Abstract
Integrin-linked kinase (ILK) is an adaptor protein required to establish and maintain the connection between integrins and the actin cytoskeleton. This linkage is essential for generating force between the extracellular matrix (ECM) and the cell during migration and matrix remodelling. The mechanisms by which ILK stability and turnover are regulated are unknown. Here we report that the E3 ligase CHIP–heat shock protein 90 (Hsp90) axis regulates ILK turnover in fibroblasts. The chaperone Hsp90 stabilizes ILK and facilitates the interaction of ILK with α-parvin. When Hsp90 activity is blocked, ILK is ubiquitinated by CHIP and degraded by the proteasome, resulting in impaired fibroblast migration and a dramatic reduction in the fibrotic response to bleomycin in mice. Together, our results uncover how Hsp90 regulates ILK stability and identify a potential therapeutic strategy to alleviate fibrotic diseases.
Keywords: extracellular matrix, fibrosis, Hsp90, integrin, integrin-linked kinase
Introduction
The extracellular matrix (ECM) is a dynamic structure that is produced by cells, assembled into a precise supramolecular structure and constantly remodelled. The ECM provides structural support for tissues and also actively guides tissue morphogenesis and homeostasis by regulating cell adhesion, migration, and growth factor activity. Therefore, tight regulation of ECM remodelling is a prerequisite for tissue function (Hynes and Naba, 2012). Excessive and progressive accumulation of ECM results in tissue fibrosis leading to scarring and destruction of normal tissue architecture and ultimately to a failure of organ function (Wynn and Ramalingam, 2012).
The major cell surface receptors that bind and assemble the ECM are integrins. Integrins comprise 24 α/β-heterodimeric transmembrane proteins with overlapping substrate specificity and cell type-specific expression patterns (Hynes, 2002; Humphries et al, 2006). Signals arising from integrins regulate multiple aspects of cell behaviour, including cell migration, survival, proliferation, and differentiation. Binding of integrins to their ECM ligands leads to the recruitment of filamentous (F-) actin to integrin adhesion sites and remodelling of the actin cytoskeleton. The linkage of integrins to F-actin is central for the precise spatiotemporal control of cell migration, as well as for remodelling of the ECM, as it allows the cell to generate contractile forces and to regulate maturation and turnover of cell–matrix adhesions (Schwartz, 2010; Wickström et al, 2011). Integrins do not bind actin directly, but establish and maintain this linkage with the help of actin-binding and -regulatory proteins. A large body of structural, biochemical, and genetic evidence has identified the integrin-linked kinase (ILK)–PINCH–parvin (IPP) complex as a central regulator of the integrin–actin linkage (Wickström et al, 2010b). ILK is a ubiquitously expressed pseudokinase consisting of three structurally distinct domains. The N terminus consists of five ankyrin repeats followed by a pleckstrin homology (PH)-like domain and a C-terminal kinase-like domain. The ankyrin repeats mediate the interaction with PINCH, whereas the C-terminal kinase-like domain binds parvin. The importance of these proteins in the integrin–actin connection is demonstrated by deletion studies in mice, where loss of ILK expression results in severe defects in F-actin organization at adhesion sites, failure in epiblast polarization and peri-implantation lethality (Sakai et al, 2003). At the cellular level, ILK deficiency leads to aberrant remodelling of the actin and microtubule cytoskeletons and decreased force generation, which compromises cell adhesion, migration, and ECM remodelling (Sakai et al, 2003; Stanchi et al, 2009; Wickström et al, 2010a). The precise molecular mechanisms by which the IPP complex exerts these functions are not clear. The stability of each individual component of the IPP complex depends on the assembly of the complex, which occurs in the cytoplasm and thus before it is recruited to integrin adhesion sites. Depletion of any component of the IPP complex leads to a decrease in the protein levels of the other two complex members (Fukuda et al, 2003; Li et al, 2005). How the assembly and stability of the IPP complex is regulated is not known.
The covalent attachment of ubiquitin to target proteins is one of the most prevalent post-translational modifications regulating the functional diversity and dynamics of the proteome. In addition to regulating DNA repair, transcription, signalling and endocytosis, ubiquitination targets proteins to proteasomal and lysosomal degradation (Komander and Rape, 2012). Carboxyl terminus of Hsp70-interacting protein (CHIP, also known as Stub1) is an enzyme with E3/E4 ubiquitin ligase activity. CHIP consists of two functional domains; the N-terminal tetratricopeptide domain (TPR) that binds chaperones, such as Hsc70/Hsp70 and Hsp90, and the C-terminal U-Box domain that executes ubiquitin ligase activity. The ability of CHIP to link chaperones to the ubiquitin proteasome system allows CHIP to play an important role in protein quality control (Schulman and Chen, 2005). While the chaperone Hsc70 binds all misfolded proteins, Hsp90 interacts with so called client proteins that play central regulatory roles within the cell, such as protein kinases, transcription factors and receptors. Hsc70 and Hsp90 can simultaneously bind to HOP (Hsp70-Hsp90 organizing protein), which enables the passage of client proteins from Hsc70 to Hsp90 followed by final folding and maturation (Taipale et al, 2010). CHIP can bind to either Hsc70 or Hsp90. However, CHIP has been shown to preferentially bind to Hsc70 and ubiquitinate Hsc70-bound proteins. Since misfolded proteins that cannot be refolded back to the native state spend more time in a complex with Hsc70, they are more likely to become ubiquitinated by CHIP and subsequently degraded by proteasome (Stankiewicz et al, 2010). Under normal conditions, proteins are found in a complex with Hsc70–HOP–Hsp90 and the balance is shifted towards the folding process. However, upon inhibition of Hsp90 by geldanamycine or 17AAG, the folding of client proteins cannot be completed, their residence time in complex with Hsc70 increases, and as a consequence they become ubiquitinated by CHIP and degraded (Kundrat and Regan, 2010).
To address the molecular mechanisms by which the stability of ILK and the IPP complex are regulated, we analysed the ubiquitination of ILK and the functional consequences of this modification. We report that the binding of Hsp90 to ILK stabilizes ILK, and inhibition of Hsp90 activity promotes polyubiquitination of ILK by the E3 ligase CHIP, leading to its proteasomal degradation. Furthermore, Hsp90 promotes the interaction of ILK with α-parvin, thereby facilitating cellular force generation, ECM remodelling, and cell migration. As a consequence, inhibition of Hsp90 efficiently blocks the fibrotic response to bleomycin in mouse skin.
Results
ILK is modified by polyubiquitination
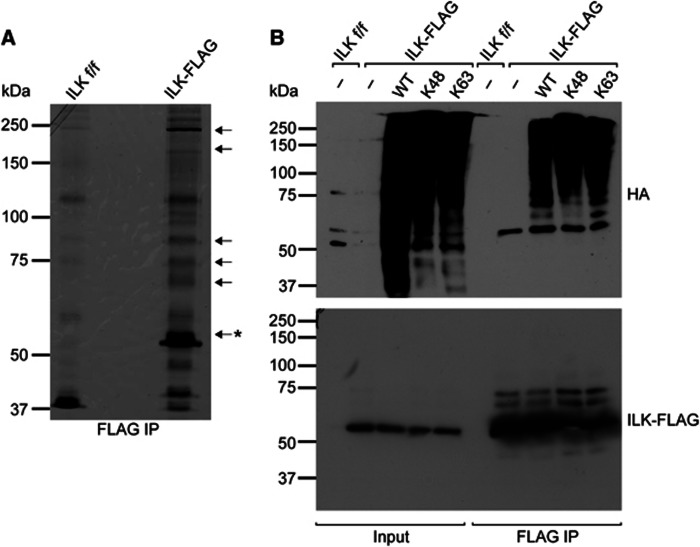
To investigate the potential post-translational modifications on ILK, we performed FLAG immunoprecipitation experiments of ILK−/− fibroblasts stably reconstituted with FLAG-tagged ILK (from here on referred to as ILK-FLAG cells) under reducing conditions. ILK f/f fibroblasts, the parental cell line from which both ILK−/− and ILK-FLAG cells were generated, were used as negative control for the immunoprecipitation. We separated the immunoprecipitates with SDS–polyacrylamide gel electrophoresis (SDS–PAGE), excised bands that were enriched in respect to the negative control, and analysed the protein content using mass spectrometry (Figure 1A). All bands contained ILK, suggesting that it is modified to generate higher molecular weight forms. In addition, all excised bands contained ubiquitin. To further determine if ILK was indeed ubiquitinated and to assess which type of ubiquitin chains were attached to ILK, we performed in vivo ubiquitination assays where ILK-FLAG cells where transfected with HA-tagged ubiquitin or ubiquitin mutants containing only a single lysine (K) K48 or K63 with all other lysines mutated to arginines (R). The analysis revealed the typical ladder pattern of higher molecular weight ILK bands indicative of post-translational modifications (Figure 1B). In addition, the immunoblot against the HA tag confirmed the presence of ubiquitin on ILK. Furthermore both K48- and K63-linked HA ubiquitins were present on ILK, showing that ILK is modified by both K48- and K63-linked polyubiquitin chains.
Figure 1.
ILK is modified by polyubiquitination. (A) Silver staining of FLAG immunoprecipitates from ILK-FLAG expressing fibroblasts. ILK f/f cells were used as a negative control. Arrows indicate bands analysed by mass spectrometry; asterisk indicates the expected molecular weight of unmodified ILK. ILK and ubiquitin were identified from all bands analysed. (B) Western blot analysis of FLAG immunoprecipitates from ILK−/− fibroblasts expressing ILK-FLAG and HA-tagged ubiquitin or ubiquitin mutants containing a single lysine K48 or K63. Both K48- and K63-linked ubiquitin chains are detected.
Source data for this figure is available on the online supplementary information page.
