Abstract
We report 24 unrelated individuals with deletions and 17 additional cases with duplications at 10q11.21q21.1 identified by chromosomal microarray analysis. The rearrangements range in size from 0.3 to 12 Mb. Nineteen of the deletions and eight duplications are flanked by large, directly oriented segmental duplications of >98% sequence identity, suggesting that nonallelic homologous recombination (NAHR) caused these genomic rearrangements. Nine individuals with deletions and five with duplications have additional copy number changes. Detailed clinical evaluation of 20 patients with deletions revealed variable clinical features, with developmental delay (DD) and/or intellectual disability (ID) as the only features common to a majority of individuals. We suggest that some of the other features present in more than one patient with deletion, including hypotonia, sleep apnea, chronic constipation, gastroesophageal and vesicoureteral refluxes, epilepsy, ataxia, dysphagia, nystagmus, and ptosis may result from deletion of the CHAT gene, encoding choline acetyltransferase, and the SLC18A3 gene, mapping in the first intron of CHAT and encoding vesicular acetylcholine transporter. The phenotypic diversity and presence of the deletion in apparently normal carrier parents suggest that subjects carrying 10q11.21q11.23 deletions may exhibit variable phenotypic expressivity and incomplete penetrance influenced by additional genetic and nongenetic modifiers.
Keywords: CHAT, SLC18A3, genomic rearrangement, array CGH
Introduction
Segmental duplications comprise approximately 5% of the human genome [Bailey et al., 2002]. Misalignment of segmental duplications during meiosis can cause genomic instability through nonallelic homologous recombination (NAHR) [Lupski, 1998; Mefford and Eichler, 2009; Shaffer et al., 2001; Stankiewicz and Lupski, 2002]. In NAHR, improper crossing-over between nonallelic, yet homologous, segments (such as low-copy repeats [LCRs]) on sister chromatids or on homologous chromosomes produces microdeletions, microduplications, and inversions of the intervening genomic sequence, depending on the orientation of the segmental duplications [Koolen et al., 2006; Shaffer and Lupski, 2000; Sharp et al., 2006; Shaw-Smith et al., 2006; Stankiewicz and Lupski, 2002; Mefford and Eichler, 2009]. Chromosomal rearrangements associated with segmental duplications include deletions at 3q29 (MIM# 609425) [Willatt et al., 2005] and their reciprocal duplications (MIM# 611936) [Ballif et al., 2008a]; Williams-Beuren syndrome (MIM# 194050) and deletions at 7q11.23 and their reciprocal duplications (MIM# 609757) [Berg et al., 2007; Somerville et al., 2005]; Angelman (MIM# 105830) and Prader-Willi syndromes (MIM# 176270) and maternally and paternally derived deletions, respectively, of 15q11q13; deletions at 16p11.2p12.2 (MIM# 613604) and the reciprocal duplications [Ballif et al., 2007]; Smith-Magenis syndrome (MIM# 182290) and deletions at 17p11.2 and their reciprocal Potocki-Lupski syndrome duplications (MIM# 610883) [Potocki et al., 2007, 2000]; and duplication at 17p12 in Charcot-Marie-Tooth disease type 1A (MIM# 118220) and the reciprocal deletion causing hereditary neuropathy with liability to pressure palsies (MIM# 162500) [reviewed in Stankiewicz and Lupski, 2002]. Such recurrent syndromes are termed “genomic disorders” and usually meet several criteria: the rearrangement has breakpoints in flanking segmental duplications, is usually de novo in affected individuals and rarely observed in controls, and patients with the same rearrangement have similar, consistent phenotypes [Lupski, 1998; Mefford and Eichler, 2009]. The underlying genomic architecture in each of the genomic disorders identified to date is similar: a stretch of unique sequence (50–10 Mb) flanked by large (>10 kb), highly homologous (>95%) segmental duplications that provide the substrate for NAHR.
To date, interstitial deletions involving 10q11.2 have been reported in over 10 patients with variable abnormal phenotypes, individuals with a normal phenotype, and two prenatal cases, one with a normal and the other with an abnormal phenotype [Bisgaard et al., 2007; Fewtrell et al., 1994; Fryns et al., 1991; Ghai et al., 2011; Holden and MacDonald, 1985; Kirchhoff et al., 2005; Lobo et al., 1992; Shapiro et al., 1985; Zenger-Hain et al., 1993]. The only clinical feature common to a majority of subjects was intellectual disability (ID)/developmental delay (DD); whereas epilepsy, ataxia, clinodactyly, and bowel obstruction were all reported in one subject. However, because all but three of these individuals were identified by standard G-banded chromosome analysis, the precise deletion intervals are unknown.
Here, we report the clinical and molecular characterization of 24 individuals with deletion at 10q11.21q21.1 and 17 individuals with duplications of the same region.
Materials and Methods
Patient Ascertainment
Individuals with 10q11.21q21.1 deletions and duplications reported here were identified after referral for chromosomal microarray analysis to clinical laboratories, including Signature Genomic Laboratories (SGL) (patients 8–14, 17, 18, 24, 30–38), Baylor College of Medicine (BCM) (patients 1–6, 19–23, 25–29), Washington University School of Medicine (patients 7, 39–41), Children's Hospital of Philadelphia (patient 15), and Service de Génétique Médicale CHUV, Lausanne, Switzerland (patient 16). Common indications for study included DD, ID, dysmorphic features, and congenital anomalies. We reevaluated DNA samples by sequencing for patients 1–7, 11, 15, 16, and 18–20 after obtaining informed consent via protocols approved by Institutional Review Board (IRB) for Human Subject Research at BCM and IRB, Spokane.
Initial Microarray Analysis
Deletions and duplications were initially ascertained through microarray-based comparative genomic hybridization (aCGH) using either BAC-based whole-genome arrays (SignatureChipWG v1.0.1 [designed and manufactured by SGL; patients 9–10, 13–14, 24, 29, 31–32]) or oligonucleotide-based whole-genome arrays (44K BCM V6 [patients 1–3], 105K BCM V7 [patients 4–6, 21, 22], 180K BCM V8 [patients 19–20, 22, 23, 25–28], 105K SignatureChip OS v1.0 [SGL, patient 8], 135K SignatureChip OS v2.0 [SGL, patients 11–12, 17–18, 24, 33–38], or 244K off-the-shelf array [Agilent Technologies, Santa Clara, CA; patient 16]). The deletion in patients 7 and 15 and duplications in patients 39–41 were initially identified using the genome-wide human single nucleotide polymorphism (SNP) array 6.0 (Affymetrix, Santa Clara, CA). DNA samples from both parents of patient 16 were tested using Agilent oligoNT 180K (Agilent Technologies, Santa Clara CA, USA). The BCM oligonucleotide arrays and the SignatureChip OS v 1.0 are custom designed by BCM Medical Genetics Laboratory (MGL) [Lu et al., 2007; Ou et al., 2008] and Signature Genomics, respectively, and manufactured by Agilent Technologies. The SignatureChip OS v2.0 is custom designed by SGL and manufactured by Roche NimbleGen (Madison, WI). Studies were performed according to the manufacturer's instructions or previously described methods [Ballif et al., 2008a,b; Cheung et al., 2005; Duker et al., 2010].
