Abstract
EMBO J (2013) 32 10, 1381–1392. doi:; DOI: 10.1038/emboj.2013.42
A new study by Bevington and Boyes reports that expression of the IRF4 transcription factor is sufficient to prematurely activate assembly of immunoglobulin light chain loci in pro-B cells. Using this in vivo model, they provide evidence that transcription-coupled disruption of nucleosome cores may be a key event in unlocking DNA substrates for recognition and assembly by V(D)J recombinase.
Immunoglobulin (Ig) and T-cell receptor (Tcr) diversity is generated in developing lymphocytes via somatic recombination of V (variable), D (diversity) and J (joining) gene segments, which are splayed out on seven distinct antigen receptor loci. This assembly process is mediated by a V(D)J recombinase machinery consisting of RAG1 and RAG2 as its core enzymatic components (Cobb et al, 2006). The RAG1/2 complex targets conserved recombination signal sequences (RSSs) that directly flank all V, D and J gene segments. The selection and recombination of gene segments is tightly regulated throughout lymphocyte development, restricting the assembly of Ig or Tcr loci to a single lineage (B or T cells, respectively) and to a specific stage within that lineage (Feeney, 2009). Much of this regulation relies on changes in the accessibility of chromatin associated with RSSs, permitting or preventing their recognition by V(D)J recombinase.
Similar to other forms of gene regulation, changes in RSS accessibility involve revisions to local patterns of histone modifications and reconfiguration of nucleosomes, both of which are linked to the transcriptional activity of a region (Abarrategui and Krangel, 2006; Osipovich et al, 2007). At Ig and Tcr loci, transcription and rearrangement of gene segments are coordinated by a collection of cis-acting regulatory elements, including transcriptional enhancers and promoters. Transcription of unrearranged gene segments is accompanied by the deposition of tri-methylated histone 3 lysine 4 (H3K4me3), a covalent modification that attracts the RAG complex (Liu et al, 2007; Matthews et al, 2007). Transcription also augments the local accessibility of chromatin to nuclear factors via the clearance and/or remodelling of nucleosomes; however, the precise mechanisms for generating accessibility of chromosomal RSSs to V(D)J recombinase remain unclear.
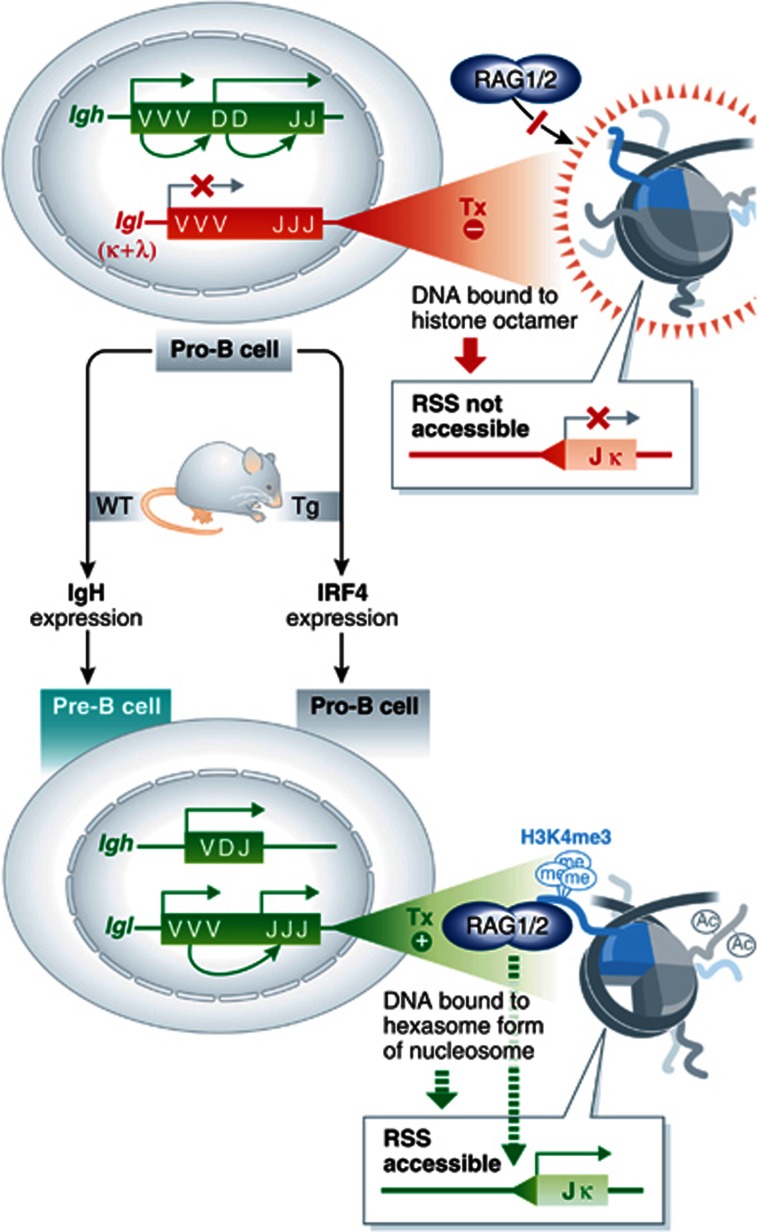
Bevington and Boyes (2013) studied the molecular requirements for activation of Ig light chain loci, which are normally silent in early-stage pro-B cells but become prime targets for V(D)J recombinase in a later stage, called pre-B cells, following functional assembly of an Ig heavy chain gene. Developmentally appropriate targeting of the recombination machinery to Ig kappa (Igk) and lambda (Igl) light chain loci depends on the association of stage-specific transcription factors with distal regulatory elements. Prior studies have shown that the pro- to pre-B cell transition is blocked in Interferon Regulatory Factor 4 (IRF4)/IRF8 double-knockout mice, but can be rescued by expression of only IRF4 (Lu et al, 2003; Ma et al, 2006). The developmental consequences of these manipulations to IRF4/8 expression are largely linked to their effects on induction of Igk and Igl regulatory elements, which, in turn, control light chain gene recombination. In normal animals, expression of IRF4 is upregulated during the pro- to pre-B cell transition, suggesting that IRF4 may be a gatekeeper, restricting Ig light chain gene assembly to pre-B cells (Figure 1). To test this hypothesis, Bevington and Boyes create transgenic mice that force premature IRF4 expression in pro-B cells and find that, indeed, IRF4 is sufficient to induce early light chain transcription and rearrangement in this progenitor subset (Figure 1). Moreover, enforced IRF4 expression breaks the normal order of Ig light chain rearrangement, with more efficient Igl recombination compared with Igk in the transgenic pro-B cells.