ILK interacts with E3 ligase CHIP
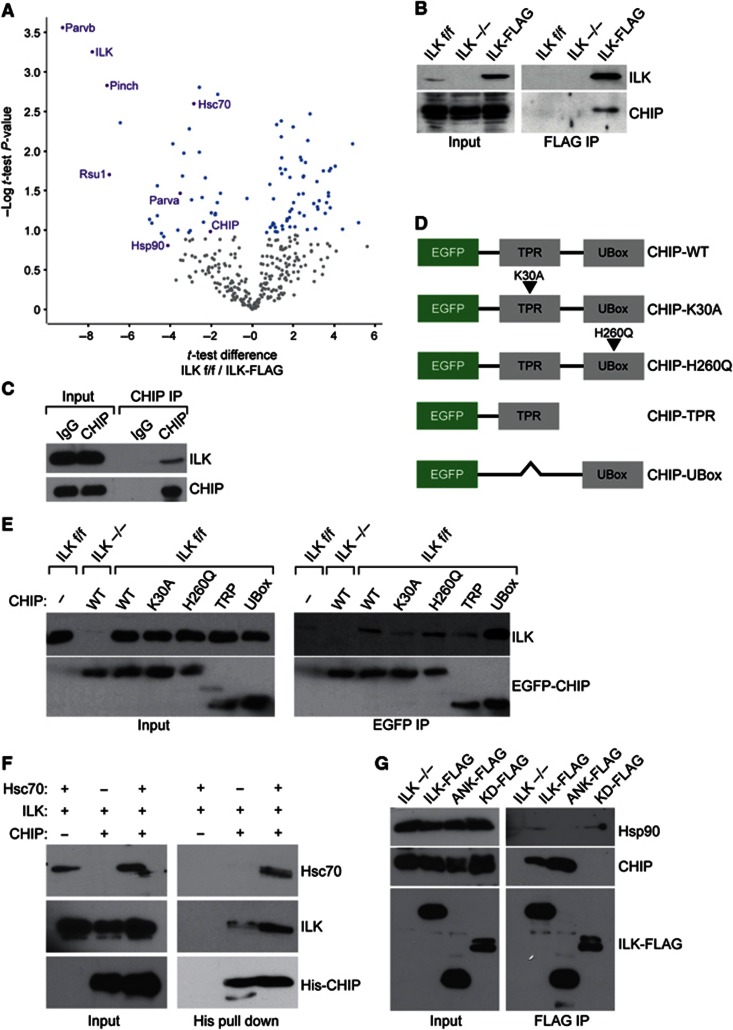
We next sought to identify the E3 ligase responsible for the ubiquitination of ILK. To this end we performed FLAG immunoprecipitations of ILK under non-reducing conditions to preserve protein–protein interactions and subjected the immunoprecipitates to mass spectrometry. The immunoprecipitates were performed in biological triplicates and quantitative analysis of the precipitates was performed using a label-free method (Hubner et al, 2010). The analyses identified several known major interacting partners of ILK, such as PINCH1, α- and β- parvin as well as RSU-1, confirming the validity of the assay. Interestingly, the analyses also revealed significant binding to a chaperone complex consisting of Hsc70, Hsp90, and the E3 ligase CHIP (Figure 2A; Supplementary Table S1). We confirmed the interaction between ILK and CHIP by detecting CHIP in ILK immunoprecipitates using western blotting (Figure 2B). In addition, this interaction also occurred between endogenous proteins, as endogenous ILK was co-precipitated with endogenous CHIP (Figure 2C).
Figure 2.
ILK interacts with the E3 ligase CHIP. (A) A volcano plot representing results of label-free FLAG pull downs of ILK. The differences in t-test of ILK f/f control versus ILK-FLAG are plotted against negative logarithmic P-values of the t-test performed from triplicates. The curve separates proteins specifically enriched in either condition (blue dots) from background (grey dots). Proteins specifically enriched in ILK-FLAG pull downs are found in the upper left quadrant of the plot. Known interacting partners of ILK as well as novel interacting partners CHIP, Hsc70 and Hsp90, are marked in purple. Names of all proteins specifically interacting with ILK-FLAG are reported in Supplementary Table S1. (B) FLAG pull down from ILK−/− fibroblasts expressing ILK-FLAG. CHIP is detected in the ILK immunoprecipitate. (C) Lysates from ILK f/f cells were subjected to immunoprecipitation with antibodies against CHIP. Endogenous ILK is detected in CHIP immunoprecipitates. Rabbit IgG was used as a negative control. (D) Schematic illustration of various EGFP-CHIP constructs. (E) GFP pull down from fibroblasts expressing EGFP-CHIP or mutants. Endogenous ILK is detected in immunoprecipitates of full-length EGFP-CHIP and EGFP-U-Box. K30A mutation of CHIP abrogates ILK binding. (F) Western blot of His pull down with recombinant His-tagged CHIP and ILK. ILK co-precipitates with CHIP and presence of Hsc70 enhances this interaction. (G) FLAG pull down from ILK−/− fibroblasts expressing ILK-FLAG, ANK-FLAG, and KD-FLAG. CHIP interacts with ANK-FLAG, whereas Hsp90 binds the KD.
Source data for this figure is available on the online supplementary information page.
To analyse this interaction in more detail, we cloned a full-length EGFP-tagged CHIP as well as several truncations and point mutations of the protein (Figure 2D). These included a point mutation of K30 to alanine (K30A) that abolishes the interaction of CHIP with its co-chaperones Hsc70 and Hsp90, a point mutation of histidine 260 to glutamine (H260Q) that abolishes the E3 ligase activity of CHIP, and deletions of the TPR and the U-Box domains (Ballinger et al, 1999; Okiyoneda et al, 2010). All constructs were transiently transfected into ILK f/f fibroblasts and immunoprecipitated with an anti-EGFP antibody. These experiments showed that CHIP interacts with endogenous ILK, that the interaction occurs via the U-Box of CHIP, and that the interaction is not dependent on the catalytic activity of CHIP, as the H260Q mutation did not affect ILK binding. Interestingly, however, the K30A mutation present in the TPR domain attenuated the interaction, suggesting that Hsc70/Hsp90 is involved in ILK binding to CHIP (Figure 2E). To confirm this hypothesis, we performed in vitro binding assays using recombinant ILK, Hsc70, and His-tagged CHIP. We used the His tag to pull down CHIP in the presence of ILK and in the presence or absence of Hsc70. These analyses revealed that ILK can bind CHIP directly and that Hsc70 enhances the binding (Figure 2F), confirming the in vivo results showing that CHIP interacts with ILK and that this interaction is facilitated by an intact TPR domain (Figure 2E). Finally, we sought to identify the domain within ILK that mediates the interaction with CHIP as well as to investigate the potential interaction with Hsp90, the other chaperone identified to interact with ILK in our mass spectrometry screen. To this end we transiently expressed full-length ILK-FLAG, the N-terminal ankyrin repeats (ANK-FLAG), or the C-terminal kinase-homology domain (KD-FLAG) of ILK in ILK−/− cells. This experiment revealed that CHIP interacts with the ankyrin repeats of ILK, whereas Hsp90 binds the kinase-homology domain (Figure 2G). Taken together these experiments identify the CHIP-Hsp90-Hsc70 chaperone machinery as novel interaction partners of ILK.
CHIP ubiquitinates ILK in vitro and in vivo
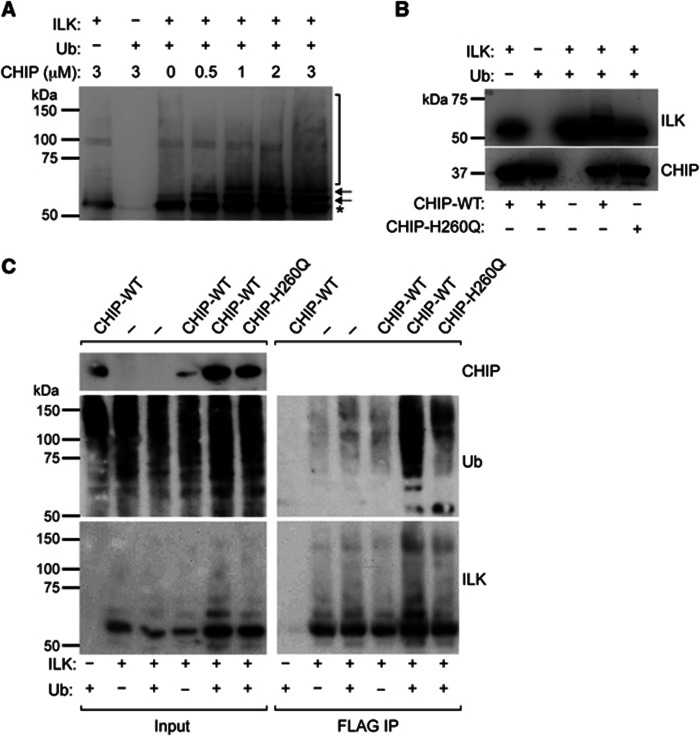
Next we investigated whether CHIP binding to ILK leads to ILK ubiquitination. We first performed in vitro ubiquitination assays with recombinant ILK and CHIP using constant concentrations of ILK, ubiquitin, E1 and E2 enzymes, as well as Hsc70, but increasing concentrations of CHIP. The immunoblot for ILK revealed a ladder-like appearance of higher molecular weight bands of ILK typical for ubiquitination. The intensity of the high-molecular weight bands correlated with the CHIP concentration, indicating an increase in the efficiency of the ubiquitination with increasing amounts of CHIP in the reaction (Figure 3A). The specificity of the reaction was further confirmed with an in vitro ubiquitination assay using wild-type CHIP (CHIP-WT) and the E3 ligase-dead mutant CHIP-H260Q. CHIP-WT showed robust ILK ubiquitination, while CHIP-H260Q induced no ubiquitination (Figure 3B).
Figure 3.
CHIP ubiquitinates ILK in vitro and in vivo. (A) Western blot of in vitro ubiquitination assay. Addition of recombinant CHIP induces multiple higher molecular weight bands recognized by ILK antibody (arrows, bracket) in a dose-dependent manner. ILK band of expected molecular weight is marked by asterisk. (B) Western blot of in vitro ubiquitination assay. CHIP-WT but not ligase-dead H260Q mutant CHIP ubiquitinates ILK. (C) Western blot analysis of FLAG immunoprecipitates from CHO cells expressing ILK-FLAG, CHIP, CHIP-H260Q, and ubiquitin-HA. Expression of CHIP but not ligase-dead H260Q mutant enhances ubiquitination of ILK.
Source data for this figure is available on the online supplementary information page.
To assess whether CHIP is also capable of ubiquitinating ILK in vivo, we performed in vivo ubiquitination assays in CHO cells transiently transfected with ILK-FLAG together with CHIP-WT or CHIP-H260Q and ubiquitin. Immunoblotting with antibodies against ILK and ubiquitin revealed a stronger ladder pattern and ubiquitin signal in the immunoprecipitates of cells transfected with CHIP-WT compared with cells transfected with CHIP-H260Q (Figure 3C). This indicated that increased expression of CHIP-WT leads to an increase in ILK ubiquitination, whereas overexpression of CHIP-H260Q mutant does not enhance the basal level of ILK ubiquitination. Taken together these experiments demonstrate that CHIP is capable of ubiquitinating ILK both in vitro and in vivo.
