Abstract
Pteridine reductase (PTR1) is a potential target for drug development against parasitic Trypanosoma and Leishmania species. These protozoa cause serious diseases for which current therapies are inadequate. High-resolution structures have been determined, using data between 1.6 and 1.1 Å resolution, of T. brucei PTR1 in complex with pemetrexed, trimetrexate, cyromazine and a 2,4-diaminopyrimidine derivative. The structures provide insight into the interactions formed by new molecular entities in the enzyme active site with ligands that represent lead compounds for structure-based inhibitor development and to support early-stage drug discovery.
1. Introduction
The protozoan parasites Trypanosoma and Leishmania species are the causal agents of serious diseases such as human African trypanosomiasis (HAT; sleeping sickness) and the different forms of leishmaniasis (Barrett et al., 2007; Reithinger et al., 2007). The drugs in current use, for example the arsenical compound called melarsoprol, are toxic, possess limited efficacy and are costly. In addition, the available treatments are being compromised by the increase in drug-resistant parasites (Croft et al., 2006; Delespaux & de Koning, 2007). New drugs are therefore urgently sought and the significant advances in understanding the biology and metabolism of the protozoans holds the promise of identifying new targets for the application of structure-based inhibitor approaches to underpin early-stage drug discovery (Hunter, 2009).
One promising target is pteridine reductase (PTR1; EC 1.5.1.33), an enzyme unique to trypanosomatid parasites that supports the provision of reduced biopterins necessary for metacyclogenesis (Cunningham et al., 2001) and which has been implicated in resistance to reactive oxygen and nitrogen species in Leishmania (Moreira et al., 2009). This NADPH-dependent short-chain dehydrogenase/reductase (SDR) is a broad-spectrum pterin reductase that is able to catalyze four reductions (Nare et al., 1997): biopterin to dihydrobiopterin (H2B), H2B to tetrahydrobiopterin (H4B), folate to dihydrofolate (H2F) and H2F to tetrahydrofolate (H4F). The last of these reductions is the same reaction as is catalyzed by dihydrofolate reductase (DHFR; EC 1.5.1.3; Blakley, 1995), a key enzyme of folate metabolism and a validated target for antifolate drugs such as methotrexate (MTX), pyrimethamine and trimethoprim (McGuire, 2003). PTR1, since it is able to reduce H2F, contributes to antifolate drug resistance by providing a bypass of DHFR inhibition (Hardy et al., 1997).
We have previously identified distinct molecular scaffolds to support the design of new PTR1 inhibitors based on structural data for the T. brucei (TbPTR1) and L. major (LmPTR1) enzymes (Gourley et al., 2001; McLuskey et al., 2004; Schüttelkopf et al., 2005; Dawson et al., 2006; Gibson et al., 2009; Mpamhanga et al., 2009) and have shown that dual DHFR–PTR1 inhibition may provide a successful treatment for trypanosomatid infections (Cavazzuti et al., 2008; Tulloch et al., 2010). Potent DHFR inhibitors are already known and we seek to design or discover novel PTR1 inhibitors, concentrating on the enzyme from T. brucei (TbPTR1) since this organism causes the disease with the greatest unmet medical need, HAT. We identified ligands of interest (Fig. 1) and predicted that the three largest compounds, each of which has a well defined pharmacology and proven bioavailability, would be likely to be potent inhibitors of the enzyme. These compounds carry a number of new molecular features, e.g. dimethoxyphenyl and trimethoxyphenyl groups, that would serve to inform about what interactions are possible in the PTR1 active site.
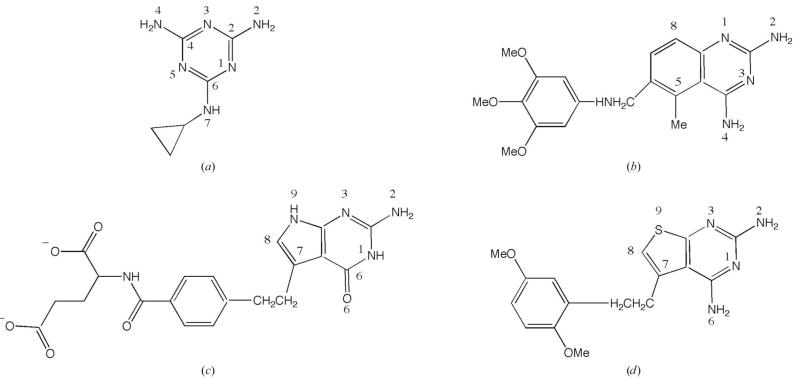
Figure 1.
The chemical structures and atomic numbering of the four ligands. (a) Cyromazine, N-cyclopropyl-1,3,5-triazine-2,4,6-triamine. (b) Trimetrexate, 5-methyl-6-[(3,4,5-trimethoxyphenyl)aminomethyl]quinazoline-2,4-diamine. (c) Pemetrexed, 2-{4-[2-(4-amino-2-oxo-3,5,7-triazabicyclo[4.3.0]nona-3,8,10-trien-9-yl)ethyl]benzoyl}aminopentanedioic acid. (d) PY848, 2,4-diamino-5-[2-(2,5-dimethoxyphenyl)ethyl]thieno[2,3-d]-pyrimidine.