Figure 1.
(Left) Developmental control of Ig gene rearrangements. Pro-B cells normally undergo Igh recombination (active, green), but repress assembly of Igk and Igl light chain genes (inactive, red). Upon functional assembly and expression of Igh, wild-type (WT, left arrow) pro-B cells differentiate into pre-B cells and activate Igk/l recombination. Bevington and Boyes show that the developmental block in Igk/l activation can be overcome by transgene-driven expression of IRF4 in pro-B cells (Tg, right arrow). (Right) Authors’ model of recombinase accessibility. RSSs (triangle) flanking Igk/l gene segments are transcriptionally inert (Tx –) in wild-type pro-B cells (top) and associate with conventional nucleosome octamers, which block access to the RAG complex. In pre-B cells or pro-B cells expressing IRF4, transcriptional activation decorates histone tails with acetylation (Ac) and H3K4me3, the latter of which serves as a platform for stable docking of RAG complexes. Transcription also leads to the expulsion of histone dimers, resulting in a hexasome form of nucleosomes, which may unmask RSSs for cleavage by RAG.
With these observations in hand, Bevington and Boyes use B-cell precursors from their in vivo model to explore mechanistic aspects of RSS accessibility. First, they show that preferential assembly of Igl versus Igk genes correlates with relative levels of transcription, but not with H3K4me3 at the composite gene segments. From the latter observation, the authors conclude that forced activation of recombination can be uncoupled from simple recruitment of RAG by its binding to H3K4me3. Instead, Bevington and Boyes proceed to test whether transcription-coupled reconfiguration of the nucleosomes that are associated with gene segments is a primary requirement for targeting by RAG. Using a combination of molecular and biochemical approaches, they provide evidence that transcription mediates transient access of the RAG complex to RSSs, which is accompanied by the partial loss of histone H2B from resident nucleosomes. This process is reminiscent of transcription-coupled eviction of H2A–H2B dimers from nucleosomes during the passage of RNA polymerase, transiently leaving a ‘hexasome’ form of nucleosomes on transcribed regions. The ephemeral hexasomes are thought to enhance regional accessibility to other nuclear factors for several minutes (Thiriet and Hayes, 2006).
To further support their proposed model of recombinase accessibility, the authors show that RSS substrates assembled into hexasomes are more efficiently cleaved by recombinant RAG proteins in vitro than when the same substrates are assembled with conventional nucleosome octamers (Figure 1). This observation raises the interesting possibility that the shorter footprint of hexasome-bound DNA generated during transcription of gene segments is a key mechanism for promoting RSS accessibility to recombinase. Moreover, the octamer to hexasome conversion may underlie transcription-coupled reorganization or depletion of nucleosomes reported for recombinase-accessible loci (Kondilis-Mangum et al, 2010). However, future in vivo studies are required to clarify the relative importance of nucleosome depletion, remodelling and histone eviction in preparing Ig and Tcr gene segments for recombination.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abarrategui I, Krangel MS (2006) Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol 7: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Bevington S, Boyes J (2013) Transcription-coupled eviction of histones H2A/H2B governs V(D)J recombination. EMBO J 32: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM (2006) Accessibility control of V(D)J recombination. Adv Immunol 91: 45–109 [DOI] [PubMed] [Google Scholar]
- Feeney AJ (2009) Genetic and epigenetic control of V gene rearrangement frequency. Adv Exp Med Biol 650: 73–81 [DOI] [PubMed] [Google Scholar]
- Kondilis-Mangum HD, Cobb RM, Osipovich O, Srivatsan S, Oltz EM, Krangel MS (2010) Transcription-dependent mobilization of nucleosomes at accessible TCR gene segments in vivo. J Immunol 184: 6970–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S (2007) A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Medina KL, Lancki DW, Singh H (2003) IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev 17: 1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Turetsky A, Trinh L, Lu R (2006) IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development. J Immunol 177: 7898–7904 [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo A.J, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA (2007) RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM (2007) Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol 8: 809–816 [DOI] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ (2006) Histone dynamics during transcription: exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation. Results Probl Cell Differ 41: 77–90 [DOI] [PubMed] [Google Scholar]