Interaction of ILK and CHIP is modulated by the cytoskeleton
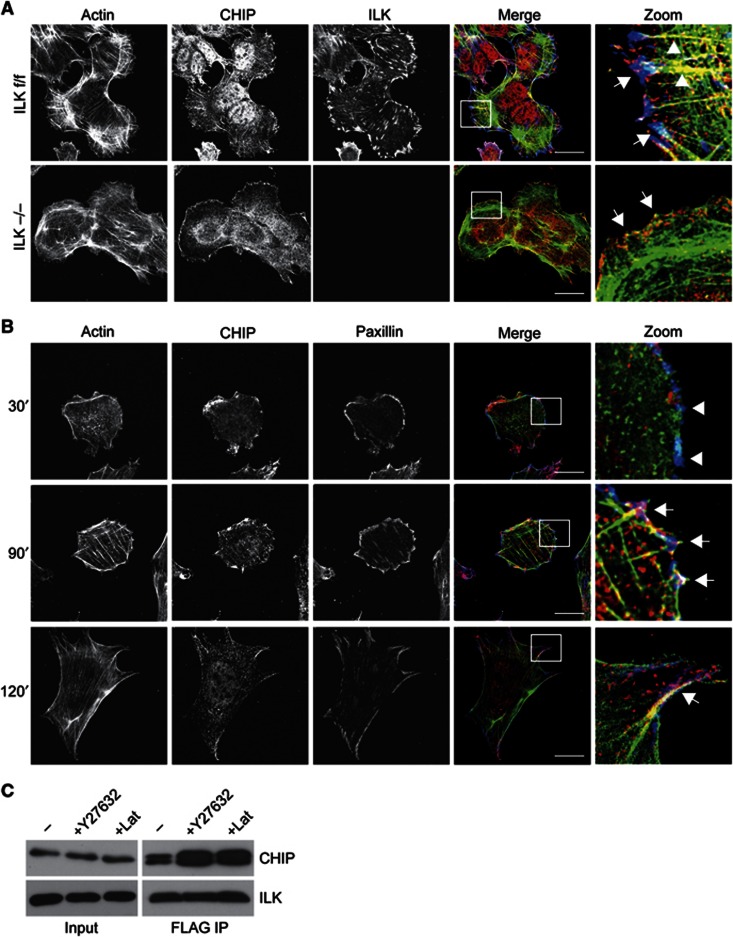
Next we performed immunofluorescence analyses to identify the subcellular compartment in which the ILK–CHIP interaction occurs. To this end we stained ILK f/f cells with antibodies against ILK and CHIP, and observed that CHIP was found in cytoplasmic filaments where it co-localized with F-actin stress fibres but not with ILK. In addition, CHIP staining was found in the cell periphery where it partly co-localized with ILK in focal adhesions (Figure 4A). This localization was, however, independent of ILK as a peripheral localization in focal adhesions and membrane ruffles was also found in ILK−/− cells (Figure 4A, Supplementary Figure S1). Since this type of localization has not been observed previously, we further confirmed the finding by analysing the localization of EGFP-CHIP. When expressed at low levels in ILK f/f fibroblasts, EGFP-CHIP showed a similar partial co-localization with peripheral adhesions and F-actin stress fibres (Supplementary Figure S2).
Figure 4.
Interaction of ILK and CHIP is modulated by the cytoskeleton. (A) Immunofluorescence analysis of endogenous CHIP, ILK, and phalloidin to stain the actin cytoskeleton in ILK f/f and ILK−/− fibroblasts. Right panels show an enlargement of the area indicated by the white rectangle. Note co-localization in of ILK and CHIP in focal adhesions (arrows) and localization of CHIP along actin stress fibres (arrowheads). CHIP co-localizes with actin in the cell periphery in the absence of ILK. Scale bars 20 μm. (B) Immunofluorescence analysis of endogenous CHIP, paxillin, and phalloidin to stain for the actin cytoskeleton in ILK f/f fibroblasts during 30, 90, and 120 min of cell spreading. CHIP localizes to focal adhesions during late stages of cell spreading. Right panels show an enlargement of the area indicated by the white rectangle. Arrowheads indicate immature focal complexes lacking CHIP and actin stress fibres. Arrows indicate co-localization of CHIP, paxillin, and actin in focal adhesions. Scale bars 30 μm. (C) FLAG pull down from ILK−/− fibroblasts expressing ILK-FLAG and treated with Y27632 or Lat. Increased levels of CHIP are detected in immunoprecipitates treated with Y27632 or Lat compared to control.
Source data for this figure is available on the online supplementary information page.
To address the kinetics by which CHIP localizes to adhesion sites, we performed immunofluorescence analyses of spreading cells using paxillin as a marker for focal adhesions and phalloidin to stain F-actin. The analyses confirmed the partial localization of CHIP to focal adhesions and stress fibres during late stages of spreading (90–120 min). Interestingly, the assay also revealed that at early stages of spreading (30 min) CHIP was absent from small, immature adhesions containing paxillin but lacking a linkage to F-actin stress fibres (Figure 4B). To assess whether the interaction of ILK and CHIP is regulated by Rho-kinase (ROCK)/myosin-II-mediated actomyosin contractility, we treated cells with the ROCK inhibitor Y27632. Alternatively, we treated cells with Latrunculin (Lat) that blocks actin polymerization and thus stress fibre formation and adhesion maturation, and determined the interaction of ILK and CHIP. In response to either treatment the amount of CHIP bound to ILK increased (Figure 4C). These results show that the interaction between ILK and CHIP does not depend on actomyosin contractility or an intact actin cytoskeleton, and rather suggest that disassembly of mature, force-bearing focal adhesions in response to Y27632 or Lat leads to increased interaction of these two proteins.
ILK is stabilized through its interaction with Hsp90
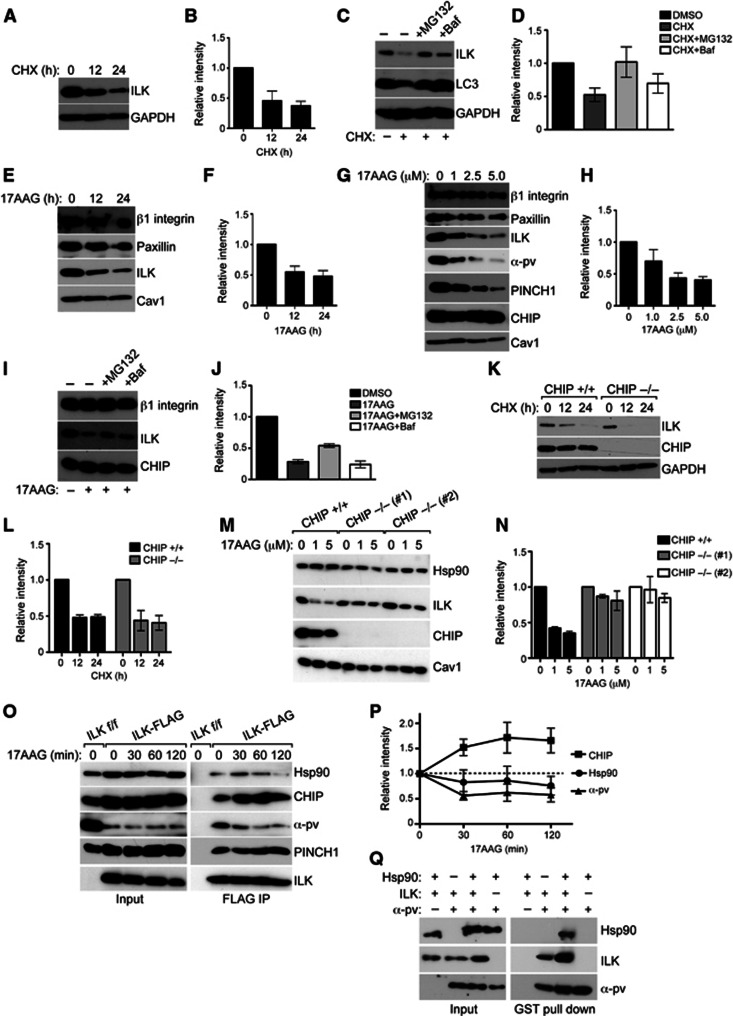
A major consequence of K48-linked ubiquitination is proteasomal degradation of the target protein (Komander and Rape, 2012). Since we observed that ILK is modified by K48-linked polyubiquitin chains (Figure 1B), we decided to investigate the consequences of this ubiquitination in more detail. To this end we analysed the half-life of the ILK protein by treating fibroblasts with cycloheximide (CHX) to inhibit protein synthesis. Western blot analyses showed that the majority of ILK was degraded in 24 h (Figure 5A and B). To identify the degradation pathway, we inhibited the proteasome or lysosome using MG132 or bafilomycin (Baf), respectively, in CHX-treated cells. Both MG132 and Baf attenuated ILK degradation, suggesting that both pathways contribute to degradation of ILK (Figure 5C and D).
Figure 5.
Hsp90 stabilizes ILK and protects it from CHIP-mediated degradation. (A) Western blot of fibroblasts treated with CHX to stop protein synthesis for time points indicated. Majority of ILK is degraded in 24 h. GAPDH is used as loading control. (B) Quantification of ILK levels from experiments from panel A. Data are presented as mean±s.e.m., n=3. (C) Western blot of fibroblasts treated with CHX to stop protein synthesis for time points indicated. Inhibition of proteasome with MG132 or inhibition of lysosome with Baf retards ILK degradation. LC3 is a positive control for inhibition of lysosome; GAPDH is used as loading control. (D) Quantification of ILK levels from experiments from C. Data are presented as mean±s.e.m., n=3. (E) Western blot of fibroblasts treated with 5 μM 17AAG to inhibit Hsp90 for time points indicated. Inhibition of Hsp90 leads to downregulation of ILK levels. Other focal adhesion proteins such as paxillin or β1 integrin are unaffected. Caveolin1 (Cav1) is used as a loading control. (F) Quantification of ILK levels from experiments from E. Data are presented as mean±s.e.m., n=3. (G) Western blot of fibroblasts treated with increasing concentrations of 17AAG. Inhibition of Hsp90 leads to dose-dependent downregulation of ILK levels together with PINCH1 and α-parvin. Paxillin or β1 integrin are unaffected. (H) Quantification of ILK levels from experiments from G. Data are presented as mean±s.e.m., n=3. (I) Western blot of fibroblasts treated with 17AAG in the presence of MG132 or Baf. Blocking the proteasome with MG132 inhibits 17AAG-induced degradation of ILK. (J) Quantification of ILK levels from experiments from I. Data are presented as mean±s.e.m., n=3. (K) Western blot of CHIP+/+ and CHIP−/− fibroblasts treated with CHX for time points indicated. ILK levels are reduced to a comparable extent in CHIP+/+ and CHIP−/− fibroblasts. (L) Quantification of ILK levels from experiments from K. Data are presented as mean±s.e.m., n=3. (M) Western blot of CHIP+/+ and CHIP−/− fibroblasts from two independent isolations treated with 17AAG. Inhibition of Hsp90 leads to downregulation of ILK levels. This effect is attenuated in CHIP−/− cells. (N) Quantification of ILK levels from experiments from M. Data are presented as mean±s.e.m., n=3. (O) Western blot of FLAG immunoprecipitates from ILK-FLAG expressing cells treated with 17AAG. Inhibition of Hsp90 leads to an increase in the interaction of ILK with CHIP and a dissociation of ILK from Hsp90 and α-parvin. Interaction with PINCH1 is not affected. (P) Quantification of experiments from O. Data are presented as mean±s.e.m., n=3. (Q) Western blot of GST pull down with recombinant GST-tagged α-parvin together with recombinant ILK and Hsp90. ILK co-precipitates with α-parvin, whereas Hsp90 co-precipitates with α-parvin only in the presence of ILK. Presence of Hsp90 enhances the interaction between ILK and α-parvin.