We now report high-resolution crystal structures of these TbPTR1 ligand complexes. Cyromazine (N-cyclopropyl-1,3,5-triazine-2,4,6-triamine) is a cyclopropyl derivative of melamine that is used in veterinary medicine as an ectoparasiticide but with an unknown mode of action. The structure has been reported at 2.0 Å resolution previously (Tulloch et al., 2010) and we now extend that to 1.1 Å resolution, revealing a level of detail not previously observed. Pemetrexed (2-{4-[2-(4-amino-2-oxo-3,5,7-triazabicyclo[4.3.0]nona-3,8,10-trien-9-yl)-ethyl]benzoyl}aminopentanedioic acid) is a folate anti-metabolite (Avendaño & Menendez, 2008) which inhibits thymidylate synthase, DHFR and glycinamide ribonucleotide formyltransferase, enzymes that are involved in both purine and pyrimidine biosynthesis. This compound is in clinical use for the treatment of lung cancer (Baldwin & Perry, 2009). Trimetrexate {5-methyl-6-[(3,4,5-trimethoxyphenyl)aminomethyl]quinazoline-2,4-diamine} is a potent DHFR inhibitor that is used in the treatment of pneumocystis pneumonia and is being investigated as an anticancer agent (Fishman, 1998; Purcell & Ettinger, 2003). Finally, there is the pyrimidine derivative 2,4-diamino-5-[2-(2,5-dimethoxyphenyl)ethyl]thieno-[2,3-d]-pyrimidine (PY848; Rosowsky et al., 1993). To complement the structural data, Ki values were determined.
2. Materials and methods
2.1. Protein purification and storage
Recombinant TbPTR1 was purified by established methods (Dawson et al., 2006) and was concentrated to 22 mg ml−1 in 20 mM Tris–HCl pH 8.0. Aliquots were frozen in liquid nitrogen and stored at 253 K until required.
2.2. Ligand sourcing and inhibition assay
Cyromazine was a gift from Novartis Animal Health Inc. (Basel, Switzerland); the other compounds were obtained from the Center for Organic and Medicinal Chemistry, Research Triangle International (North Carolina, USA) and were used as supplied. The Ki values for these ligands were determined using a previously published method, with MTX providing a control (data not shown; Dawson et al., 2006).
2.3. Crystal preparation
Crystals of TbPTR1 in a binary complex with oxidized cofactor (NADP+) were grown using sitting-drop vapour diffusion in drops consisting of 10 μl protein solution (6 mg ml−1 TbPTR1, 1 mM NADP+ and 1 mM DTT in 20 mM Tris–HCl pH 8.0) and 10 μl reservoir solution (2–3 M sodium acetate and 20–100 mM sodium citrate pH 4.5). The conditions were stored at room temperature (approximately 291 K) and monoclinic plate-like crystals formed overnight and grew to 2 mm in length over several days. The ligands pemetrexed, trimetrexate and PY848 (2.5 μl at a concentration of 10 mM) were added to drops containing one or two large crystals and the samples were left for 24 h. The final concentration of the ligand was approximately 1 mM.
Crystals of the TbPTR1–NADPH–cyromazin complex were grown in hanging drops consisting of 1.5 μl protein solution [TbPTR1 at 6–10 mg ml−1, 1 mM NADPH, 3 mM cyromazin, 1%(v/v) DMSO and 20 mM DTT] and an equal volume of reservoir solution (1.5–3 M sodium acetate, 10–50 mM sodium citrate in the pH range 4.5–6.0) at approximately 291 K. The largest crystals grew to approximately 0.5 × 0.3 × 0.1 mm over a few days. Crystals grown in excess of 2.6 M sodium acetate were flash-cooled in liquid nitrogen directly from the mother liquor. Those obtained at a lower salt concentration were cryoprotected with 3 M sodium acetate prior to flash-cooling.
2.4. X-ray data collection and structure determination
X-ray data were collected at the European Synchrotron Radiation Facility (ESRF), Grenoble, France on beamline ID14-4 for the pemetrexed, trimetrexate and PY848 complexes and on beamline ID29 for the cyromazine complex. Both beamlines were equipped with an ADSC Q315 CCD detector. X-ray images from the pemetrexed and trimetrexate complexes were integrated and scaled with XDS and XSCALE (Kabsch, 2010). MOSFLM (Leslie, 2006) and SCALA (Evans, 2006) were used to process the data from the PY848 and cyromazine complexes. All of the complex structures are isomorphous, with four subunits in the asymmetric unit, and refinement started using a tetramer of TbPTR1 (Dawson et al., 2006; PDB code 2c7v) for rigid-body refinement in REFMAC5 (Murshudov et al., 1999). Several rounds of restrained refinement, still using REFMAC5, were carried out together with inspection of electron-density and difference density Fourier maps (Collaborative Computational Project, Number 4, 1994), model manipulation and identification of multiple conformers, water molecules, ions and ligands with Coot (Emsley & Cowtan, 2004) using similar protocols to those previously published (Tulloch et al., 2010). Noncrystallographic symmetry restraints were not used at any stage in these analyses. In all four cases, the initial refinement calculations included isotropic thermal parameters and these were subsequently changed to anisotropic thermal parameters for the cyromazine, pemetrexed and trimetrexate complexes. With the high-resolution diffraction data available for the cyromazine and pemetrexed complexes, it was decided to switch to the refinement program SHELX (Sheldrick, 2008). C—H and main-chain amide N—H H atoms were included in the final stages of refinement with anisotropic thermal parameters refined for all non-H atoms using appropriate restraints. Figures were generated with PyMOL (DeLano, 2002). Crystallographic statistics are given in Table 1.