High-Resolution Microarray Analysis to Refine the Breakpoints
Patients whose deletions were initially ascertained on BAC array had subsequent higher resolution studies done on whole-genome oligonucleotide arrays (105K SignatureChip OS v1.0, patient 9, or 135K SignatureChip OS v2.0, patients 10, 13, 14, and 27). These arrays have coverage of the unique sequence in the region (proximal to LCR 10q11.2A, between LCR 10q11.2A and B, between LCR 10q11.2B and C, between LCR 10q11.2C and D, between LCR 10q11.2D and E, between LCR10q11.2E and F, and distal to LCR10q11.2F).
Custom region-specific high-resolution 72K oligonucleotide arrays (Roche NimbleGen; median probe spacing 193 bp) were designed to determine the more precise location of the breakpoints, and DNA samples from patients 2–6 were reanalyzed on this array. Whole-genome high-resolution oligonucleotide microarray CGH analysis was also performed with the NimbleGen arrays 385K (patient 1) and 2.1M (patients 2, 3, and 4), in accordance with the manufacturer's instructions.
Fluorescence in situ Hybridization (FISH) Analysis
FISH was performed using BAC clones RP11-70E21 (patients 1– 4, 6, 20, 22, 25, 26, and 28), RP11-563N6 (patients 5, 19, and 22), RP11-541M12 (patients 7, 40, and 41), RP11-635L5 (patients 8, 9, 24, 29, and 35), RP11-100M24 (patients 10, 11, 33, and 36), RP11-140C5 (patients 12, 17, and 18), RP11-1005F22 (patients 13, 14, 30, 31, and 38), RP11-165A4 (patient 27), RP11-10N24 (patient 32), RP11-219K22 (patient 34), and RP11-168P8 (patient 37) from the 10q11.21q21.1 region to confirm and visualize the abnormalities using standard methodology.
DNA Sequencing of CHAT
Overlapping amplicons covering the entire coding region of 17 exons from all isoforms of CHAT (MIM# 118490) were amplified and sequenced in patients 1–7, 9, 11, 15, 16, 18, and 19 by conventional Sanger di-deoxynucleotide sequencing (Lone Star Labs, Houston, TX). DNA sequences were analyzed by comparison with reference sequence (NM_001451.2) with the use of Sequencher V4.8 (GeneCodes, Ann Arbor, MI). Individual primer sequences and polymerase chain reaction (PCR) conditions are available on request.
Bioinformatics and In Silico Sequence Analysis
Genomic sequences of 1 Mb in size for the region between 45 and 57 Mb (10q11.21q21.1) were downloaded from the UCSC genome browser (Build hg18, UCSC genome browser, March 2006) and masked using Repeatmasker. The repeat-masked sequences were then analyzed using Blast2, and the sequences with >90% identity and alignments of more than 400 bp were aligned according to the coordinates and their orientation.
To assess the chromosome architecture causing deletions and duplications in the 10q11.2 region, we evaluated the presence of LCRs using the ICAass (v 2.5) algorithm. The graphical display was performed using Miropeats (v 2.01) (The Genome Institute at Washington University, St Louis, MO) [Parsons, 1995a, b]. The program was run using two thresholds of 1,000 and 5,000 bp. For ease of computation, the 8.0 Mb interval was analyzed in two nonover-lapping blocks of 3.9 Mb each. The hg18 coordinates for these blocks are chr10: 45,300,000–49,200,000 and chr10: 49,300,000–53,200,000.
Results
Molecular Analysis
We identified 24 unrelated individuals with microdeletions at 10q11.21q11.23 by chromosomal microarray analysis. The deletions range in size from ~1.9 to ~10.9 Mb (Tables 1 and 2; Figs. 1–3).
Table 1.
Summary of the Results of Microarray Studies of 20 Unrelated Individuals with Microdeletions in 10q11.21q11.23
| Patient | Coordinates (hg18) | Size (Mb) | Parental origin | Microarray platform | FISH verification | Possible mechanism of formation |
Additional copy number changes |
|---|---|---|---|---|---|---|---|
| 1 | 45,979,000–56,907,000 | ~10.9 | Paternal | 72K (NimbleGen) | RP11-70E21 | ? | - |
| 2 | 45,512,000–51,585,000 | ~6 | Paternal | 72K (NimbleGen) | RP11-70E21 | NAHR | - |
| 3 | 48,941,000–52,218,000 | ~3.3 | Maternal | 72K (NimbleGen) | RP11-70E21 | ? | arr 13q22.3(77,299,714–77,471,991)×3mat |
| 4 | 45,480,000–51,585,000 | ~6 | Paternal | 72K (NimbleGen) | RP11-70E21 | NAHR | - |
| 5 | 45,612,000–51,585,000 | ~6 | Maternal | 72K (NimbleGen) | RP11-563N6 | NAHR | arr 3q13.12q13.32(108,208,150–120,229,969)×1dn |
| 6 | 45,612,000–51,585,000 | ~6 | Maternal | 72K (NimbleGen) | RP11-70E21 | NAHR | - |
| 7 | 45,947,671–51,263,703 | ~5.3 | Paternal, grandmaternal | Affymetrix SNP 6.0 | RP11-541M12 | NAHR | arr 12q23.1q23.2(chr12: 95,227,874–100,061,773)×3mat |
| 8 | 46,400,346–51,237,832 | ~4.8 | Unknown | 135K (SignatureChipOS v2.0) | RP11-635L5 | NAHR | - |
| 9 | 46,400,346–51,237,832 | ~4.8 | Maternal | 135K (SignatureChipOS v2.0) | RP11-635L5 | NAHR | arr 6p25.3p22.1(115,226–126,640,914)×3 der(7)t(6;7)(p22.1;p22.3) |
| 10 | 46,400,346–51,237,832 | ~4.8 | Paternal | 135K (SignatureChipOS v2.0) | RP11-100M24 | NAHR | - |
| 11 | 46,400,346–51,237,832 | ~4.8 | Maternal | 135K (SignatureChipOS v2.0) | RP11-100M24 | NAHR | - |
| 12 | 46,400,346–51,237,832 | ~4.8 | Unknown | 135K (SignatureChipOS v2.0) | RP11-140C5 | NAHR | arr 14q31.3q32.11(88,853,671–88,965,421)×1 |
| 13 | 49,062,854–52,062,367 | ~3.0 | Unknown | 135K (SignatureChipOS v2.0) | RP11-1005F22 | ? | - |
| 14 | 49,062,854–52,062,367 | ~3.0 | Paternal | 135K (SignatureChipOS v2.0) | RP11-1005F22 | ? | - |
| 15 | 45,927,753–51,581,847 | ~5.7 | de novo | Affymetrix SNP 6.0 | None | NAHR | ? |
| 16 | 48,871,525–50,765,047 | ~1.9 | de novo | 244K Agilent | None | NAHR | Confined placental mosaicism: arr cgh 2(×3) |
| 17 | 46,400,346–51,237,832 | ~4.8 | Unknown | 135K (SignatureChipOS v2.0) | RP11-140C5 | NAHR | - |
| 18 | 46,400,346–51,237,832 | ~4.8 | Unknown | 135K (SignatureChipOS v2.0) | RP11-140C5 | NAHR | - |
| 19 | 46,384,979–52,085,077 | ~5.7 | Unknown | BCM V8.0 OLIGO | RP11-563N6 | ? | arr 15q21.2q21.3(49,846,193–51,236,840)×3 |
| 20 | 46,384,979–51,265,056 | ~4.9 | Maternal | BCM V8.0 OLIGO | RP11-70E21 | NAHR | arr 9q34.3(139,662,925–140,138,901)×3mat |
Table 2.