Source data for this figure is available on the online supplementary information page.
Hsp90 is a chaperone that associates with specific client proteins and assists them in acquiring and maintaining their native conformation. Inhibition of Hsp90 with geldanamycin or 17AAG leads to destabilization and subsequent degradation of client proteins (Taipale et al, 2010). As we observed an interaction between ILK and Hsp90, we next assessed whether inhibition of Hsp90 would trigger ILK degradation in fibroblasts. To this end we treated the cells with 17AAG for 12 and 24 h and observed a time-dependent decrease in ILK levels. Other focal adhesion proteins such as β1 integrin and paxillin were not affected showing that the inhibitor does not target adhesions in general and that ILK is a specific client protein (Figure 5E and F). To further confirm the specificity of the inhibitor treatment, we treated fibroblasts with increasing concentrations of 17AAG. We observed a dose-dependent decline of ILK levels, accompanied by a decrease in the levels of the other IPP complex constituents, PINCH1 and α-parvin. Again, β1 integrin and paxillin were not affected (Figure 5G and H). To assess the mechanism by which ILK is degraded upon inhibition of Hsp90, we blocked the proteasome and the lysosome in the presence of 17AAG. Only MG132 attenuated ILK degradation, showing that inhibition of Hsp90 leads to proteasomal degradation of ILK (Figure 5I and J). To address whether basal turnover of ILK was mediated by CHIP, we isolated fibroblasts from CHIP+/+ and−/− mice (Dai et al, 2003), and blocked protein synthesis by CHX. ILK levels were reduced to the same extent in the presence or absence of CHIP, showing that CHIP does not regulate the basal turnover of ILK (Figure 5K and L). However, when cells were treated with 17AAG, ILK was efficiently degraded in CHIP+/+ cells, whereas the degradation was significantly attenuated in two independent CHIP−/− cell lines (Figure 5M and N), suggesting that when Hsp90 is blocked, CHIP is required to target ILK for proteasomal degradation. To further confirm this finding, we performed ILK immunoprecipitations and analysed the interaction between ILK and CHIP/Hsp90. As expected, inhibition of Hsp90 led to a rapid increase in the interaction between ILK and CHIP, whereas the interaction between ILK and Hsp90 was slightly decreased (Figure 5O and P). Interestingly, the interaction between ILK and α-parvin also rapidly declined, whereas the interaction between ILK and PINCH1 was not significantly affected (Figure 5O and P).
This, together with the finding that Hsp90 binds the kinase-homology domain, led us to hypothesize that Hsp90 could be important in folding the kinase-like domain, thereby stabilizing or facilitating the interaction between ILK and α-parvin. To test this hypothesis, we performed in vitro GST pull-down assays between GST-tagged α-parvin and ILK in the presence of Hsp90. Interestingly, Hsp90 was only pulled down with α-parvin when ILK was present in the reaction, indicating that Hsp90 interacts with ILK, but not with α-parvin, and that Hsp90 is capable of binding ILK when ILK is also bound to α-parvin (Figure 5Q). In addition, the presence of Hsp90 in the reaction enhanced ILK binding to α-parvin (Figure 5Q), confirming that Hsp90 stabilizes the interaction of the two proteins.
Inhibition of Hsp90 leads to the reorganization of the F-actin cytoskeleton and focal adhesions
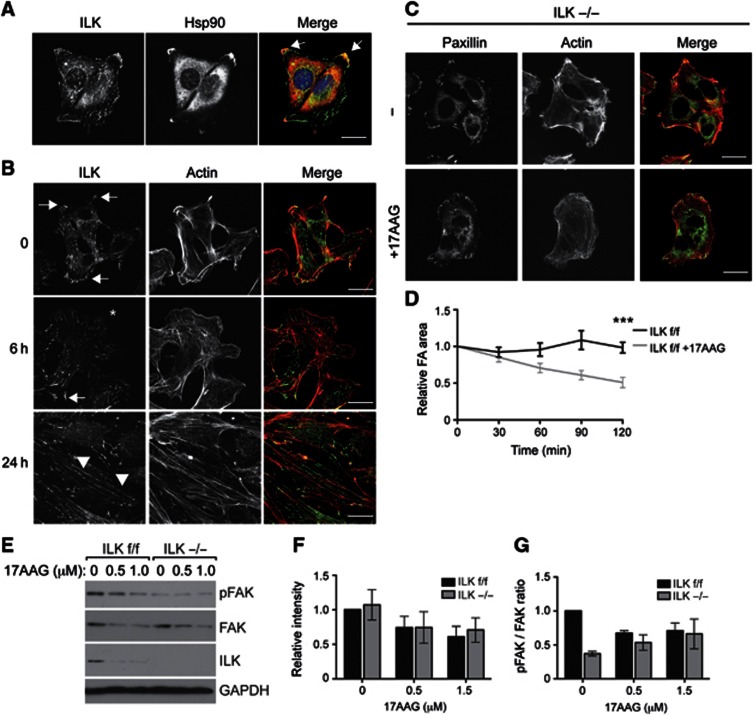
We next addressed the functional consequences of the ILK–Hsp90 interaction. We observed that Hsp90 is capable of binding ILK, which enhances binding to α-parvin. On the other hand the IPP complex localizes to focal adhesions, where it plays an essential role in linking integrins to the actin cytoskeleton. In addition, recent proteomic screens have identified Hsp90 as a component of the integrin adhesome (Humphries et al, 2009; Schiller et al, 2011). Therefore, we first tested whether Hsp90 was present in focal adhesions. Indeed, when we stained fibroblasts with antibodies against Hsp90 and ILK, we found ILK in peripheral focal complexes, mature focal adhesions, and central fibrillar adhesions. Hsp90 was detected in mature focal adhesions and diffusely around the nucleus (Figure 6A). To determine whether Hsp90 influences the functional properties of integrin adhesion sites, we analysed whether Hsp90 inhibition affects ILK localization and the morphology of the different adhesion structures. Interestingly, 17AAG treatment led to the gradual loss of ILK from large peripheral focal adhesions but not from the thin central fibrillar adhesions. This effect was visible already at 6 h and complete at 24 h. In addition, the actin cytoskeleton underwent dramatic reorganization leading to the appearance of long, thin stress fibres and elongation of cells (Figure 6B). Interestingly, the focal adhesion protein paxillin, which is not a client of Hsp90, also showed increased localization to fibrillar adhesions, suggesting that 17AAG induced a more global rearrangement of adhesion structures (Supplementary Figure S3). The absence of such rearrangements in ILK−/− cells that displayed a less spread morphology, absence of fibrillar adhesions, and less abundant actin stress fibres both in the presence and absence of 17AAG, suggests that the morphological changes are mainly driven by ILK (Figure 6C).
Figure 6.
Inhibition of Hsp90 leads to reorganization of the F-actin cytoskeleton and focal adhesions. (A) Immunofluorescence analysis of ILK and Hsp90 in fibroblasts. Note co-localization of ILK and Hsp90 in large peripheral focal adhesions (arrows), but not in cytoplasmic fibrillar adhesions or focal complexes. Scale bar, 20 μm. (B) Immunofluorescence analysis of ILK and actin in fibroblasts treated with 17AAG. Note loss of ILK from focal adhesions (arrows, asterisk), an increase in fibrillar adhesions (arrowheads), and reorganization of the actin cytoskeleton. Scale bars 20 μm. (C) ILK−/− fibroblasts were treated with 17AAG after which they were fixed and stained with antibodies against paxillin and with phalloidin to visualize the actin cytoskeleton. No major change in focal adhesion or the actin cytoskeleton was observed upon 17AAG treatment. Scale bars 20 μm. (D) Quantification of relative focal adhesion area from live-cell imaging experiments on cells treated with 17AAG. Note that focal adhesions are disassembled as indicated by a decrease in focal adhesion area in 17AAG-treated cells. Data are presented as mean±s.e.m., n=20 movies from three independent experiments, ***P≤0.0001 (Student’s t-test). (E) Western blot of ILK f/f and ILK−/− fibroblasts treated with 17AAG. 17AAG treatment leads to a decrease in total FAK levels in ILK f/f and ILK−/− cells, and to a decrease in FAK phosphorylation (pFAK) in ILK f/f cells. ILK−/− cells show reduced basal levels of pFAK. (F) Quantification of total FAK levels from experiments shown in E. Data are presented as mean±s.e.m., n=3. (G) Quantification of pFAK to total FAK ratio from experiments shown in E. Data are presented as mean±s.e.m., n=3.
Source data for this figure is available on the online supplementary information page.
To analyse the effect of Hsp90 inhibition on focal adhesion dynamics in more detail, we performed live-cell imaging analyses of cells transfected with Paxillin-Cherry and treated with 17AAG. These analyses showed that in response to 17AAG treatment, large peripheral focal adhesions were disassembled, whereas more thin and elongated cytoplasmic fibrillar adhesions were still generated (Supplementary Movie 1). The net effect of this change was a decrease in focal adhesion area over time (Figure 6D).
To investigate the mechanism by which Hsp90 and ILK could regulate focal adhesion dynamics in more detail, we analysed the levels and activity of focal adhesion kinase (FAK). FAK is a central regulator of focal adhesion turnover, plays an important role in generating and sensing of cellular forces (Mitra et al, 2005), acts downstream of ILK, and was shown to be a client of Hsp90 (Lorenz et al, 2007; Schwock et al, 2009). In line with a previous report, we also observed that Hsp90 inhibition with 17AAG reduced the levels of FAK. This occurred to a comparable extent in ILK f/f and ILK−/− cells, and no difference in basal levels of FAK was observed (Figure 6E and F). Importantly, however, we observed that FAK activity, assessed by levels of tyrosine 397 phosphorylated FAK, was decreased in ILK−/− cells compared to controls (Figure 6E and G). In addition, whereas the ratio between FAK and phosphorylated FAK decreased in wild-type cells treated with 17AAG, this ratio remained constant in ILK−/− cells (Figure 6G). This result confirms that ILK acts upstream of FAK and that 17AAG targets FAK both directly as well as indirectly by decreasing its activity through upstream regulators.