Table 1.
Crystallographic statistics. Values in parentheses are for the highest resolution shell.
| Ligand | Cyromazin | Pemetrexed | PY848 | Trimetrexate |
|---|---|---|---|---|
| PDB code | 3×9n | 2×9g | 3mcv | 2×9v |
| Resolution range (Å) | 45.13–1.15 | 41.80–1.10 | 44.90–1.70 | 31.05–1.30 |
| Space group | P21 | P21 | P21 | P21 |
| Unit-cell parameters | ||||
| a (Å) | 74.50 | 74.72 | 74.50 | 74.56 |
| b (Å) | 90.00 | 90.57 | 89.82 | 90.71 |
| c (Å) | 82.40 | 83.58 | 82.26 | 82.46 |
| β (°) | 115.0 | 115.6 | 115.5 | 115.7 |
| Scaling | ||||
| No. of measurements | 1358806 | 1704349 | 598600 | 1337387 |
| No. of unique measurements | 385887 | 396089 | 105672 | 239044 |
| Multiplicity | 3.4 (3.2) | 4.3 (3.8) | 5.7 (5.5) | 5.6 (5.5) |
| Completeness (%) | 96.2 (93.7) | 98.7 (96.7) | 98.5 (97.4) | 98.8 (98.6) |
| 〈I/σ(I)〉 | 15.3 (4.3) | 15.6 (4.4) | 9.8 (2.4) | 15.9 (4.6) |
| Rmerge† (%) | 6.1 (23.0) | 5.2 (29.2) | 6.1 (30.8) | 8.7 (40.1) |
| Refinement | ||||
| Rwork‡ (%)/No. of reflections | 12.58/368821 | 12.06/378868 | 13.5/100335 | 12.1/226950 |
| Rfree§ (%)/No. of reflections | 15.26/16882 | 14.68/17155 | 16.7/5310 | 14.8/12094 |
| R.m.s.d. bond lengths (Å) | 0.016 | 0.014 | 0.012 | 0.010 |
| R.m.s.d. bond angles (°) | 2.24 | 2.12 | 1.454 | 1.405 |
| DPI¶ | 0.03 | 0.04 | 0.09 | 0.04 |
| Ramachandran plot analysis | ||||
| Favoured (%) | 96.8 | 96.5 | 96.5 | 96.5 |
| Outliers (%) | 0.0 | 0.0 | 0.0 | 0.0 |
| Model | ||||
| Total amino acids | 992 | 1002 | 1000 | 1000 |
| Overall B factor (Å2) | 15.4 | 15.8 | 10.1 | 15.3 |
| Residues in subunit A | 248 | 251 | 251 | 250 |
| Residues in subunit B | 248 | 250 | 251 | 251 |
| Residues in subunit C | 249 | 252 | 248 | 249 |
| Residues in subunit D | 247 | 249 | 250 | 250 |
| B factor, subunit A (Å2) | 11.5 | 14.8 | 12.0 | 11.9 |
| B factor, subunit B (Å2) | 12.5 | 13.4 | 12.1 | 11.8 |
| B factor, subunit C (Å2) | 12.8 | 14.9 | 13.1 | 12.8 |
| B factor, subunit D (Å2) | 12.6 | 14.3 | 13.0 | 12.8 |
| Additional groups | ||||
| Solvent | ||||
| No. | 1382 | 1372 | 793 | 1202 |
| Average B factor (Å2) | 32.4 | 32.2 | 24.6 | 34.9 |
| NADP+ | ||||
| B factor, subunit A (Å2) | 7.4 | 9.1 | 8.8 | 8.6 |
| B factor, subunit B (Å2) | 9.0 | 7.7 | 9.0 | 8.4 |
| B factor, subunit C (Å2) | 8.4 | 7.9 | 11.6 | 8.5 |
| B factor, subunit D (Å2) | 9.2 | 8.5 | 10.2 | 9.8 |
| Ligand | ||||
| B factor, subunit A (Å2) | 9.8 | 14.2 | 14.4 | 16.5 |
| B factor, subunit B (Å2) | 22.3 | 17.7 | 18.8 | 13.4 |
| B factor, subunit C (Å2) | 21.8 | 18.2 | 19.4 | 15.3 |
| B factor, subunit D (Å2) | 23.7 | 19.2 | 19.5 | 16.7 |
| DTT | ||||
| B factor, subunit A (Å2) | 26.1 | — | — | — |
| B factor, subunit B (Å2) | 33.2 | — | — | — |
| B factor, subunit C (Å2) | 31.7 | — | — | 34.1 |
| B factor, subunit D (Å2) | 20.8 | — | — | — |
| Acetate | ||||
| No. | 1 | 1 | 4 | 6 |
| B factor (Å2) | 12.4 | 13.3 | 21.8 | 19.7 |
| Na+ | ||||
| No. | 2 | |||
| B factor (Å2) | 29.1 |
Rmerge = Σhkl Σi|Ii(hkl) − 〈I(hkl)〉|/Σhkl Σi Ii(hkl), where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
Rwork = Σhkl | |FobsI − |Fcalc| |/Σhkl | Fobs |, where Fobs is the observed structure-factor amplitude and Fcalc is the structure-factor amplitude calculated from the model.
