. Cells were transfected with poly(I:C) (2 μg/ml), pppRNA (1.33 μg/ml), ISD (2 μg/ml) and c-diGMP (8.66 μg/ml) using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. LPS (200 ng/ml) and CMA were directly added to the medium. Human PBMCs were transfected as described above, at a density of 4 × 106 per ml, human fibroblasts at a density of 1.5 × 105 per ml. For western blot experiments, cells were lysed after 2 h, if not indicated otherwise. For cytokine assays, supernatants were collected after 18–20 h. For RNA isolation, cells were lysed after 4 h.
Enzyme-linked immunosorbent assay
Cell culture supernatants were assayed for human IP-10 (BD Biosciences), human IL-6 (BD Biosciences), human IFNα (eBioscience), mouse IFNβ (BioLegend), mouse IP-10 (R&D Systems) or mouse IL-6 (BD Biosciences), according to the manufacturer’s instructions.
Quantitative real-time PCR analysis
RNA from macrophages was reverse transcribed using the RevertAid First Strand cDNA Synthesis kit (Fermentas) and quantitative PCR analysis was performed on an ABI 7900HT. All murine gene expression data are presented as relative expression to HPRT1. Primer sequences are available upon request.
Luminescence assays
pIFNβ-FFLuc macrophages (Lienenklaus et al, 2009) were stimulated as indicated and were lysed with passive lysis buffer 18–20 h after stimulation, to determine luminescence. For VSV*ΔG(Luc) replicon assays, macrophages were treated with ligands or supernatants as indicated and subsequently infected at an MOI of 10 with the VSV*ΔG(Luc) replicon virus particles. Firefly-luciferase activity was measured in the lysates using a 2104 EnVision Multilabel reader from Perkin-Elmer.
Plasmid overexpression experiments
293T cells were seeded with a density of 2 × 105 cells per ml at 100 μl 96-well plate. IFNβ (12.5 ng) promoter–reporter plasmid pIFNβ–GLuc or pELAM-GLuc, and STING constructs were transfected using GeneJuice according to the manufacturer’s instructions. For titration experiments, empty pCI vector was used as a stuffer. After 24 h, the cells were transfected with c-diGMP or were stimulated with CMA. After further 20 h, GLuc activity was measured in the supernatants using coelenterazine as a substrate.
Cell viability assays
Cell viability was assessed using CellTiter-Blue (Promega) according to the manufacturer’s instructions. The assay was performed immediately after (Gaussia) or before (Firefly) luciferase measurement.
Differential scanning fluorimetry
Thermal shift assays were carried out using a CFX 96TM Real-Time System (Biorad) for recording the fluorescence signal (HEX: Ex/Em: 450–490/560–580 nm) as a function of temperature. The temperature gradient was set from 15 to 80°C, with an increment of 0.5°C and incubation steps of 15 s. Each 20 μl reaction (buffer: 20 mM Tris, 150 mM NaCl, (10% DMSO for reactions with CMA), pH 7.5), with or without 2 mM/1 mM/0.5 mM of c-diGMP/c-diAMP/CMA contained 1 mg/ml of STING and a dilution of 1:500 of SYPRO Orange dye (Invitrogen).
Cloning of human and mouse STING constructs for protein expression
Human STING AA139–379 (R220H+H232R) was cloned from a human macrophage cDNA library into pET28-SUMO1-eGFP vector via BamHI and NotI restriction sites. The mouse STING constructs AA138–378 and AA138–341 were cloned from a mouse lung tissue cDNA library into pET28-SUMO1-eGFP vector via AgeI and NotI restriction sites. These plasmids were used to transform E. coli Rosetta (DE3) protein expression strain cells (Novagen).