Hsp90-mediated stabilization of ILK promotes force generation, matrix assembly, and migration
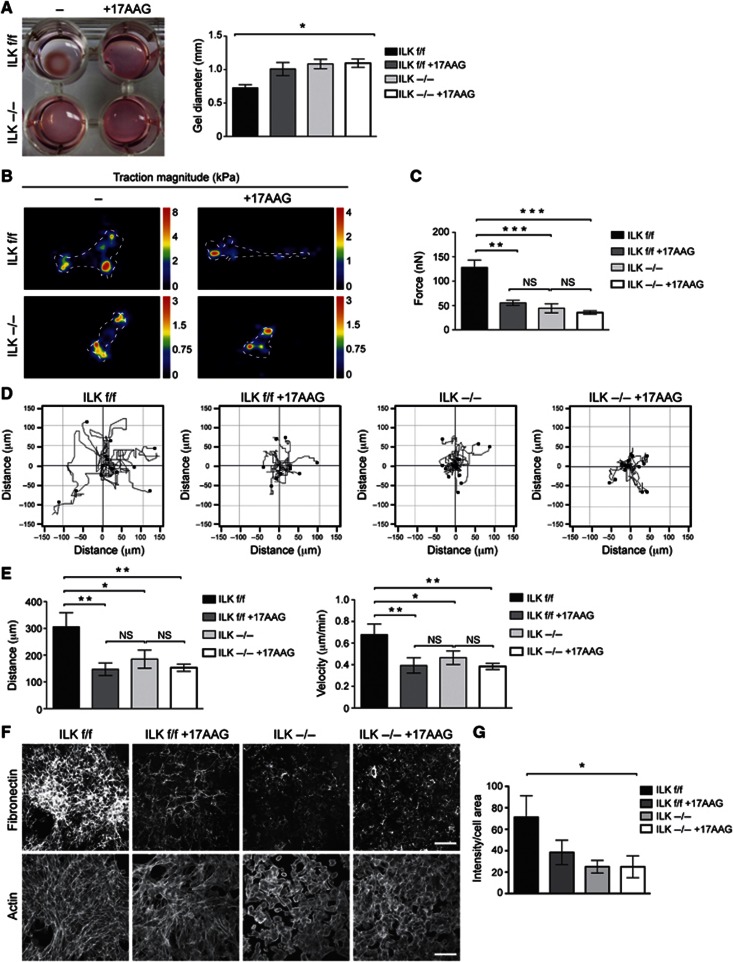
As focal adhesions and their linkage to the actin cytoskeleton are required for cells to be able to generate forces on their substrates and subsequent ECM remodelling, we analysed the role of Hsp90 in this process using a collagen gel contraction assay. Wild-type fibroblasts embedded in a 3D collagen matrix exerted contractile forces on the collagen matrix, which resulted in the contraction of the collagen gel. The ability to contract collagen matrices was lost in cells treated with 17AAG or in cells lacking ILK (Figure 7A). Treatment of ILK-deficient cells with 17AAG did not induce an additive effect (Figure 7A).
Figure 7.
Hsp90-mediated stabilization of ILK promotes force generation, matrix assembly, and migration. (A) Gel contraction assay with ILK f/f and−/− fibroblasts treated with 17AAG. 17AAG or deletion of ILK impairs the ability of fibroblasts to contract collagen gels. 17AAG in ILK−/− cells does not induce an additive effect. Data are presented as mean±s.e.m., n=3, *P=0.0174 (Friedman’s ANOVA). (B) Heat-scale map of traction stress magnitudes. The colour code indicates local traction in kPa. Cell outlines are indicated by dotted lines. (C) Quantification of total cellular traction force. Data are presented as mean±s.e.m., n>30, ***P≤0.0001 (Kruskal–Wallis), NS=not significant. (D) Tracks of ILK f/f and−/− fibroblasts migrating in 3D collagen. 17AAG or deletion of ILK impairs the ability of fibroblasts to migrate. (E) Quantification of migration distance (left panel) and velocity (right panel) from 3D migration assays. Data are presented as mean±s.e.m., n=5, **P=0.002 (repeated measures ANOVA), NS=not significant. (F) Immunofluorescence staining of the fibronectin matrix and the actin cytoskeleton (phalloidin) to visualize cell area. Note decreased matrix deposition in ILK f/f cells treated with 17AAG, and in ILK−/− cells. Scale bars 100 μm. (G) Quantification of the integrated intensity of the fibronectin matrix staining normalized to total cell surface area. Data are presented as mean±s.e.m., n=4, *P=0.0214 (Kruskal–Wallis).
To address whether ILK and Hsp90 are indeed required for cellular force generation, we measured forces exerted by single cells on the ECM by traction force microcopy (Sabass et al, 2008). Measurement of total cellular traction force revealed that both inhibition of Hsp90 with 17AAG or deletion of ILK resulted in decreased cellular force generation (Figure 7B and C). Force generation is a prerequisite for cell migration in a 3D environment. Indeed, 17AAG-mediated inhibition of Hsp90 and cell contractility led to a dramatic decrease in the ability of fibroblasts to migrate within 3D collagen gels (Figure 7D and E, Supplementary Movies 2, 3, 4, 5). A similar defect was observed in cells lacking ILK, whereas 17AAG treatment in ILK-deficient cells did not induce an additive effect (Figure 7D and E).
Collagen gel contraction is controlled by cellular forces and remodelling of the matrix. Therefore, we addressed whether ILK also regulates the deposition of the fibronectin matrix, a process tightly dependent on force generation by integrins. We plated ILK f/f and ILK−/− fibroblasts on coverslips and allowed them to deposit fibronectin matrix for 24 h in the presence or absence of 17AAG. Immunofluorescence analysis showed that ILK f/f cells had deposited an extensive fibrillar network of fibronectin. The abundance of fibrillar fibronectin was significantly reduced in ILK f/f cells treated with 17AAG. ILK−/− cells both with and without 17AAG completely lacked the ability to deposit fibronectin fibrils and displayed mainly patches of fibronectin in the extracellular space and punctate staining inside the cell (Figure 7F and G). No difference in fibronectin protein expression was observed (Supplementary Figure S4). Altogether these results show that the stability of ILK, regulated through Hsp90 activity, is critical for the architecture of focal adhesions and the actin cytoskeleton. This in turn allows the cells to transduce the necessary force for cell migration and ECM remodelling.
Inhibition of Hsp90 attenuates bleomycin-induced skin fibrosis
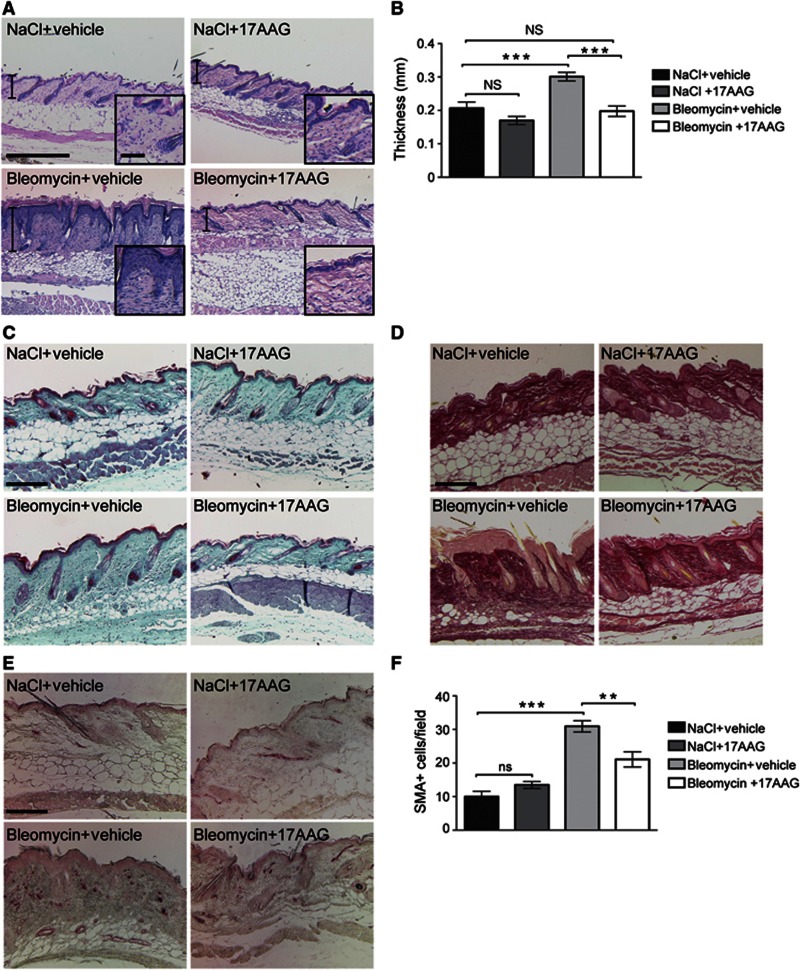
We have previously observed that ILK is essential for myofibroblast differentiation as well as ECM production and remodelling in the skin (Blumbach et al, 2010). This pro-fibrotic activity suggests that degrading ILK by inhibiting Hsp90 might represent a suitable therapeutic strategy to treat fibrosis. To induce a state resembling human inflammation-induced fibrosis in mice, we injected bleomycin intradermally into 6-week-old mice for 2 weeks, with or without parallel intraperitoneal administration of 17AAG (60 mg/kg). Bleomycin-treated mice exhibited extensive skin fibrosis characterized by thickening of the dermis (Figure 8A and B) and enhanced accumulation of collagens in the dermis and subcutis (Figure 8 C and D). Strikingly, mice treated with bleomycin together with 17AAG were significantly protected from bleomycin-induced fibrosis, as shown by reduced dermal thickening and reduced collagenous matrix content as compared to animals treated with bleomycin only (Figure 8A–D). To further quantify the extent of the fibrotic response, we implemented blinded histopathological scoring of the abundance of the collagenous matrix in the dermis and subcutaneous fat, ranging from 0 (no fibrosis) to 4 (significant fibrosis). As expected, bleomycin-treated mice received high scores (3.9±0.1) compared to mice treated with NaCl (1.0±0.3) or NaCl and 17AAG (0.9±0.2). In contrast, the clinical score of bleomycin +17AAG-treated mice (1.1±0.2) did not differ from NaCl-treated controls. Importantly, ILK levels were found to be decreased in tissue extracts from mice treated with bleomycin+17AAG compared to mice treated with bleomycin only (Supplementary Figure S5A).
Figure 8.