Summary of the Results of Microarray Studies and Phenotypic Features of Additional Unrelated Individuals with Microdeletions of 10q11.21q11.23
| Patient | Age | Gender | Indication | Coordinates (hg18) | Size (Mb) | FISH verification | Parental origin | Microarray platform | Additional copy number changes |
|---|---|---|---|---|---|---|---|---|---|
| 21 | 4 years | F | Mild DD/ID, DF | 49,121,974–50,641,724 | ~1.5 | - | Unknown | BCM V7.1 OLIGO | - |
| 22 | 1 year | F | N/A | 48,102,606–50,641,752 | ~2.5 | RP11-563N6 | Unknown | BCM V7.4 OLIGO | arr 9q34.2(135,752,638–135,912,152)×3 |
| 23 | 8 years | M | DD | 46,384,979–51,672,034 | ~5.3 | - | Unknown | BCM V8.1 OLIGO | - |
| 24 | 11 years | F | Cleft palate | 46,400,346–51,237,832 | ~4.8 | RP11-635L5 | Paternal | 135k (SignatureChipOS v2.0) | - |
DD, developmental delay; DF, dysmorphic features; ID, intellectual disability; N/A, not available.
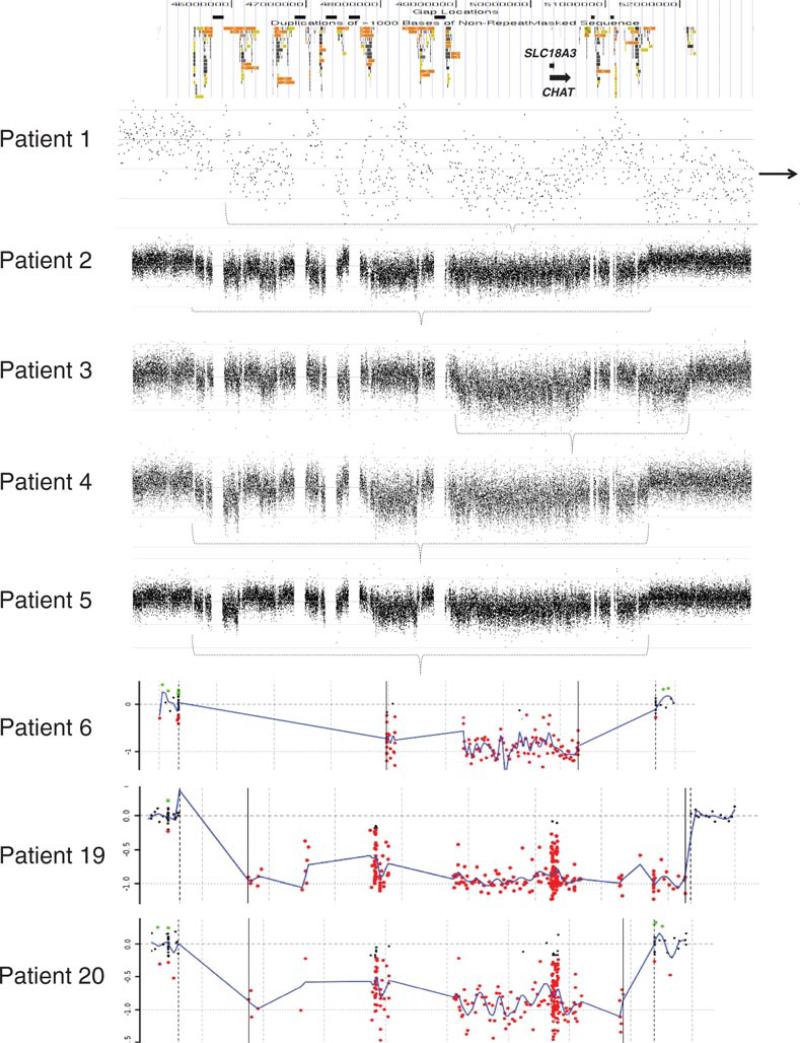
Figure 1.
Chromosomal microarray results for eight samples analyzed by BCM, using RocheNimbleGen oligonucleotide-based microarrays. At the top is a schematic of the genomic architecture of the region. Black boxes represent sequence gaps. Colored boxes represent segmental duplications. For each plot, probes are arranged on the X-axis according to physical mapping positions, with the most proximal 10q11.21 probes to the left and the most distal 10q11.23 probes to the right. Values along the Y-axis represent log2 ratios of patient:control signal intensities. Deletion intervals are represented by a dotted bracket (patients 1–5) or vertical, solid black lines (patients 6, 19–20).