Rfree is the same as Rwork except calculated with a 5% subset of data that were excluded from refinement calculations.
Diffraction-component precision index (Cruickshank, 1999).
3. Results and discussion
3.1. TbPTR1 structure and organization of the active site
PTR1 is a tetrameric SDR with a single α/β-domain subunit that is a Rossmann-fold repeat (Gourley et al., 2001; Fig. 2). An elongated active site is primarily formed by a single subunit, with one end blocked by the C-terminus of a partner subunit. A loop formed by residues 207–215 in TbPTR1 forms part of the active site and since it interacts with substrates this is called the substrate-binding loop (Tulloch et al., 2010; Fig. 2). The enzyme structure is well conserved in TbPTR1–ligand complexes (Tulloch et al., 2010) and this observation also applies to the four complexes now reported. For example, the Cα atoms of the four subunits in the TbPTR1–pemetrexed complex overlay with an r.m.s.d. in the range 0.17–0.22 Å. Given the high degree of noncrystallographic symmetry and the conservation of the interactions formed in the four active sites in the asymmetric unit, it is only necessary to detail one and subunit A has been chosen arbitrarily. Some structural variations are observed in the TbPTR1–cyromazine structure involving the propyl substitutent and this is detailed below.
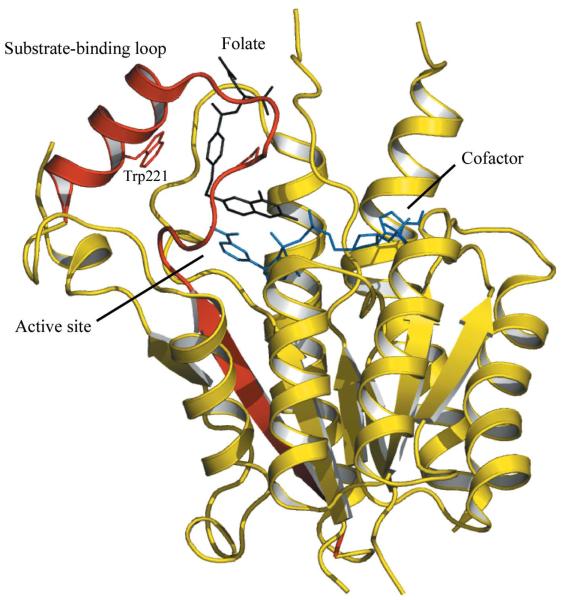
Figure 2.
Ribbon diagram of the TbPTR1 subunit architecture and the position of the active site. The β6–loop–α6 segment is coloured red. The cofactor is depicted as blue sticks and the substrate, folate, as black sticks. The side chain of Trp221, on the edge of the active site, is shown as red sticks. Figs. 2 and 3 were derived using PDB entry 3bmc (Tulloch et al., 2010).
The binding of the cofactor is essential to create both the catalytic centre and the substrate-binding site and explains why PTR1 displays an ordered sequential mechanism with first NADPH binding and then substrate. The product, a reduced pterin, is released followed by NADP+ before another reduction can occur (Luba et al., 1998). The pterin substrates and inhibitors bind in a π-sandwich between the nicotinamide and Phe97 and also interact with the β-phosphate group. Here, the ribose and a phosphate of the cofactor, Arg14, Ser95, Asn127, Lys178 and two catalytically important residues, Asp161 and Tyr174, are positioned to organize the active site and interact with ligands (Fig. 3).
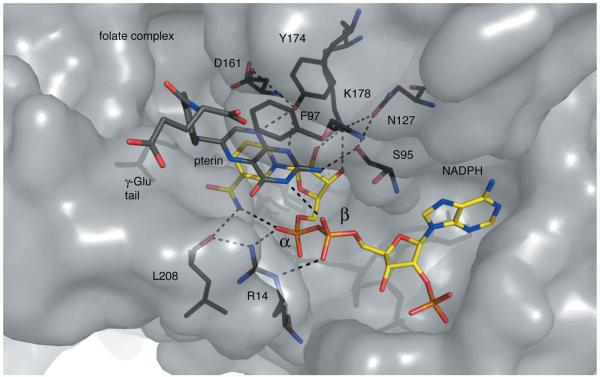
Figure 3.