Bleomycin-induced skin fibrosis is blocked by inhibition of Hsp90. (A) Haematoxylin/eosin staining of bleomycin-treated skin shows characteristic fibrosis of the skin with dermal thickening. This effect is not visible in mice treated with 17AAG together with bleomycin. Scale bar, 150 μm (large panel), 20 μm (inset). (B) Quantification of dermal thickness from bleomycin-treated mice. Data are presented as mean±s.e.m., n=5/5/8/5, ***P=0.0004 (ANOVA), NS=not significant. (C) Trichrome staining of bleomycin-treated skin shows increased density of the extracellular matrix in the dermis. This effect is not visible in mice treated with 17AAG together with bleomycin. Scale bar, 200 μm. (D) Picosirius red staining of bleomycin-treated skin shows enhanced abundance of collagenous matrix in the dermis and subcutaneous fat. This effect is not visible in mice treated with 17AAG together with bleomycin. Scale bar, 150 μm. (E) α-SMA staining identifies elevated numbers of activated fibroblasts in the dermis of bleomycin-treated mice. This effect is attenuated in mice treated with 17AAG together with bleomycin. (F) Quantification of α-SMA-positive cells. Data are presented as mean±s.e.m., n=5/5/8/5, ***P<0.0001, **P<0.005 (ANOVA), NS=not significant.
Activation of fibroblasts is a critical step in fibrosis. This activation has been shown to depend on the presence of TGF-β as well as mechanical tension (Tomasek et al, 2002), which are both modulated by ILK (Blumbach et al, 2010, Figure 7B and C). As expected, abundant numbers of activated, α-smooth muscle actin (α-SMA) expressing fibroblasts were found in the dermis of bleomycin-treated mice that were not treated with 17AAG. In contrast, the numbers of activated fibroblasts were severely attenuated in mice treated with 17AAG and bleomycin (Figure 8E and F). The amount of mast cells and F4/80-positive macrophages was not significantly altered between bleomycin− and bleomycin+17AAG-treated mice (Supplementary Figure S5B–E), indicating that the primary effect of 17AAG is on fibroblasts. Taken together these results show that 17AAG efficiently attenuates the fibrotic response and targets fibroblast activation in this process.
Discussion
In the present study we show that the stability of ILK is regulated by the Hsp90–Hsc70 chaperone machinery, and that inhibition of Hsp90 leads to polyubiquitination of ILK by the E3 ligase CHIP resulting in proteasomal degradation. We further demonstrate that the stabilization of ILK by Hsp90 is required to allow the interaction of ILK with the key binding partner α-parvin. Inhibition of Hsp90 chaperone activity leads to removal of ILK from focal adhesions, resulting in severe reduction of cellular force generation and migration. Furthermore, we show that blocking Hsp90 activity in vivo efficiently attenuates fibroblast activation and the development of skin fibrosis in mice.
Coordinated assembly and disassembly of focal adhesions is required for effective cellular force generation and cell shape changes during locomotion and ECM remodelling. To understand the detailed molecular mechanisms underlying focal adhesion dynamics, it is critical to dissect the molecular regulation of individual focal adhesion proteins. As ILK is a major scaffold protein downstream of β1 integrins, alterations in its stability and turnover are likely to affect the functions of focal adhesions and thereby cellular behaviour on a more global level. We observed that ILK is polyubiquitinated via K48- and K63-linked ubiquitin chains. Using mass spectrometry and subsequent biochemical analyses, we identified CHIP as the E3 ligase inducing K48-linked poly-ubiquitin chains on ILK. Our studies revealed that CHIP is not involved in regulating basal turnover of ILK, while it induced proteasomal degradation of ILK after chemical inhibition of Hsp90. The consequence of the K63-linked polyubiquitin of ILK remains to be analysed in future studies. Interestingly, CHIP has been shown to be an essential component of the chaperone machinery for cytoskeletal proteins such as myosin and filamin (Nyamsuren et al, 2007; Arndt et al, 2010). However, the activity of CHIP seems to be compensated, at least partially, by other E3 ligases such as Parkin in vivo, which is best demonstrated by the relatively mild phenotype of CHIP-deficient mice that do not show typical cell migration-associated defects (Dai et al, 2003).
Treatment of cells with the Hsp90 inhibitor 17AAG led to an almost complete depletion of ILK protein demonstrating that ILK is an Hsp90 client protein. A similar reduction of ILK protein levels after Hsp90 inhibition has already previously been reported to operate in several cell lines (Aoyagi et al, 2005). Furthermore, our studies also confirmed that Hsp90 binds ILK through the kinase-like domain. Interestingly, most reported Hsp90 clients are kinases, and recent studies show that 60% of the mammalian kinases are client proteins of Hsp90 (Taipale et al, 2012). Although ILK does not possess kinase activity (Fukuda et al, 2009; Lange et al, 2009), it is likely that the kinase-like fold provides the structural basis for its recognition by Hsp90. This hypothesis is supported by a recent study showing that Hsp90 binds to residues located in the αE helix of kinases, and in addition it requires a particularly long αD–αE loop (Taipale et al, 2012). These regions do not significantly differ between ILK and bona fide kinases (Fukuda et al, 2009). Another unifying feature of Hsp90 clients is their inherent instability (Taipale et al, 2012). The expression of the ILK kinase-like domain has been reported to be technically difficult due to aggregation, clearly indicating that the kinase-like domain of ILK is highly instable (Fukuda et al, 2009). Furthermore, several mutations that were initially described to disrupt the putative kinase activity were later shown to actually severely abrogate the structure and stability of ILK, as well as to reduce its ability to bind to α-parvin (Fukuda et al, 2009; Lange et al, 2009). Therefore, it will be interesting to assess whether these mutations might influence the ability of Hsp90 to recognize and stabilize the structure of the kinase-like domain.
Our data show that Hsp90 activity is required for the stability of the ILK–α-parvin interaction. The interaction between ILK and its key partner α-parvin is critical for the function of both proteins within the focal adhesions. It has been shown that α-parvin binds to the pseudoactive site of ILK, but it is not clear how the association and dissociation of α-parvin from ILK is regulated. Interestingly, Hsp90 has been suggested not to be involved in de novo protein folding, but to assist refolding of partially unfolded proteins and intriguingly, to stabilize the hydrophobic ligand-binding cleft of proteins (Pratt et al, 2008). Therefore, it is possible that Hsp90 facilitates the interaction between ILK and α-parvin by stabilizing the kinase-like domain of ILK in a conformation in which it is capable of binding α-parvin. In line with recent proteomic screens (Humphries et al, 2009; Schiller et al, 2011), we find that Hsp90 is present at cell–matrix adhesion sites, particularly in large peripheral focal adhesions that are known to be exposed to high traction forces (Hoffman et al, 2011). It is intriguing to speculate that at these sites ILK might be susceptible for force-induced unfolding and that this is prevented by Hsp90. This is supported by the finding that ILK disappears from large peripheral adhesions upon Hsp90 inhibition. In this respect, Hsp90 activity, through its stabilizing effect on the ILK–α-parvin interaction, might represent a novel mechanism by which adhesion turnover is regulated.
Fibroblasts orchestrate ECM deposition and remodelling under both physiological and pathological conditions. To carry out these functions, fibroblasts need to migrate to the sites of injury, convert to myofibroblasts, and exert force on the ECM to allow its contraction and remodelling. Therefore, fibroblasts play a crucial role in all fibrotic diseases (Tomasek et al, 2002). We have previously demonstrated that ILK is a key regulator of fibroblast migration, myofibroblast conversion, and ECM remodelling (Sakai et al, 2003; Stanchi et al, 2009; Blumbach et al, 2010). Our in vitro studies now further indicate that targeting ILK stability by inhibiting Hsp90 activity induces functional defects resembling ILK ablation in fibroblasts. Blocking Hsp90 leads to loss of force-bearing focal adhesions, inability of cells to exert force on the ECM, and subsequently to a severe defect in cell migration and ECM remodelling. This led us to test and demonstrate that targeting Hsp90 using 17AAG represents an effective therapeutic strategy in fibrosis. 17AAG has been extensively tested in terms of toxicology and pharmacokinetics, and it is currently in phase II studies for several types of cancer. It has already demonstrated clinical benefits against Her2-positive breast cancer (Neckers and Workman, 2012). Although our in vitro studies implicate that ILK is a major Hsp90 target in the processes of cellular force generation and cell migration, it is very likely that other Hsp90 clients also play an important role in the pathogenesis of fibrosis. For example, TGF-β signalling that plays a central role in myofibroblast activation has been shown to be regulated by Hsp90 (Wrighton et al, 2008). This probably explains the very high efficacy of 17AAG treatment in blocking skin fibrosis, and highlights the potential of this drug as a successful treatment strategy of this complex and difficult-to-treat disease. Taken together our data provide a strong rationale for using Hsp90 inhibition as a therapeutic strategy for the treatment of fibrotic diseases.
Materials and methods
Plasmid constructs
Full-length ILK cDNA, ankyrin repeats (ANK: amino acids 1–180 of murine ILK), or kinase domain (KD; amino acids 181–452 of murine ILK) was cloned into p3xFLAG plasmid and subsequently into pCMLFG retroviral vector (Naviaux et al, 1996) to generate ILK-3xFLAG-pCLMFG. CHIP cDNA was obtained by performing RT PCR (iScript Select cDNA Synthesis Kit, Bio-Rad) from fibroblast RNA. The PCR fragment was cloned into a TOPO vector (Invitrogen), from where it was subsequently subcloned into pEGFP-C1 vector (Clontech). The TPR and U-Box domains were amplified from the full-length cDNA using the following primers: 5′-GAATTCGCCCTTCGCCATGAAG-3′ (TPR forward) and 5′-TGGATCCTCAATCATCTTCATGACCCTCG-3′ (TPR reverse); 5′-TGAATTCGAAGAAGCGCTGGAACAGTATCG-3′ (U-Box forward) and 5′-TGGATCCTCAATAGTCCTCTACCCAG-3′ (U-Box reverse). The K30A and H260Q point mutations were generated by performing site-directed mutagenesis PCR using the following primers: 5′-CGAGTGCGCAAGAGCTCGCGGAGCAGGGAAACCGGC-3′ (K30A forward) and 5′-GCCGGTTTCCCTGCTCCGCGAGCTCTTGCGCACTCG-3′ (K30A reverse); 5′-GGACATTGAGGAGCAACTGCAGCGTGTGGGC-3′ (H260Q forward) and 5′-GCCCACACGCTGCAGTTGCTCCTCAATGTCC-3′ (H260Q reverse). The ubiquitin constructs pRK5-HA-Ubiquitin-WT (plasmid no. 17608), pRK5-HA-Ubiquitin-K48 (plasmid no. 17605), and pRK5-HA-Ubiquitin-K63 (plasmid no.17606) were from Addgene.