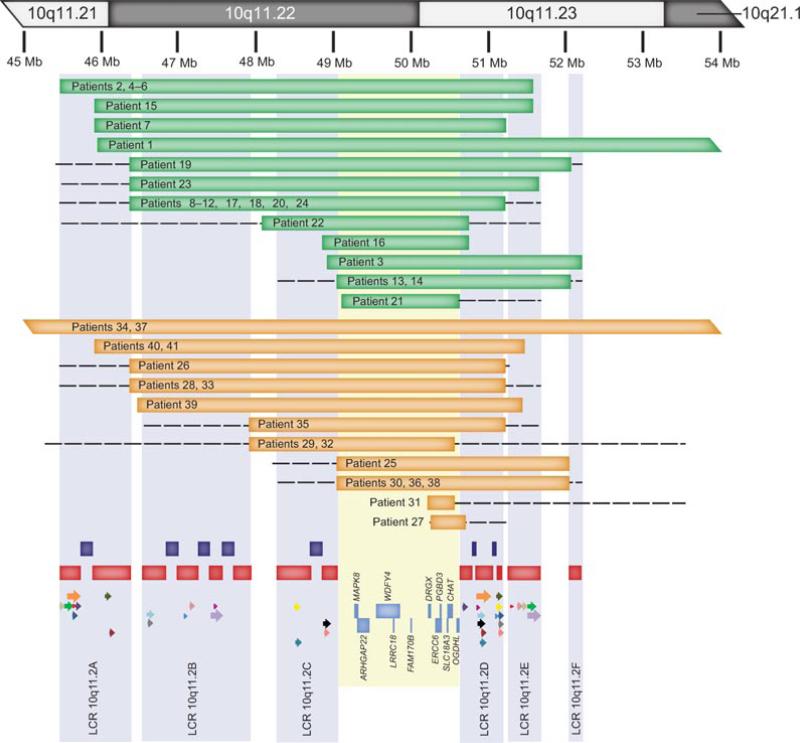
Figure 3.
Summary of chromosomal microarray analysis of individuals with deletions and duplications 10q11.21q21.1. At the top of the figure is a partial idiogram showing chromosome bands 10q11.21q21.1 with genomic coordinates corresponding to the hg18 build of the human genome. Green bars represent the deletions and the orange bars the duplications. Red bars represent segmental duplications. Only the directly oriented paralogous LCR subunits (18) between LCR10q11.2s A–C and D–F, that have a potential for NAHR, are shown for illustrative purposes as the colored arrows. Each of the 18 paralogous pairs is shown in different color (Supp. Table S1). Note that NAHR might have mediated rearrangements in patients 2, 4–12, 15, 17–18, 20, 23, 24, 26, 28, 33, 40, and 41 (between LCR10q11.2A and LCR10q11.2E), 16 and 21 (between LCR10q11.2C and LCR10q11.2D), 22, 29, and 32 (between LCR10q11.2A–B and LCR10q11.2D–E), and 35 (between LCR10q11.2B and LCR10q11.2E). Dark blue bars depict seven genomic gaps. Semitransparent light blue shading represents the collapsed LCR clusters A–F. Semitransparent yellow shading represents the smallest region of unique sequence shared by all deletion patients.
Twenty deletions were visualized by FISH, and parental testing in 15 subjects showed that the deletions were inherited from the mother in six patients (3, 5, 6, 9, 11, and 20), from the father in seven patients (1, 2, 4, 7, 10, 14, and 24), and were apparently de novo in patients 15 and 16 (Tables 1 and 2). Parental samples for the remaining nine patients (8, 12, 13, 17–19, and 21–23) were unavailable.
We have also identified 17 individuals with reciprocal microduplications involving 10q11.21q21.1, ranging in size from ~0.3 to ~12 Mb (Table 3; Fig. 3). Parental testing showed that the duplications were inherited from the mother in six patients and from the father in three patients. Parental samples for the other eight patients were unavailable for testing (Table 3).
Table 3.
Summary of the Results of Microarray Studies and Phenotypic Features of 17 Unrelated Individuals with Microduplications of 10q11.21q11.23
| Patient | Age | Gender | Indication | Coordinates (hg18) | Size (Mb) | FISH verification | Parental origin | Microarray platform | Additional copy number changes |
|---|---|---|---|---|---|---|---|---|---|
| 25 | 4 years | M | Autism, speech delay | 49,062,900–52,085,077 | ~3.0 | RP11-70E21 | Unknown | BCM V8.1 OLIGO | arr 12p13.2(11,897,593–12,310,534)×3 |
| 26 | 4 years | M | Moderate DD, ASDs | 46,384,979–51,249,165 | ~4.9 | RP11-70E21 | Maternal | BCM V8.1 OLIGO | - |
| 27 | 3 months | F | Hypotonia, abnormal brain MRI | 50,265,332–50,721,613 | ~0.5 | RP11-165A4 | Maternal | BCM V8.1 OLIGO | - |
| 28 | 12 years | M | Moderate DD, ASDs, seizures, DF, MCA | 46,384,979–51,265,056 | ~4.9 | RP11-70E21 | Unknown | BCM V8.2 OLIGO | - |
| 29 | 14 years | F | T-cell primary immunodeficiency | 47,928,933–50,581,445 | ~2.7 | RP11-635L5 | Paternal | SignatureChipWG v1 .0.1 | arr Xq25(RP11-229G9)×1 pat |
| 30 | 24 years | M | DD, DF | 49,151,859–52,085,217 | ~2.9 | RP11-1005F22 | Maternal | 135K (SignatureChipOS v1.1) | arr Xp22.33/Yp11.32(463,937–543,186)×3 pat |
| 31 | 15 years | M | DD, Mild ID, autistic features | 50,239,677–50,581,445 | ~0.3 | RP11-1005F22 | Maternal | SignatureChipWG v1.0.1 | - |
| 32 | 8 years | M | DD | 47,928,933–50,581,445 | ~2.7 | RP11-10N24 | Unknown | SignatureChipWG v1.0.1 | - |
| 33 | 14 years | F | Encephalopathy | 46,400,346–51,237,832 | ~4.8 | RP11-100M24 | Unknown | 135K (SignatureChipOS v2.0) | - |
| 34 | 6 years | M | DD, DF | 43,288,081–55,324,785 | ~12 | RP11-219K22 | Unknown | 135K (SignatureChipOS v2.0) | arr 10q21.1(56,542,636–56,802,473)×1 |
| 35 | 35 years | M | MCA | 47,921,367–51,237,832 | ~4.7 | RP11-635L5 | Paternal | 135K (SignatureChipOS v2.0) | - |
| 36 | 14 years | M | DF | 49,062,854–52,062,367 | ~3.0 | RP11-100M24 | Maternal | 135K (SignatureChipOS v2.0) | - |
| 37 | 3 years | F | DD, DF, seizure disorder | 43,288,081–55,324,785 | ~12.0 | RP11-168P8 | Unknown | 135K (SignatureChipOS v2.0) | arr 15q13.3(29,816,893–30,226,405)×1 |
| 38 | 7 years | F | DD, seizure disorder, skeletal anomalies | 49,062,854–52,062,367 | ~3.0 | RP11-1005F22 | Unknown | 135K (SignatureChipOS v2.0) | - |
| 39 | 4 years | M | DD | 46,497,745–51,472,379 | ~5.0 | N/A | Unknown | Affy SNP 6.0 | - |
| 40 | 22 months | F | Global DD | 45,947,671–51,510,978 | ~5.6 | RP11-541M12 | Maternal | Affy SNP 6.0 | - |
| 41 | 4 months | M | DD | 45,947,671–51,492,393 | ~5.5 | RP11-541M12 | Paternal | Affy SNP 6.0 | - |
ASDs, autistic spectrum disorders; DD, developmental delay; DF, dysmorphic features; ID, intellectual disability; MCA, multiple congenital anomalies; N/A, not available.