Expression and purification of human and mouse STING constructs
For all used STING constructs, the expression and purification procedure was the following: E. coli Rosetta (DE3) cells were grown in 3 l of LB media supplemented with Kanamycin (50 mg/l) and Chloramphenicol (34 mg/l) at 37°C for 3 h, to OD600=0.8. Expression of N-terminal His6-SUMO1-tagged STING was induced by adding IPTG (Roth) to a final concentration of 0.2 mM. Expression was done overnight at 18°C. Cells were collected by centrifugation and were resuspended in lysis buffer (50 mM Tris, 500 mM NaCl, 10 mM imidazole, 5% glycerol, 2 mM β-mercaptoethanol, pH 7.5) and lysed by sonication. The soluble His6-SUMO1-STING was purified by Ni-affinity chromatography. The His6-SUMO1-tag was removed by proteolytic cleavage with SenP2 protease during dialysis overnight (20 mM Tris, 150 mM NaCl, 3% glycerol, 2 mM β-mercaptoethanol, pH 7.5) and a second Ni-affinity chromatography step. To remove additional contaminants, a HiTrap Q FF (GE Healthcare) purification was applied. Finally, the STING containing flow-through was used in a HiLoadTM 26/60 Superdex 75 prep grade (GE Healthcare) size-exclusion chromatography step (20 mM Tris, 150 mM NaCl, pH 7.5). Purified STING was concentrated (10–15 mg/ml) with a 10 kDa cut-off centrifugal concentrator device (Millipore) and was flash frozen in liquid nitrogen for storage (−80°C).
Crystallization of mmSTING AA149–348 with CMA
A protein solution of mmSTING (8 mg/ml) was saturated with CMA (by adding solid powder due to its low solubility in aqueous solution) and incubated on ice for 1 h. Prior to crystallization, the protein solution was centrifugated and filtered to remove solid CMA. Crystals were grown using the hanging drop vapour diffusion method with drops of 1:1 ratio protein:reservoir. The reservoir solution contained 0.1 M HEPES, pH 7, 1.65 M ammonium sulphate, 2% PEG 400 (v/v) and saturating amounts of CMA. mmSTING crystals appeared after ∼4 days at 20°C. For cryoprotection, the crystals were soaked in reservoir solution containing 12% (±)-1,3-butanediol (v/v) before flash freezing in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were collected at beamline X06SA at the Swiss Light Source (Villigen, Switzerland). Data processing was carried out using XDS (Kabsch, 2010). The structure was solved by molecular replacement with PHASER (McCoy et al, 2007) from the ccp4 package (Winn et al, 2011), using a mouse search model created from the human STING structure (4EMT (Shu et al, 2012)) by ClustalW2 alignment (Goujon et al, 2010; Larkin et al, 2007) and modelling with the SWISS-MODEL structure homology-modelling server (Peitsch, 1995; Arnold et al, 2006; Kiefer et al, 2009). The structure was refined by rounds of manual model building carried out with COOT (Emsley and Cowtan, 2004) and refinement using Phenix (Adams et al, 2010). The final structure with R/Rfree=21/23.7 shows good stereochemistry and no outliers in the Ramachandran plot. Figures were created with PyMOL (Schrödinger, 2010). Coordinates and structure factors have been deposited with the Protein Data Bank (accession number 4JC5).
Supplementary Material
Supplementary figures source data
Acknowledgments
We thank Dr U Kalinke for providing us MAVS-deficient macrophages; Dr S Akira for providing us TBK1/IKKε-deficient MEFs, and MyD88- and TRIF-deficient macrophages; Dr S Weiss for providing us pIFNβ-FFLuc mice; Dr R Vance for providing us goldenticket (Gt) mice; and Dr G Zimmer for providing us VSV*ΔG(Luc) replicon particles. We thank EMBL for supplying the pET28-SUMO1-eGFP vector and the SenP2 vector. We thank Stefan Emming for help with cloning and protein purification, and Dr Gregor Witte for help with model refinement. We thank the Swiss Light Source and the European Synchrotron Radiation Facility for generous beam time and excellent on-site support. This work was supported by grants from the German Research Foundation (SFB670), the European Research Council (ERC–2009–StG 243046) to VH, NIH grant U19AI083025 to K-PH and a BIF fellowship to TC. VH is a member of the excellence cluster ImmunoSensation.