Residues and hydrogen bonds organize the TbPTR1 active site. PTR1 is shown as a grey semi-transparent van der Waals surface. Specific residues are coloured: C, black; N, blue; O, red. NADPH is depicted with C atoms coloured yellow and P atoms orange and folate with C atoms in dark grey. Dashed lines represent potential hydrogen-bonding interactions.
A series of hydrogen bonds help to position key functional groups to interact with ligands. On one side of the active site, Lys178 NZ donates hydrogen bonds to the nicotinamide ribose O2′ and O3′ groups and to Asn127 OD1. Asn127 ND2 donates two hydrogen bonds to the carbonyl groups of Ala96 and Leu123 (data not shown) and so the side chain is well fixed. Since Asn127 OD1 then accepts a hydrogen bond from Ser96 OG, the serine hydroxyl group is aligned to function as a hydrogen-bond acceptor in the active site. The ribose O2′ is aligned to function as a hydrogen-bond donor. Nearby, Tyr174 OH is within hydrogen-bonding distance of Asp161 OD2. On the other side of the active site, the Arg14 guanidinium group helps to organize the start of the substrate-binding loop that is adjacent to the nicotinamide by donating hydrogen bonds to the main-chain carbonyl groups of Leu208 and Leu209 (not shown). In addition, Arg14 also forms hydrogen bonds to both the α-phosphate and β-phosphate groups of NADPH. The nicotinamide N7 donates hydrogen bonds to the carbonyl group of Leu208 and the α-phosphate O2. These interactions contribute to the placement of the β-phosphate O2 to accept hydrogen bonds donated by ligands.
3.2. The TbPTR1–NADPH–cyromazine complex
The structure of the complex of TbPTR1–NADP+ and cyromazine (Ki > 35 μM) has previously been reported at 2.0 Å resolution (Tulloch et al., 2010) and this is now extended to 1.1 Å by virtue of being able to grow more ordered larger crystals combined with the use of an undulator as opposed to a bending-magnet beamline for diffraction measurements. The placement of the ligand in the active site and key interactions are depicted in Fig. 4. The use of NADPH and a higher concentration of DTT appears to have helped the crystallization process. In the lower resolution structure Cys168 at one side of the active site has been oxidized to sulfenic acid. In this higher resolution structure the cysteine is present as a thiol, but nearby is an oxidized circular DTT that adopts two conformations. This DTToccupies what in other structures is a water-filled cavity. We see no difference in the structure of or the interactions with the different states of the cofactor.
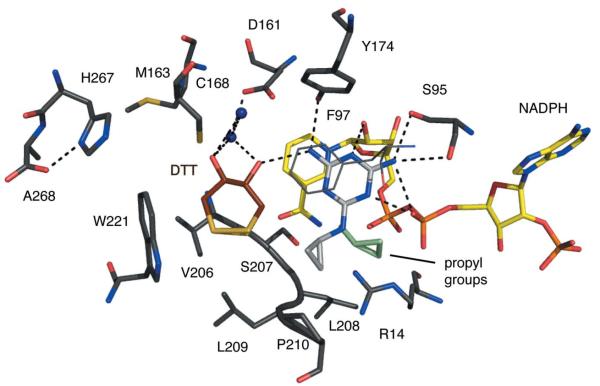
Figure 4.
TbPTR1 in complex with cyromazine. Atomic positions are coloured: N, blue; O, red; P, orange; S, dark yellow; C of PTR1, dark grey; C of cofactor, yellow; C of DTT, brown. The C atoms of cyromazine are shown in light grey but for the second conformer of the propyl group they are shown in light green. The side chain of Phe97 is directly over the cyromazine-binding position and for clarity is shown in thin dark grey lines. Potential hydrogen bonds are depicted as dashed lines and water molecules are shown as blue spheres.
The triazine group binds as previously reported. The proximity of the NADPH β-phosphate O2 to N1 suggests that the ligand is protonated here, as observed when the archetypal antifolate MTX binds DHFR (Bennett et al., 2006) or PTR1 (Gourley et al., 2001). The amino N2 group donates a hydrogen bond to Ser95 OG and the geometry at this high resolution is consistent with a second hydrogen bond: a bifurcated interaction (Leonard et al., 1995) with the Ser95 main-chain carbonyl and the β-phosphate O2. N3 accepts a hydrogen bond from the ribose O2′ and N4 donates one to Tyr174 OH. N7 participates in a water-mediated interaction with O2 of the β-phosphate. The position of the propyl groups varies between the four subunits. In subunit A only one orientation is observed, with van der Waals interactions formed to Arg14, Phe97 and Pro210. In subunits B, C and D the propyl group is disordered over two positions: in addition to the subunit A-type orientation, the propyl group is also observed pointing away from the active-site pocket. Refinement indicated relative occupancies of 0.33:0.67 for these two orientations (data not shown). In the lower resolution structure only one orientation of the propyl group, corresponding to that observed in subunit A, was modelled (Tulloch et al., 2010).