Smallest Region of Overlap
A comparison of the approximately 1.9 Mb smallest region of deletion overlap in the 24 deletions reported here (between 48.9 and 50.8 Mb) (Fig. 3) to locations of benign copy number variants (CNVs) in the Database of Genomic Variants revealed the presence of a small, unique sequence overlap of approximately 1.5 Mb (between 49.2 and 50.6 Mb; LCR10q11.2C to LCR10q11.2D) that has not been reported to be deleted in control individuals.
DNA Sequencing of CHAT
To determine whether the second allele of CHAT has a point mutation that could be unmasked by the 10q11.2 deletion, we sequenced all coding exons of CHAT in a cohort of 13 subjects (patients 1–7, 9, 11, 15, 16, 18, and 19) with heterozygous deletions of 10q11.2 that included CHAT. We did not identify any change in 221 amplicons analyzed.
Computational Analysis of the 10q11.21q11.23 Region
Using the hg18 build of the UCSC genome browser [Bailey et al., 2001] and the hg17 build of the Human Genome Segmental Duplication Database, we identified a complex arrangement of six segmental duplication clusters in the 10q11.21q11.23 region, labeled LCR 10q11.2A-LCR10q11.2F (Fig. 3). These segmental duplications range in size from 32 to 427 kb and have a complex evolutionary structure [Deloukas et al., 2004]. More than half of the analyzed 6.5 Mb region in 10q11.21q11.23 is occupied by these LCR clusters and seven genomic gaps. We identified 18 paralogous pairs of directly oriented LCR subunits between LCR10q11.2A–C and LCR10q11.2D–F (Fig. 3; Supp. Table S1). Sixteen similar sized approximately 6 Mb deletions (patients 2, 4–12, 15, 17–18, 20, 23, and 24) and five duplications (patients 26, 28, 33, 40, and 41) are flanked by LCR10q11.2A and LCR10q11.2E that harbor subunits approximately 130 kb in size with greater than 98.2% DNA sequence identity and in direct orientation with respect to each other. Rearrangements in six individuals might have been caused also by NAHR between LCR10q11.2C and LCR10q11.2D (patients 16 and 21) and LCR10q11.2A–B and LCR10q11.2D–E (patients 22, 29, and 32), and by LCR10q11.2B and LCR10q11.2E (patient 35). Five deletions (patients 1, 3, 13, 14, and 19) and nine duplications (25, 27, 30, 31, 34, 36–38, and 39) are not flanked by directly oriented LCR subunits and represent atypical breakpoints (Fig. 3; Supp. Table S1).
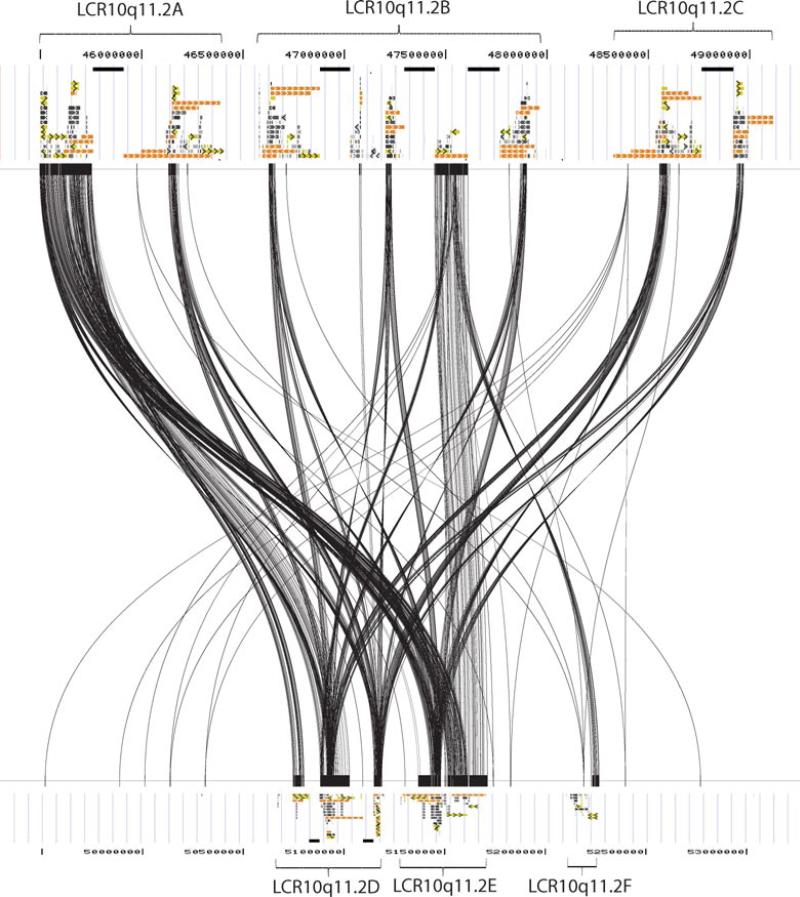
Using Miropeats program, we have identified the paralogous LCR subunits in the proximal and distal LCR10q11.2 that can serve as a substrate to NAHR (Figs. 3 and 4; Supp. Table S1). Those directly oriented overlap with the defined breakpoint regions and can serve as the substrates for NAHR (Fig. 3).
Figure 4.
DNA sequence homology between the proximal (chr10: 45,300,000–49,200,000) and distal (chr10: 49,300,000–53,200,000) LCR10q11.2 clusters for the paralogous subunits larger than 1 kb in size (hg18) using Miropeats program analysis. The upper and bottom panels depict the UCSC segmental duplication track.
Identification of Additional Copy Number Changes in Patients
Nine of the 24 individuals with deletions have secondary copy number alterations. Three of the additional copy number changes in patients 3, 7, and 20 were inherited from phenotypically normal mothers. Patient 5 has an approximately 12 Mb additional, de novo deletion in 3q13, which likely confounds the clinical phenotype associated with the 10q deletion. The parental origins of the additional copy number changes in patients 9, 12, 19, and 22 are unknown. Patient 16 had mosaic trisomy 2 confined to the placenta identified by aCGH and confirmed by FISH in 15% of cells (Table 1). Uniparental disomy (UPD) testing was not performed.