Cell culture and transfection
Primary kidney fibroblasts were isolated from 3-week-old ILK f/f, CHIP+/+ and CHIP−/− mice, and immortalized with SV40 virus. Single cell clones from ILK f/f fibroblasts were subsequently obtained, from which the ILK gene was deleted by adenoviral expression of Cre-recombinase to generate ILK−/− cells. Single cell clones of ILK−/− fibroblast were subsequently retro- or lentivirally infected with FLAG-tagged ILK to establish stable cell lines (Azimifar et al, 2012). All fibroblasts were cultured in DMEM supplemented with 10% FCS.
Retroviral particles were produced by transient transfection of HEK293 cells. Viral supernatant was harvested 24 and 48 h after transfection and passed through a 0.45-μm filter. The filtrates were centrifuged at 50 000 g for 2 h at 17°C, combined, mixed with 2 ml of 20% sucrose, and centrifuged at 42 000 g for 2 h. Pellets were resuspended in 50–100 μl HBSS. Ten to twelve microliters of the virus preparation was added onto 1.5–2 × 106cells grown on six-well plates and incubated over night.
Transient transfections were performed using Lipofectamine 2000 reagent (Roche) according to the manufacturer’s instructions.
Chemical treatments
The 17AAG was generously provided by the National Cancer Institute, NIH. Lat and Y27632 were from Sigma.
Preparation of cell extracts and western blot
Cells grown on polystyrene dishes were rinsed in phosphate-buffered saline (PBS), suspended in lysis buffer (50 mM Tris–HCl buffer (pH 8.0) containing 150 mM NaCl, 1% Triton-X 100, 0.05% sodium deoxycholate, 10 mM EDTA, protease, and phosphatase inhibitors), and cleared by centrifugation. The lysates were then reduced in Laemmli sample buffer at 95°C, separated by PAGE in the presence of SDS, transferred onto PVDF membranes and subjected to western blot analyses using the standard protocols. The following antibodies were used: mouse anti-ILK (BD Transduction Laboratories), rabbit anti-ILK (Cell Signaling), anti-PINCH (Transduction Laboratories), anti-parvin (Cell Signaling), anti-GAPDH (Calbiochem), anti-paxillin (BD Transduction Laboratories), anti-CHIP (C-terminal ab; Sigma), anti-FLAG M2 (Sigma), anti-ubiquitin (Cell Signaling), anti-HA (Roche) anti-Hsp90 (Cell Signaling), anti-Hsp70 (Cell Signaling), anti-LC3 (Cell Signaling), anti-β1 integrin (self made polyclonal in rabbit), anti-caveolin-1 (Santa Cruz), anti-fibronectin (Abcam), anti-FAK (Cell Signaling), anti pFAK (Y397; Invitrogen), and anti-mouse/rabbit HRP (Bio-Rad Laboratories). Western blots were quantified using densitometry (ImageJ software) from three independent experiments.
Immunoprecipitation experiments
Cells grown on polystyrene dishes were lysed in lysis buffer as described above. FLAG immunoprecipitation was carried out according to the manufacturer’s protocol (Sigma). GFP immunoprecipitation was performed using Miltenyi Biotec MultiMACS GFP Isolation Kit according to the manufacturer’s protocol.
For endogenous CHIP immunoprecipitation, 1 mg of total cell lysate was homogenized by sonication. Subsequently, 5 μg of anti-CHIP C-terminal and 2 μg of anti-CHIP N-terminal antibody (both from Sigma) were added to the lysate and incubated for 1 h at 4°C. Seven micrograms of total rabbit IgG was used as control. Twenty microliters of protein-G agarose (Roche) was then added and incubation was continued for 30 min. The agarose was pelleted by centrifugation and rinsed once with lysis buffer and twice with Tris-buffered saline. The immunocomplexes were eluted in Laemmli sample buffer by boiling and analysed by western blotting.
Mass spectrometry
FLAG immunoprecipitates from ILK-FLAG cells were analysed by quantitative mass spectrometry using ILK f/f fibroblasts as a negative control. Immunoprecipitates were trypsin digested on beads followed by peptide concentration and purification using StageTips. Peptides were then separated by reversed-phase liquid chromatography coupled online to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). Data were analysed using the label-free quantification algorithm in the MaxQuant software. Statistical significance was analysed from three biological replicates.
In vivo ubiquitination assay
Cells grown on polystyrene dishes were rinsed in PBS, suspended in 100 μl denaturing lysis buffer (1% SDS, 50 mM Tris (pH 7.4), 5 mM EDTA, 25 mM N-ethylmaleimide) and boiled for 5 min at 95°C. The lysates were then diluted 1:10 in non denaturing lysis buffer (1% Triton-X 100, 300 mM NaCl, 50 mM Tris (pH 7.4), 5 mM EDTA, 25 mM N-ethylmaleimide), incubated on ice for 5 min, homogenized using a needle, and cleared by centrifugation. FLAG immunoprecipitation was then carried out according to the manufacturer’s protocol (Sigma).
GST and His pull downs
ILK-FLAG was expressed in CHO cells and purified with the FLAG affinity resin (Sigma). α-Parvin cDNA was subcloned into pGEX-4T1 (GE Healthcare) for the expression of GST-fusion protein and CHIP cDNA in pET-16b (Millipore) for the expression of His-tagged protein. Recombinant α-parvin and CHIP were subsequently expressed and purified from BL21 E. coli under reducing conditions. Recombinant Hsp90-His and Hsc70 were purchased from Abcam and Enzo Life Sciences, respectively.
His pull downs were performed by incubating 0.1 nmol of recombinant proteins in His reaction buffer (20 mM Tris (pH 8.0), 150 mM NaCl, 10 mM imidazole, 0.25 mM ATP) for 1 h at room temperature. Proteins were precipitated with TALON Metal Affinity resin (Clontech) for 1 h and washed extensively with 20 mM Tris (pH 8.0), 150 mM NaCl, 15 mM imidazole. Bound proteins were eluted in 20 mM Tris (pH 8.0), 150 mM NaCl and 200 mM imidazole, and reduced by boiling in Laemmli sample buffer, followed by western blot analysis.
For GST pull downs 1 μg of recombinant proteins were incubated in GST reaction buffer buffer (50 mM Tris (pH 7.4), 100 mM NaCl, 2 mM MgCl2, 0.25 mM ATP) for 45 min at room temperature. Proteins were then precipitated with glutathione beads (Novagen) for 30 min, washed extensively with reaction buffer, eluted from beads using Laemmli sample buffer, and analysed by western blotting.
In vitro ubiquitination assay
In vitro ubiquitination assays were performed by incubating 0.5 μg recombinant human ILK (Origene), 0.1 μM E1 (Calbiochem), 8 μM UbcH5a (Sigma), 4 μM CHIP-His, 100 nM Hsp40 and1 μM Hsc70 (gifts from J. Höhfeld), and 5 mM ATP, 2.5 mg/ml Ubiquitin (Boston Biochem) in 20 μl of reaction buffer (20 mM MOPS (pH 7.2), 100 mM KCl, 5 mM MgCl2, 10 mM DTT, 1 mM PMSF) for 2 h at 30°C. Reactions were stopped by adding Laemmli sample buffer, after which samples were analysed by western blot.
Immunofluorescence
For cellular immunostaining cells were grown on glass coverslips and fixed in 4% paraformaldehyde. Nonspecific protein binding sites were saturated with 3% bovine serum albumin (BSA) and 5% goat serum in PBS, followed by incubation with primary antibodies in blocking solution and subsequent incubation with secondary antibodies. Finally, the coverslips were washed and mounted on glass slides with elvanol. The following primary antibodies were used: mouse anti-ILK (1:500; BD Transduction Laboratories), anti-paxillin (1:300; BD Biosciences), phalloidin Alexa488 (1:500; Invitrogen), anti-CHIP N-terminus (1:500; Sigma), and anti-Hsp90 (1:250; Cell Signaling). Secondary antibodies were conjugated with Alexa488, Alexa546 (Invitrogen) or Cy3 (Jackson ImmunoResearch Laboratories). Fluorescent images were collected by laser scanning confocal microscopy (TCS SP5X; Leica) with a × 100 or × 20 oil objective using LAS software. All images were recorded sequentially and averaged at least twice. Acquired confocal images in Figure 4 were subsequently deconvoluted using the Huygens Professional software (Scientific Volume Imaging Inc).
For quantification of the fibronectin matrix, four random fields were collected with a confocal microscope using a × 20 objective. Matrix structures were detected by intensity-based thresholding using ImageJ software. Fluorescent intensity from this area was then measured and integrated over total matrix area. Subsequently, total cell surface area was defined from phalloidin staining of the corresponding fields and the integrated fibronectin intensity was normalized over the total cell surface area.
Live-cell imaging
Fibroblasts were grown on glass bottom imaging plates and transfected with Paxillin-Cherry. Live images were captured with an Axio Observer (Zeiss) microscope, a CSU10 spinning-disc confocal scanhead (Yokogawa), and an Evolve EMCCD camera (Photometrics), with a × 100 oil objective. Acquisition was controlled by Metamorph software. Images were collected at 37°C, 5% CO2, and processed by low pass filtering. Quantification of focal adhesion areas was performed using Metamorph software. First, focal adhesions marked by Paxillin-Cherry were identified using intensity-based thresholding. Subsequently, focal adhesion surface area was measured as a function of time using the integrated morphometric analysis tool. The total focal adhesion surface area at time point 0 was then set as 1, and surface area at 30, 60, 90, and 120 min of imaging was normalized to the value at time point 0. This normalization yielded the relative surface area of focal adhesion over time. Twenty movies/condition from three independent experiments were analysed.
3D migration assay
Cells (1 × 106) were resuspended in 1 ml of growth medium and mixed with type I collagen (PureCol; 3 mg/ml) in 1:2 ratio. The gels were loaded in custom-made invasion chambers and allowed to harden at 37°C for 30 min. DMEM supplemented with 10% FCS and 1 mg/ml EGF, as well as additional inhibitors, when required, were subsequently applied on top of the gel. The chambers were sealed and imaged using a Zeiss Axiovert 40C microscope (× 5 objective) equipped with a CCD camera (Prosilica, GC2450), heated stage (37°C) with 4.7% CO2 with 1 min/frame for 20 h. Cell migration was tracked and quantified using the Chemotaxis and Migration plug-in tool of ImageJ software. Quantifications represent five independent experiments.
3D gel contraction assay
Fibroblasts (2 × 106) were suspended in 800 μl of 5 × Hank’s Balanced Salt solution and 50% fetal calf serum, and mixed with 3 ml of type I collagen (4 mg/ml), avoiding air bubble formation. The gels were loaded in 24-well plates (500 μl) and allowed to harden at 37°C for 30 min. DMEM supplemented with 10% FCS was subsequently applied on top of the gel. After allowing the cells to spread for 16 h, 1 μM 17AAG or the vehicle DMSO was added into the growth medium. After 3 days of culture, gel contraction was determined by measuring gel diameter. Experiments were performed in triplicates and quantifications represent three independent experiments.