3.3. The TbPTR1–NADP+–pemetrexed complex
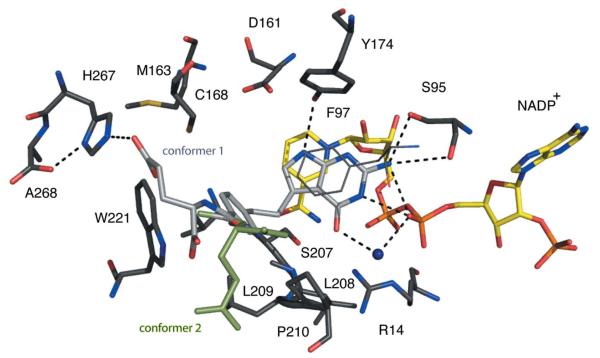
Pemetrexed (Ki of ~30 nM) binds adopting a substrate-like orientation with the pyrimidine positioned in the same way as observed in the complexes with biopterin, H2B and H2F (Fig. 5; Gourley et al., 2001; Tulloch et al., 2010). N1 donates a hydrogen bond to the β-phosphate O2, and N2 forms hydrogen bonds to Ser95, including a bifurcated interaction with the β-phosphate O2. N3 and N9 accept and donate hydrogen bonds to the ribose O2′ and Tyr174 OH, respectively. O6 accepts hydrogen bonds donated from Arg14 and a water molecule, which also interacts with the β-phosphate O2. The benzyl group and linking region form van der Waals interactions with Phe197, Val206, Leu209, Pro210, Met213 (not shown) and Trp221. The glutamate tail is directed out of the active site, is solvent-accessible and adopts two conformations. In one conformation, hydrogen bonds are formed to His267 from a partner subunit in the tetramer and to the main-chain amide of Met169 (data not shown). The other conformation allows van der Waals interactions with Pro99 (data not shown).
Figure 5.
TbPTR1 in complex with pemetrexed. The same colour scheme as Fig. 4 is adopted, with C atoms of the pemetrexed coloured light grey. In addition, the C atoms of conformer 1 of the glutamate tail are shown in light grey and those of conformer 2 are shown in light green.
3.4. The TbPTR1–NADP+–trimetrexate complex
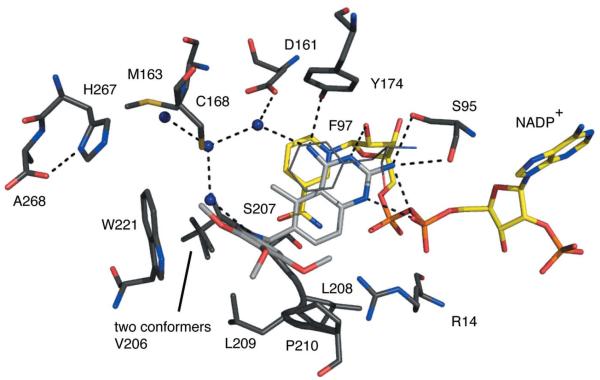
Trimetrexate (Ki of ~70 nM) binds with the quinazoline adopting an orientation similar to that observed for the pterin of MTX (Fig. 6; Gourley et al., 2001). N1 accepts a hydrogen bond from the ribose O2′ and N2 forms the same hydrogen-bonding interactions to Ser95 and the β-phosphate as seen in the other two complexes. N4 donates a hydrogen bond to Tyr174 OH and to a water molecule that also interacts with Asp161. Tyr174 OH and Asp161 OD2 share a hydrogen bond donated by the hydroxyl group. N1 is the site of protonation that allows interaction with the β-phosphate O2. The methyl substituent at C7 is directed into the solvent-filled cavity. The amide in the linker region between the quinazoline and the trimethoxy-substituted phenyl group is directed towards the solvent-filled cavity and donates a hydrogen bond to a well ordered water molecule. This water then forms a hydrogen bond to the thiol group of Cys168. The trimethoxyphenyl group makes van der Waals interactions with Cys168, Phe171 (not shown), Leu209, Pro210, Met213 (not shown) and Trp221. The methoxy groups are all exposed to solvent and interactions with ordered water molecules are observed.
Figure 6.
TbPTR1 in complex with trimetrexate. The same colour scheme as Fig. 4 is adopted, with C atoms of the trimetrexate coloured light grey.
3.5. The TbPTR1–NADP+–PY848 complex
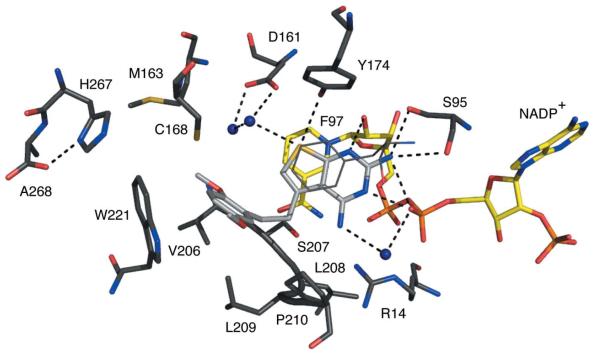
PY848 (Ki of approximately 70 nM) binds in a similar orientation to that seen for pemetrexed despite the difference of the N6 hydrogen-bond-donating amino group compared with the hydrogen-bond-accepting O6 of the latter (Fig. 1). N1, N2 and N3 form similar interactions as noted for cyromazine and pemetrexed (Fig. 7). N6, like N7 of cyromazine, forms a water-mediated link to the β-phosphate O2. S9 accepts hydrogen bonds donated from Tyr178 OH and water that in turn contribute to a network of hydrogen-bonded solvent molecules near Asp161. The dimethoxyphenyl group forms van der Waals interactions with Phe97, Cys168, Leu209, Pro210 and Trp221. One of the methoxy groups is tucked down near Val206; the other is directed out of the active site and is solvent-accessible.