Deletion and Duplication Frequency in Affected and Control Populations
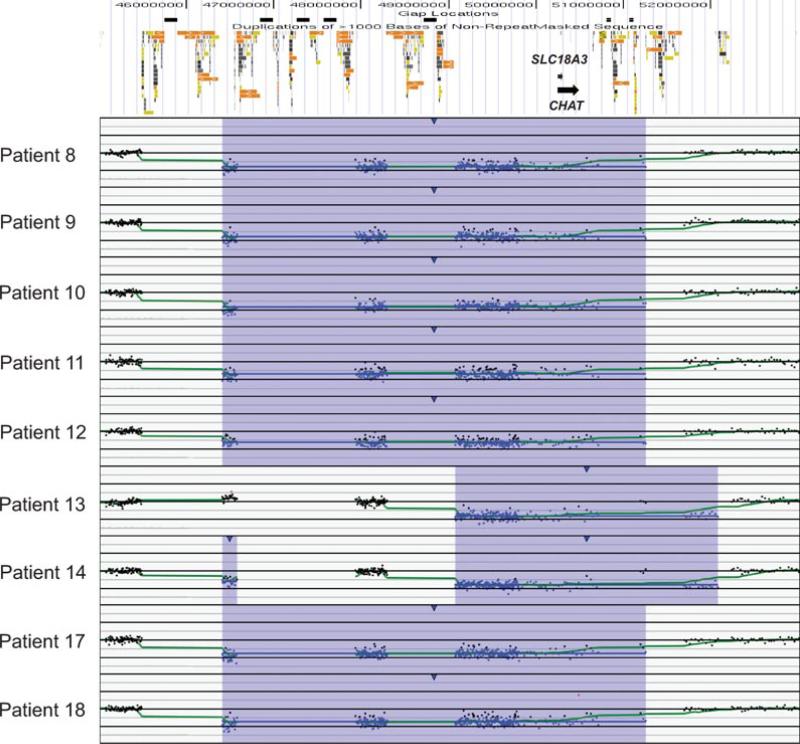
We compared the frequency of the 10q11.21q11.23 rearrangements among our study population to the frequency in controls to determine whether the deletion predisposes individuals to an abnormal phenotype. No complete deletions or duplications of the unique sequence between LCR 10q11.2C and LCR 10q11.2D were found in six control groups consisting of 2,792 individuals [Kirov et al., 2009], 2,493 individuals [Itsara et al., 2009], 2,026 individuals [Shaikh et al., 2009], 1,152 individuals [Zogopoulos et al., 2007], 450 individuals [Conrad et al., 2010], and 270 individuals [Redon et al. 2006], although several small CNVs within this region were reported in two of the control populations [Itsara et al., 2009; Shaikh et al., 2009]. CNVs overlapping the proximal LCRs are also more frequent in these control populations, such as that seen in patient 14 (Fig. 2). Combining these populations yields a frequency of 0/9,183 LCR10q11.2C to LCR10q11.2D deletions in healthy controls.
Figure 2.
Chromosomal microarray results for patient samples analyzed by SignatureChip oligonucleotide-based microarray. At the top is a schematic of the genomic architecture of the region. Black boxes represent sequence gaps. Colored boxes represent segmental duplications. For each plot, probes are arranged on the X-axis according to physical mapping positions, with the most proximal 10q11.22 probes to the left and the most distal 10q11.23 probes to the right. Values along the Y-axis represent log2 ratios of patient:control signal intensities.
To determine the frequency of the rearrangements in our study populations, we have combined the frequency of the deletions and duplications (involving the entire 1.9 Mb region between LCR10q11.2C and D), respectively, detected at BCM (11/25,354 and 4/25,354) with the frequency in Signature Genomics’ patient database (10/32,821 and 8/32,821). Thus, the combined frequencies of deletions and duplications in our study populations are 21/58,175 and 12/58,175, respectively. The differences in deletion and duplication prevalences between the affected and the control populations are statistically significant (P = 0.046, Fisher Exact test) for deletions and not significant (P = 0.17, Fisher Exact test) for duplications, when considering the number of deletion and duplication alleles in diploid individuals or the number of individuals harboring deletion and duplication events.
Clinical Analysis
Clinical characterization of 20 of 24 individuals with microdeletions at 10q11.21q11.23 revealed variable clinical features (Table 4). The only clinical features common to a majority of individuals were ID and DD. Other clinical features identified in two or more individuals include failure to thrive, growth retardation, or short stature (42%, 8/19), chronic constipation (37%, 7/19), hypotonia (32%, 6/19), gatroesophageal reflux (GERD, 32%, 6/19), sleep apnea (26%, 5/19), cleft or high palate (26%, 5/19), epilepsy (21%, 4/19), autism spectrum disorders (ASDs; 27%, 4/15), microcephaly (19%, 3/16), corpus callosum abnormalities (18%, 2/11), attention deficit hyperactivity disorder (ADHD; 16%, 3/19), ataxia or disco-ordination (16%, 3/19), micrognathia (16%, 3/19), vesicoureteral reflux (16%, 3/19), severe eczema (16%, 3/19), scoliosis (11%, 2/19), significant congenital heart defects (11%, 2/19), microphthalmia (11%, 2/19), dysphagia (11%, 2/19), short halluces (11%, 2/19), nystagmus (11%, 2/19), and ptosis (11%, 2/19). Interestingly, patient 1's brother with the same deletion had constipation, and Decipher patient 248902 (deletion of chr10: 49,130,990–50,638,651, including CHAT) presented with hypotonia, epilepsy, and sleep apnea.
Table 4.