Author contributions: TC performed all experiments, except the DSF studies and the crystallographic studies, which were performed by TD. AA generated constructs for the study. K-PH and VH conceived or designed the experiments. TC, TD, AA, K-PH and VH analysed the data, TC and VH wrote the manuscript, and VH conceived and supervised the study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Gutman D, Saijo S, Barber GN (2012) STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci USA 109: 19386–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18: 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Rentsch M, Zimmer G (2011) A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One 6: e25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby MC, Double JA (1993) Flavone acetic acid--from laboratory to clinic and back. Anticancer Drugs 4: 3–17 [DOI] [PubMed] [Google Scholar]
- Brehm G, Storch E, Kirchner H (1986) Characterization of interferon induced in murine macrophage cultures by 10-carboxymethyl-9-acridanone. Nat Immun Cell Growth Regul 5: 50–59 [PubMed] [Google Scholar]
- Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS Stetson DB. (2012) Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med 209: 1969–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478: 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching LM, Baguley BC (1987) Induction of natural killer cell activity by the antitumour compound flavone acetic acid (NSC 347 512). Eur J Cancer Clin Oncol 23: 1047–1050 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Futami H, Eader LA, Komschlies KL, Bull R, Gruys ME, Ortaldo JR, Young HA, Wiltrout RH (1991) Flavone acetic acid directly induces expression of cytokine genes in mouse splenic leukocytes but not in human peripheral blood leukocytes. Cancer Res 51: 6596–6602 [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M Jr., Barber GN, Stetson DB (2012) Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi S, Belluti F, Bisi A, Piazzi L, Rampa A, Zampiron A, Barbera M, Caputo A, Carrara M (2006) New derivatives of xanthenone-4-acetic acid: synthesis, pharmacological profile and effect on TNF-alpha and NO production by human immune cells. Bioorg Med Chem 14: 4101–4109 [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung RL, Young HA, Urba WJ, Wiltrout RH (1988) Immunomodulation of natural killer cell activity by flavone acetic acid: occurrence via induction of interferon alpha/beta. J Natl Cancer Inst 80: 1226–1231 [DOI] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD (2012) The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol 19: 728–730 [DOI] [PubMed] [Google Scholar]
- Isaacs A, Cox RA, Rotem Z (1963) Foreign nucleic acids as the stimulus to make interferon. Lancet 2: 113–116 [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J (1957) Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147: 258–267 [PubMed] [Google Scholar]
- Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, Talbot BG, Brouillette E, Malouin F (2007a) Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol 178: 2171–2181 [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Newstead MW, Zeng X, Hyodo M, Hayakawa Y, Bhan U, Liang H, Standiford TJ (2007b) Cyclic di-GMP stimulates protective innate immunity in bacterial pneumonia. Infec Immun 75: 4942–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman HE, Centifanto YM, Ellison ED, Brown DC (1971) Tilorone hydrochloride: human toxicity and interferon stimulation. Proc Soc Exp Biol Med 137: 357–360 [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37: D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MJ, Cleeland R, Grunberg E (1976) Antiviral activity of 10-carboxymethyl-9-acridanone. Antimicrob Agents Chemother 9: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MJ, Taylor JL, Grossberg SE (1981) Induction of interferon in mice by 10-carboxymethyl-9-acridanone. Methods Enzymol 78: 284–287 [DOI] [PubMed] [Google Scholar]
- Krueger RE, Mayer GD (1970) Tilorone hydrochloride: an orally active antiviral agent. Science 169: 1213–1214 [DOI] [PubMed] [Google Scholar]
- Lara PN Jr, Douillard JY, Nakagawa K, von Pawel J, McKeage MJ, Albert I, Losonczy G, Reck M, Heo DS, Fan X, Fandi A, Scagliotti G (2011) Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. J Clin Oncol 29: 2965–2971 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lienenklaus S, Cornitescu M, Zietara N, Lyszkiewicz M, Gekara N, Jablonska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, Weiss S (2009) Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol 183: 3229–3236 [DOI] [PubMed] [Google Scholar]