Figure 7.
TbPTR1 in complex with PY848. The same colour scheme as Fig. 4 is adopted, with C atoms of PY848 coloured light grey.
Previous structures of PTR1–ligand complexes have revealed that the orientation of the substrate/product pteridines is distinct from that of compounds such as MTX, which is flipped 180° to place the chemical pattern NH2:N:NH2, as exemplified by N2–N1–N6 of trimetrexate (Figs. 1 and 6), directed towards the Ser95, ribose O2′ and Tyr174 part of the active site. Compound PY848, in either the substrate-like or MTX-like orientation, would be able to interact with the same number of hydrogen-bond contributions between the ligand and PTR1–NADPH. The compound adopts the substrate-like orientation, probably owing to the steric clash that would result between the dimethoxyphenyl group and side chains around the active site.
4. Concluding remarks
Accurate high-resolution crystal structures of four TbPTR1–ligand complexes have been determined and reveal molecular details that have not previously been observed. Common to all four structures is a matching of hydrogen-bonding capacity between the ligands and the active site, in particular forming such interactions between the ligands and the cofactor, Ser95 and Tyr174. Also shared by the four ligands is the placement of the major ring systems between Phe97 and the nicotinamide. Three of the ligands, with the exception being cyromazine, have pronounced van der Waals interactions on one side of the active site. A degree of conformational pliability exists for the propyl substitutent of cyromazine, the weakest binding inhibitor of the four, and the glutamate tail of pemetrexed and suggests areas of the active site where space is available to fill by modification of known inhibitor frameworks. The trimethoxyphenyl and dimethoxyphenyl groups of trimetrexate and PY848, respectively, are well ordered and form stabilizing van der Waals forces with residues at the periphery of the active site, in particular with Trp221. PY848 binds placing the NH2:N:NH2 pattern directed towards the β-phosphate group of the cofactor rather than, as expected, towards the Tyr174 side chain and ribose. Such an orientation prevents the steric clash of the dimethoxyphenyl group with the substrate-binding loop that would be observed with the MTX-like binding as displayed by trimetrexate. These two compounds (trimetrexate and PY848) are potent inhibitors of TbPTR1 and both display a Ki of approximately 70 nM irrespective of the different orientation of the ring systems that bind between Phe97 and the nicotinamide. Trimetrexate adopts the MTX-like mode of binding, but the Ki is improved from 140 nM for MTX to 70 nM. The presence of the trimethoxy group on trimetrexate and the increased van der Waals contacts with a hydrophobic part of the active site created by Leu209, Pro210 and Trp221 in particular may contribute to this enhanced inhibition. The complex structure certainly suggests that this substituent might be incorporated into additional scaffolds to investigate exactly what it might contribute to binding. With a Ki of around 30 nM, pemetrexate registers as a highly potent inhibitor of TbPTR1. In common with those of trimetrexate and PY848, the pemetrexate complex suggests that van der Waals interactions with a hydrophobic part of the active site are important for strong binding. The new structural models that result from our study now provide highly accurate templates that can be exploited by molecular-design approaches seeking to modify known inhibitor frameworks to incorporate new interactions in the TbPTR1 active site.
Acknowledgments
This research was funded by The Wellcome Trust (WT082596 and WT083481) and the Biotechnology and Biological Sciences Research Council (Structural Proteomics of Rational Targets; BBS/B/14434). We thank Ken Rehder, Andre Rosowsky and the Center for Organic and Medicinal Chemistry, Research Triangle International for compounds and acknowledge the ESRF for synchrotron beam time and excellent staff support.
Footnotes
PDB References: pteridine reductase complexes, 2×9n; 2×9g; 2×9v; 3mcv.