Summary of Clinical Features of Individuals with Microdeletions at 10q 11.21 q21.1
| Patient | 1a | 2 | 3 | 4 | 5 | 6b | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 18 years | 7.5 years | 11 years | 11 months | 26 months | 11 years | 12 years | 2 years 5 months | 6 years | 9 years |
| Sex | M | M | M | M | M | F | M | F | F | M |
| Height (centile) | NS | 75th–90th | 5th | NS | 90th–97th | <3rd | 84th | 5th | 5th | 90th |
| Weight (centile) | NS | 50th–75th | 10th | FTT | 50th | <3rd | 94th | 4th | <3rd | 75th–90th |
| OFC (centile) | NS | 50th | 50th | NS | 80th | 49 cm microcephalic | 55th | 19th | Microcephalic | >97th |
| Psychomotor and behavioral development | Severe ID (IQ<35), speech delay, poor eye contact | Severe DD, ASDs, anxious, severe speech delay, repetitive behaviors | ASDs, ADHD | DD | Global DD, self-injurious behaviors | DD (IQ = 58) | ADHD, Asperger syndrome, LD (IQ~75) | Global DD, at about 6 month level | DD | Normal |
| Neurology | Poor motor coordination | NS | Mild leg hyperreflexia, decline in walking skills, normal brain MRI | Seizures, hypomyelination above the tentorium | Agenesis of corpus callosum, diffuse periventricular leukomalacia, hypotonia | Asymptomatic until ~5 years of age. Cerebellar ataxia, significant intentional tremor, spastic paraparesis, increased deep tendon reflexes, central tone not significantly elevated except at ankles, abnormal MRI. CT noted small calcifications within basal ganglia, Hx of tonic/clonic seizures, fine motor impairment | Normal brain MRI | Intractable epilepsy, normal brain MRI, MRS: elevated choline: creatine, hypotonia | Dysgenesis of the corpus callosum, bilateral choroid plexus cysts | Normal |
| Dysmorphic features | Thin face, small cranium, large ears | High arched palate, thin vermilliion border, thin philtrum, narrow facies, protruding auricles | Large, overfolded ears | NS | Mild telecanthus | NS | Prominent ears, tubular nose with high nasal bridge and a bulbous tip, high arched palate | None | Round face, high forehead, low-set and posteriorly rotated ears, ptosis, short palpebral fissures, straight eyebrows, beaked nose, anteverted nostrils, mild maxillary hypoplasia, thin lips, large uvula, hypoplastic philtrum, hypodontia, pointed chin, low posterior hairline with double hair whorl, thin and webbed neck, submucous cleft palate | Epicanthal folds, mild micrognathia, mild facial asymmetry |
| Musculoskeletal | Scoliosis, hemivertebra, muscle strength 3/5 | NS | None | NS | Scoliosis | Stiffness reported | 3–4 toe cutaneous syndactyly bilaterally | NS | Leg length discrepancy, shortened halluces, arachnodactyly of the left fifth finger | Hypoplastic fifth digit phalanges |
| Other | Anal atresia, multiple impacted teeth, jaw maloclusion, chronic constipation, dysphagia | T&A for sleep apnea, chronic constipation | Fifth finger clinodactyly | Abnormal rectosigmoid ratio, apnea/hypopnea, chronic constipation | Chronic constipation, possible obstructive sleep apnea | Jaundice at 3 days, elevated liver enzymes, Hx of aniteric sclera, PE tubes, Hx of headache, apneic episode with cynaosis at 4 months. Multiple UTIs 3–5 years of age. Cystourethrogram and renal scan Nl. GERD | Pigmentary retinopathy, nystagmus, swallowing dysfunction, GERD, chronic constipation, renal reflux | Severe valvular pulmonary stenosis, severe tricuspid regurgitation, and hypertrophic right ventricle, PFO versus small atrial secundum defect, vitamin D deficiency, hepatomegaly, lacrimal duct obstruction, obstructive sleep apnea, microphthalmia, blepharophimosis, astigmatism, hypodontia, eczema, sacral dimple, renal hypoplasia and insufficiency, vesicoureteral reflux, urethrovaginal fistula, urethral fistula | Unilateral progressive NSHL, GERD, hypoplastic fifth nails |
| Patient | 11c | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 1 month | 11 years 8 months | 10 years 10 months | 26 months | 5 years 8 months | 30 week fetus | 7 years 8 months | 6 years 4 months | 20 months | 7 months |
| Sex | M | F | F | M | M | M | M | M | F | F |
| Height (centile) | 90th | 10th | <3rd | 25th–50th | <5th | 25th | 57th | 25th | 73rd | 44th |
| Weight (centile) | 97th | 25th | <3rd | 25th–50th | <5th; FTT | 3rd–10th | 58th | 25th–50th | 69th | 39th |
| OFC (centile) | 90th | 85th | NS | 50th–75th | <<3rd: -4 to -5 SD | 75th | 82nd | 0 to +1 SD | 47th | 90th |
| Psychomotor and behavioral development | Normal | DD, ADHD, dysgraphia | Global DD, at about 6 years level | DD | Global DD. Dysarthria, difficulties staying on task | NA | Global DD, regression | PDD-NOS, DD | Normal | Normal |
| Neurology | Mild intermittent ankle clonus | Normal | Myoclonic/generalized epilepsy (3–4 seizures/week since age 3 years); possible Lennox-Gastaut | Hypotonia | Abnormal brain MRI: significant hypomyelination, hypoplasia of inferior cerebellar vermis, persistent cavum septum vergae, mild ventricular prominence, ataxia, spasticity, mild dysmetria, increased DTRs (3+/4) with clonus, truncal hypotonia with some spasticity | NS | Seizures, multifocal epileptiform discharges, ESES, hypotonia, clenched muscles, toe walking, normal brain MRI | Normal brain MRI | Normal neonatal head U/S | Hypotonia |
| Dysmorphic features | None | None | Down-slanting eyes | Frontal bossing, rounded cheeks, rounded nasal tip, mildly deficient alae nasi, mild micrognathia | Short palpebral fissures, deep-set eyes, flat midface, prominent maxilla, high palate, mild micrognathia, sparse and thin hair | None | Mild ptosis, short columella | None | Round face, prominent eyes, epicanthal folds, depressed nasal bridge, arched palate | NS |
| Musculoskeletal | NS | NS | Ulnar deviation of hands, elbow contractures | NS | Delayed bone age—12–15 months at age 22 months brachydactyly | NS | Short first toes | NS | NS | Right hand anomaly (same as in father) |
| Other | Small umbilical hernia | Recurrent UTIs, severe chronic constipation, myopia, T&A | Fifth finger clinodactyly, renal reflux, resolved, seizures in paternal aunt | Micropenis, severe eczema | Congenital nystagmus, congenital cataracts, mild microphthalmia, hypermetropia, poor suck and swallow, GERD, dysphagia | Hypospadias, severe IUGR | Mild optic atrophy, aspiration, GERD, chronic constipation, diarrhea, suspected mitochondrial disorder, immune-suppressed, chronic sinusitis and otitis media | Eczema, hypopigmentation | TAPVR, tachycardia | GERD |
Patient 1's brother with the deletion has ADHD, maloclusion, and mild constipation. Patient 1's father also has ADHD.
Patient 6's mother had stroke at 25 years attributed to foramen ovale.
Patient 11's mother and maternal half brother also carry the 10q11.2 deletion; the brother has speech delay and LD; the mother has a history of speech delay and LD.
ADHD, attention deficit hyperactivity disorder; ASDs, autism spectrum disorders; DD, developmental delay; DTRs, deep tandom reflexes; ESES, electrical status epilepticus during sleep; FTT, failure to thrive; GERD, gastroesophageal reflux disease; ID, intellectual disability; IUGR, intrauterine growth retardation; LD, learning disabilities; MRS, magnetic resonance spectroscopy; NA, not applicable; NS, not specified; NSHL, neurosensory hearing loss; PDD-NOS, pervasive developmental delay-not otherwise specified; PE, pressure equalization; PFO, patent foramen ovale; SD, standard deviation; T&A, tonsillectomy and adenoidectomy; TAPVR, total anomalous pulmonary venous return; UTI, urinary tract infection.