- Mayer GD, Krueger RF (1970) Tilorone hydrochloride: mode of action. Science 169: 1214–1215 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, Fitzgerald KA, Hayakawa Y, Vance RE (2009) A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med 206: 1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2: 2212–2221 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ (2012) Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36: 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JH, Cao Z, Joseph WR, Baguley BC, Ching LM (1998) Antitumour activity of the novel immune modulator 5,6-dimethylxanthenone-4-acetic acid (DMXAA) in mice lacking the interferon-gamma receptor. Eur J Cancer 34: 1282–1289 [DOI] [PubMed] [Google Scholar]
- Patel S, Parkin SM, Bibby MC (1997) The effect of 5,6-dimethylxanthenone-4-acetic acid on tumour necrosis factor production by human immune cells. Anticancer Res 17: 141–150 [PubMed] [Google Scholar]
- Peitsch MC (1995) Protein modeling by E-mail. Nat Biotechnol 13: 658–660 [Google Scholar]
- Perera PY, Barber SA, Ching LM, Vogel SN (1994) Activation of LPS-inducible genes by the antitumor agent 5,6-dimethylxanthenone-4-acetic acid in primary murine macrophages. Dissection of signaling pathways leading to gene induction and tyrosine phosphorylation. J Immunol 153: 4684–4693 [PubMed] [Google Scholar]
- Philpott M, Ching LM, Baguley BC (2001) The antitumour agent 5,6-dimethylxanthenone-4-acetic acid acts in vitro on human mononuclear cells as a co-stimulator with other inducers of tumour necrosis factor. Eur J Cancer 37: 1930–1937 [DOI] [PubMed] [Google Scholar]
- Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA, Vogel SN (2012) 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates Stimulator of Interferon Gene (STING)- dependent innate immune pathways and is regulated by mitochondrial membrane potential. J Biol Chem 287: 39776–39788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN (2007) The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med 204: 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem Z, Cox RA, Isaacs A (1963) Inhibition of virus multiplication by foreign nucleic acid. Nature 197: 564–566 [DOI] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE (2011) The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infec Immun 79: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Xie Z, Frank S, Virreira Winter S, Mitschka S, Kolanus W, Murray A, Benenson Y (2012) Rapid hierarchical assembly of medium-size DNA cassettes. Nucleic Acids Res 40: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger LLC (2010) The PyMOL Molecular Graphics System Version 1.3 [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L (2012) Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol 19: 725–727 [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P (2012) Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 19: 722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silin DS, Lyubomska OV, Ershov FI, Frolov VM, Kutsyna GA (2009) Synthetic and natural immunomodulators acting as interferon inducers. Curr Pharm Des 15: 1238–1247 [DOI] [PubMed] [Google Scholar]
- Storch E, Kirchner H (1982) Induction of interferon in murine bone marrow-derived macrophage cultures by 10-carboxymethyl-9-acridanone. Eur J Immunol 12: 793–796 [DOI] [PubMed] [Google Scholar]
- Storch E, Kirchner H, Brehm G, Huller K, Marcucci F (1986) Production of interferon-beta by murine T-cell lines induced by 10-carboxymethyl-9-acridanone. Scand J Immunol 23: 195–199 [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA 106: 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JL, Schoenherr C, Grossberg SE (1980a) Protection against Japanese encephalitis virus in mice and hamsters by treatment with carboxymethylacridanone, a potent interferon inducer. J Infect Dis 142: 394–399 [DOI] [PubMed] [Google Scholar]
- Taylor JL, Schoenherr CK, Grossberg SE (1980b) High-yield interferon induction by 10-carboxymethyl-9-acridanone in mice and hamsters. Antimicrob Agents Chemother 18: 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinina MY, Narovlyansky AN, Deryabin PG, Amchenkova AM, Ivanova AM, Scherbenko VE, Nagurskaya EV, Bechalo VA, Timofeeva TY, Sanin AV, Ershov FI (2002) Regulation of cytokine mRNAs by interferon and interferon inducers. Russ J Immunol 7: 161–166 [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA (2010) c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328: 1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339: 826–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H (2012) Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell 46: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubaev VV, Slita AV, Krivitskaya VZ, Sirotkin AK, Kovalenko AL, Chatterjee NK (2003) Direct antiviral effect of cycloferon (10-carboxymethyl-9-acridanone) against adenovirus type 6 in vitro. Antiviral Res 58: 131–137 [DOI] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures source data