Traction force microscopy
Traction force microscopy was performed essentially as described (Dembo and Wang, 1999). Briefly, 20 kPa polyacrylamide gels (7.5% acrylamide/0.25% bis-acrylamide) containing 0.2 μm fluorescent beads (1:125; Polysciences) were casted on glass-bottomed imaging plates after which fibronectin was chemically crosslinked on gels using Sulfo-SANPAH (Pierce). Gel stiffness was measured as described (Pelham and Wang, 1997). Cells were allowed to adhere and spread, after which they were treated with 5 μM 17AAG for 6 h. Imaging was performed using a spinning disc microscope described above with a × 63 oil objective at 37°C with 5.0% CO2. Cells were imaged for 20 min at 1 min/frame rate in medium containing 0.05% trypsin to detach cells and obtain bead displacement images. Calculation of traction forces was performed using particle imaging velocimetry (PIV) and Fourier transform traction cytometry (FTTC) with regularization (9 × 10−10) using ImageJ as described previously (Maruthamuthu et al, 2011; Tseng et al, 2011). Traction forces were reconstructed at a grid spacing of 6.5 μm and total cellular force was calculated from the sum of traction magnitudes. At least 30 cells/condition were analysed.
Bleomycin-induced fibrosis
Skin fibrosis was induced by intradermal injection of bleomycin sulphate (100 μl, 1 mg/ml in 0.9% NaCl) into the back skin of anaesthetized and shaved 6-week-old female C57/Bl6 mice 5 days/week for 2 weeks as described (Yamamoto et al, 1999). Intradermal injections of NaCl (100 μl, 0.9%) were used as controls. To analyse the effect of 17AAG on the fibrotic response, four different experimental groups were composed: (1) bleomycin-injected mice with simultaneous intraperitoneal injections of 17AAG (60 mg/kg dissolved in a vehicle containing 2% egg yolk phospholipid, 5% dextrose, and 10% DMSO), (2) bleomycin-injected mice with simultaneous injection of vehicle, (3 and 4) NaCl injected mice treated with 17AAG or the vehicle, respectively. After 2 weeks, mice were sacrificed and skin lesions were subjected to histological analyses. The fibrotic response was determined by quantifying the dermal thickness within the lesion (measured as the area of dermal fibrotic tissue from the dermo-epidermal junction to the fat tissue over a defined length). All animal experiments were approved by local veterinary authorities (permit number 8.87-50.10.31.08.197).
Histological analyses
Staining of paraffin sections with haematoxylin/eosin, Masson’s trichrome, and picrosirius red was performed using standard protocols. The extent of fibrotic alteration of the skin was assessed by an experienced dermato-pathologist in a blinded fashion by scoring 2–6 picrosirius stained sections of 5–8 biopsies per group, on a scale of 0 (no fibrotic changes) to 4 (significant fibrosis). Rating took into account dermal thickness, density of collagenous connective tissue, and extent of replacement of subcutaneous adipose tissue by connective tissue. α-SMA staining was performed as described earlier (Tomasek et al, 2005) using fluorescein-conjugated mouse anti-SMA (Sigma).
Statistical analyses
Statistical analyses were performed using GraphPad Prism software (GraphPad, version 5.0). Statistical significance was determined by Friedman’s analysis of variance (ANOVA) with a Dunn’s post hoc test (Figure 7A), Kruskal–Wallis with Dunn’s (Figure 7C and G), or ANOVA with Tukey post hoc test (Figures 7E, 8B and F; Supplementary Figure S5C and E). In Figure 6 D a Student’s t-test was used. In all cases normal distribution of data was confirmed with the Kolmogorov–Smirnov test (α=0.05).
Supplementary Material
Acknowledgments
We thank Dr Jörg Höhfeld (University of Bonn, Germany) for reagents and helpful discussions, Torsten Bücher and Angelika Arora for excellent technical assistance, and Christian Pichlo for help with the in vitro binding assays and live-cell imaging. This work was supported by the Max Planck Society (to MM, RF, and SW), the Max Planck Foundation (to SW), and the Deutsche Forschungsgemeinschaft through SFB 829 (to BE, TK, and SW).
Author contributions: KR and JM performed experiments and analysed data. J-NS and KB performed and analysed the fibrosis experiments. TK and BE designed and analysed the fibrosis experiments. TG and MM designed, performed, and analysed the mass spectrometry experiments. CP provided the CHIP−/− mice. RF supervised the study, designed experiments, and analysed data. SW supervised the study, designed and performed experiments, analysed data, and wrote the paper. All authors edited the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aoyagi Y, Fujita N, Tsuruo T (2005) Stabilization of integrin-linked kinase by binding to Hsp90. Biochem Biophys Res Commun 331: 1061–1068 [DOI] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Höhfeld J (2010) Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 20: 143–148 [DOI] [PubMed] [Google Scholar]
- Azimifar SB, Böttcher RT, Zanivan S, Grashoff C, Kruger M, Legate KR, Mann M, Fässler R (2012) Induction of membrane circular dorsal ruffles requires co-signalling of integrin-ILK-complex and EGF receptor. J Cell Sci 125: 435–448 [DOI] [PubMed] [Google Scholar]
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 19: 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumbach K, Zweers MC, Brunner G, Peters AS, Schmitz M, Schulz JN, Schild A, Denton CP, Sakai T, Fässler R, Krieg T, Eckes B (2010) Defective granulation tissue formation in mice with specific ablation of integrin-linked kinase in fibroblasts—role of TGFbeta1 levels and RhoA activity. J Cell Sci 123: 3872–3883 [DOI] [PubMed] [Google Scholar]
- Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, Cyr D, Patterson C (2003) CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J 22: 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Wang YL (1999) Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J 76: 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Gupta S, Chen K, Wu C, Qin J (2009) The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol Cell 36: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Chen K, Shi X, Wu C (2003) PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J Biol Chem 278: 51324–51333 [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Grashoff C, Schwartz MA (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Bass MD, Craig SE, Pinney JW, Knight D, Humphries MJ (2009) Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal 2: ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ (2006) Integrin ligands at a glance. J Cell Sci 119: 3901–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Hynes RO, Naba A (2012) Overview of the matrisome–an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 4: a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Kundrat L, Regan L (2010) Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 49: 7428–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Wickström SA, Jakobson M, Zent R, Sainio K, Fässler R (2009) Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature 461: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Li S, Bordoy R, Stanchi F, Moser M, Braun A, Kudlacek O, Wewer UM, Yurchenco PD, Fässler R (2005) PINCH1 regulates cell-matrix and cell-cell adhesions, cell polarity and cell survival during the peri-implantation stage. J Cell Sci 118: 2913–2921 [DOI] [PubMed] [Google Scholar]
- Lorenz K, Grashoff C, Torka R, Sakai T, Langbein L, Bloch W, Aumailley M, Fässler R (2007) Integrin-linked kinase is required for epidermal and hair follicle morphogenesis. J Cell Biol 177: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V, Sabass B, Schwarz US, Gardel ML (2011) Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci USA 108: 4708–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD (2005) Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol 6: 56–68 [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM (1996) The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol 70: 5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Workman P (2012) Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res 18: 64–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyamsuren O, Faggionato D, Loch W, Schulze E, Baumeister R (2007) A mutation in CHN-1/CHIP suppresses muscle degeneration in Caenorhabditis elegans. Dev Biol 312: 193–202 [DOI] [PubMed] [Google Scholar]
- Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL (2010) Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ Jr., Wang Y (1997) Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94: 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y (2008) The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem 283: 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabass B, Gardel ML, Waterman CM, Schwarz US (2008) High resolution traction force microscopy based on experimental and computational advances. Biophys J 94: 207–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Li S, Docheva D, Grashoff C, Sakai K, Kostka G, Braun A, Pfeifer A, Yurchenco PD, Fässler R (2003) Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev 17: 926–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Friedel CC, Boulegue C, Fässler R (2011) Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep 12: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Chen ZJ (2005) Protein ubiquitination: CHIPping away the symmetry. Mol Cell 20: 653–655 [DOI] [PubMed] [Google Scholar]
- Schwartz MA (2010) Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol 2: a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwock J, Dhani N, Cao MP, Zheng J, Clarkson R, Radulovich N, Navab R, Horn LC, Hedley DW (2009) Targeting focal adhesion kinase with dominant-negative FRNK or Hsp90 inhibitor 17-DMAG suppresses tumor growth and metastasis of SiHa cervical xenografts. Cancer Res 69: 4750–4759 [DOI] [PubMed] [Google Scholar]
- Stanchi F, Grashoff C, Nguemeni Yonga CF, Grall D, Fässler R, Van Obberghen-Schilling E (2009) Molecular dissection of the ILK-PINCH-parvin triad reveals a fundamental role for the ILK kinase domain in the late stages of focal-adhesion maturation. J Cell Sci 122: 1800–1811 [DOI] [PubMed] [Google Scholar]
- Stankiewicz M, Nikolay R, Rybin V, Mayer MP (2010) CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J 277: 3353–3367 [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 11: 515–528 [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S (2012) Quantitative analysis of hsp90-client interactions reveals principles of substrate recognition. Cell 150: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA (2002) Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol 3: 349–363 [DOI] [PubMed] [Google Scholar]
- Tomasek JJ, McRae J, Owens GK, Haaksma CJ (2005) Regulation of alpha-smooth muscle actin expression in granulation tissue myofibroblasts is dependent on the intronic CArG element and the transforming growth factor-beta1 control element. Am J Pathol 166: 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Q, Wang I, Duchemin-Pelletier E, Azioune A, Carpi N, Gao J, Filhol O, Piel M, Thery M, Balland M (2011) A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab on a chip 11: 2231–2240 [DOI] [PubMed] [Google Scholar]
- Wickström SA, Lange A, Hess MW, Polleux J, Spatz JP, Kruger M, Pfaller K, Lambacher A, Bloch W, Mann M, Huber LA, Fässler R (2010a) Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev Cell 19: 574–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström SA, Lange A, Montanez E, Fässler R (2010b) The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase!. Embo J 29: 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström SA, Radovanac K, Fässler R (2011) Genetic analyses of integrin signaling. Cold Spring Harb Perspect Biol 3: a005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KH, Lin X, Feng XH (2008) Critical regulation of TGFbeta signaling by Hsp90. Proc Natl Acad Sci USA 105: 9244–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Ramalingam TR (2012) Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18: 1028–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Takagawa S, Katayama I, Yamazaki K, Hamazaki Y, Shinkai H, Nishioka K (1999) Animal model of sclerotic skin. I: local injections of bleomycin induce sclerotic skin mimicking scleroderma. J Invest Dermatol 112: 456–462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.