References
- Avendaño C, Menendez JC. Medicinal Chemistry of Anticancer Drugs. Elsevier; Amsterdam: 2008. p. 37. [Google Scholar]
- Baldwin CM, Perry CM. Drugs. 2009;69:2279–2302. doi: 10.2165/11202640-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Barrett MP, Boykin DW, Brun R, Tidwell RR. Br. J. Pharmacol. 2007;152:1155–1171. doi: 10.1038/sj.bjp.0707354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B, Langan P, Coates L, Mustyakimov M, Schoenborn B, Howell EE, Dealwis C. Proc. Natl Acad. Sci. USA. 2006;103:18493–18498. doi: 10.1073/pnas.0604977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakley RL. Adv. Enzymol. Relat. Areas Mol. Biol. 1995;70:23–102. doi: 10.1002/9780470123164.ch2. [DOI] [PubMed] [Google Scholar]
- Cavazzuti A, Paglietti G, Hunter WN, Gamarro F, Piras S, Loriga M, Allecca S, Corona P, McLuskey K, Tulloch L, Gibellini F, Ferrari S, Costi MP. Proc. Natl Acad. Sci. USA. 2008;105:1448–1453. doi: 10.1073/pnas.0704384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Acta Cryst. 1994;D50:760–763. Number 4. [Google Scholar]
- Croft SL, Sundar S, Fairlamb AH. Clin. Microbiol. Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank DWJ. Acta Cryst. 1999;D55:583–601. [Google Scholar]
- Cunningham ML, Titus RG, Turco SJ, Beverley SM. Science. 2001;292:285–287. doi: 10.1126/science.1057740. [DOI] [PubMed] [Google Scholar]
- Dawson A, Gibellini F, Sienkiewicz N, Tulloch LB, Fyfe PK, McLuskey K, Fairlamb AH, Hunter WN. Mol. Microbiol. 2006;61:1457–1468. doi: 10.1111/j.1365-2958.2006.05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. PyMOL. 2002 http://www.pymol.org. [Google Scholar]
- Delespaux V, de Koning HP. Drug Resist. Updates. 2007;10:30–50. doi: 10.1016/j.drup.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Acta Cryst. 2006;D62:72–82. [Google Scholar]
- Fishman JA. Antimicrob. Agents Chemother. 1998;42:1309–1314. doi: 10.1128/aac.42.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CL, Huggan JK, Kennedy A, Kiefer L, Lee JH, Suckling CJ, Clements C, Harvey AL, Hunter WN, Tulloch LB. Org. Biomol. Chem. 2009;7:1829–1842. doi: 10.1039/b818339b. [DOI] [PubMed] [Google Scholar]
- Gourley DG, Schüttelkopf AW, Leonard GA, Luba J, Hardy LW, Beverley SM, Hunter WN. Nature Struct. Biol. 2001;8:521–525. doi: 10.1038/88584. [DOI] [PubMed] [Google Scholar]
- Hardy LW, Matthews W, Nare B, Beverley SM. Exp. Parasitol. 1997;87:158–170. [PubMed] [Google Scholar]
- Hunter WN. J. Biol. Chem. 2009;284:11749–11753. doi: 10.1074/jbc.R800072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard GA, McAuley-Hecht K, Brown T, Hunter WN. Acta Cryst. 1995;D51:136–139. doi: 10.1107/S0907444994004713. [DOI] [PubMed] [Google Scholar]
- Leslie AGW. Acta Cryst. 2006;D62:48–57. [Google Scholar]
- Luba J, Nare B, Liang P-H, Anderson KS, Beverley SM, Hardy LW. Biochemistry. 1998;37:4093–4104. doi: 10.1021/bi972693a. [DOI] [PubMed] [Google Scholar]
- McGuire JJ. Curr. Pharm. Des. 2003;9:2593–2613. doi: 10.2174/1381612033453712. [DOI] [PubMed] [Google Scholar]
- McLuskey K, Gibellini F, Carvalho P, Avery MA, Hunter WN. Acta Cryst. 2004;D60:1780–1785. doi: 10.1107/S0907444904018955. [DOI] [PubMed] [Google Scholar]
- Moreira W, Leblanc E, Ouellette M. Free Radic. Biol. Med. 2009;46:367–375. doi: 10.1016/j.freeradbiomed.2008.10.034. [DOI] [PubMed] [Google Scholar]
- Mpamhanga CP, Spinks D, Tulloch LB, Shanks EJ, Robinson DA, Collie IT, Fairlamb AH, Wyatt PG, Frearson JA, Hunter WN, Gilbert IH, Brenk R. J. Med. Chem. 2009;52:4454–4465. doi: 10.1021/jm900414x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Acta Cryst. 1999;D55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Nare B, Hardy LW, Beverley SM. J. Biol. Chem. 1997;272:13883–13891. doi: 10.1074/jbc.272.21.13883. [DOI] [PubMed] [Google Scholar]
- Purcell WT, Ettinger DS. Curr. Oncol. Rep. 2003;5:114–125. doi: 10.1007/s11912-003-0098-3. [DOI] [PubMed] [Google Scholar]
- Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Lancet Infect. Dis. 2007;7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- Rosowsky A, Mota CE, Wright JE, Freisheim JH, Heusner JJ, McCormack JJ, Queener SF. J. Med. Chem. 1993;36:3103–3112. doi: 10.1021/jm00073a009. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. Acta Cryst. 2008;A64:112–122. [Google Scholar]
- Schüttelkopf AW, Hardy LW, Beverley SM, Hunter WN. J. Mol. Biol. 2005;352:105–116. doi: 10.1016/j.jmb.2005.06.076. [DOI] [PubMed] [Google Scholar]
- Tulloch LB, Martini VP, Iulek J, Huggan JK, Lee JH, Gibson CL, Smith TK, Suckling CJ, Hunter WN. J. Med. Chem. 2010;53:221–229. doi: 10.1021/jm901059x. [DOI] [PMC free article] [PubMed] [Google Scholar]