Although detailed clinical information was not obtained for the duplication patients, the indication provided for testing for most patients included DD/ID. The next most common manifestations included ASDs (patients 25, 26, 28, and 31) and epilepsy (patients 28, 33, 37, and 38) (Table 3).
Discussion
The conventional wisdom surrounding genomic disorders posits that they fit several criteria: the deletions/duplications are large, highly penetrant, de novo in the majority of individuals, and associated with a uniform constellation of clinical features [Mefford and Eichler, 2009]. Smith-Magenis syndrome, Prader-Willi syndrome, and Williams-Beuren syndrome are examples of such “classic” genomic disorders. In contrast to these “classic” genomic disorders, many of the more recently described recurrent genomic lesions identified in large case–control studies demonstrate apparently diverse phenotypes and are frequently inherited while showing reduced penetrance [Ensenauer et al., 2003; Hannes et al., 2008; Klopocki et al., 2007; Mefford et al., 2008; Sharp et al., 2008., Ullmann et al., 2007; Yobb et al., 2005]. These studies suggest the phenotypic effects of such copy number changes are pleiotropic and imply the existence of shared biologic pathways among multiple neurodevelop-mental conditions, which may explain the variability of neurological manifestations within some families. However, even classical deletion syndromes can present with diverse phenotypes [Shah et al., 2008].
In the present study, the only consistent clinical features present in the majority of individuals with deletions or duplications were DD and ID (Tables 1–4). However, these clinical features probably reflect an ascertainment bias, because individuals are often referred for chromosomal microarray testing for general indications such as ID/DD.
Comparison with previously reported individuals with deletions encompassing 10q11.2 [Bisgaard et al., 2007; Fewtrell et al., 1994; Fryns et al., 1991; Ghai et al., 2011; Holden and MacDonald, 1985; Kirchhoff et al., 2005; Lobo et al., 1992; Puliti et al., 1993; Shapiro et al., 1985; Zenger-Hain et al., 1993] is uninformative because most of these deletions were identified by standard cytogenetic analysis and are substantially larger than the deletions reported here. Only two of the 15 deletions in our study for which parental samples were available were de novo. In several recently described syndromes including 8p23 duplication [Barber et al., 2008], 1q21.1 microdeletion (MIM# 612474) [Mefford et al., 2008], 15q13.3 microdeletion (MIM# 612001) [Sharp et al., 2008], and 16p12.1 microdeletion (MIM# 136570) [Girirajan et al., 2010], deletions have been found in control populations as well as in unaffected family members. Recurrent microdeletions of 16p12.1 have been identified in individuals referred for genetic testing for idiopathic ID and congenital anomalies and appear to be enriched in such individuals compared to clinically normal controls. Almost all the 16p12.1 microdeletions identified have been inherited from a carrier parent; carrier parents for the 16p12.1 microdeletion are more likely to exhibit learning disability, bipolar disorder or depression, and epilepsy than noncarrier parents. The presence of varying degrees of learning disability in the adult family members suggests that some transmitted abnormalities are pathological and have an underappreciated contribution to the phenotype [Girirajan et al., 2010]. Similarly, 10q11.21q11.23 deletions are found in 10 apparently normal parents (patients 2–7, 9–10, 14, and 20) and one grandparent (patient 7); however, none have been reported in 9,183 controls. Two carrier parents (patients 1 and 11) and two carrier siblings (patients 1 and 11) are affected. The frequency of these deletions is significantly enriched in cases versus controls.
The variable expressivity resulting from haploinsufficiency of genes in the deletion region and its inheritance from apparently healthy carrier parents suggests additional modifiers, genetic and nongenetic, may influence the pathogenicity of the 10q11.21q11.23 microdeletions. Several explanations have been proposed for the variable expressivity and clinical heterogeneity in some genomic disorders. First, atypical or variable-sized copy number changes may account for the variable phenotypes in some apparently recurrent lesions. In our study population, the deletion sizes varied from 1.9 to 10.9 Mb, although there was substantial clustering of deletion intervals. In addition, five of the deletions and nine of the duplications were not flanked by directly oriented LCRs and thus are unlikely to be caused by NAHR. Atypical breakpoints have been reported for other recurrent rearrangements mediated by segmental duplications: for example, some of the rarer rearrangements of 17p11.2 associated with Smith-Magenis syndrome do not have breakpoints flanked by the typical paired segmental duplications and are not associated with known genomic architectural features [Stankiewicz et al., 2003], and some of the breakpoints in the recently identified 16p11.2p12.2 microdeletion syndrome are not flanked by segmental duplications [Ballif et al., 2007].
A “two-hit” model has also recently been proposed to account for phenotypic variability; it was first used to describe the recurrent deletion 16p12.1 [Girirajan et al., 2010]. In that study, 25% of probands carried a “second hit”—a 40-fold increase for two or more copy number changes over the general population. Furthermore, the clinical features in probands with two hits were different from those with just the second hit. Further analysis of other genomic disorders has shown clustering of two hits in copy number changes with variable phenotypes compared to syndromic lesions [Girirajan and Eichler, 2010]. One hit may be sufficient to reach a threshold that results in mild neurodevelopmental deficits, whereas a second hit is necessary for the development of a more severe neurological phenotype, including ID/DD, ASDs, or schizophrenia [Girirajan and Eichler, 2010]. Patients 5, 7, 9, 16, and 19 have additional large copy number changes, including ~12 Mb deletion 3q13.12q13.32, ~4.8 Mb duplication 12q23.1q23.2, ~11.4 Mb duplication 6p25.3p22.1, confined placental mosaicism for trisomy 2, and ~1.4 Mb duplication 15q21.2q21.3, respectively, that might have contributed additional features to their abnormal phenotypes not observed in other patients lacking additional copy number changes. In patient 19, one cannot exclude the possibility of genomic imprinting in this region also contributing to the phenotype.
A comparison of the approximately 1.9 Mb smallest region of overlap in the 24 deletions reported here to locations of benign CNVs revealed the presence of a small, unique sequence overlap of approximately 1.5 Mb (between 49.2 and 50.6 Mb, LCR 10q11.2 from C to D), where complete deletions have not been reported in control populations. This critical interval encompasses 11 RefSeq genes: MAPK8 (MIM# 601158), ARHGAP22 (MIM# 610585), WDFY4 (MIM# 613316), LRRC18, FAM170B, DRGX (MIM# 606701), ERCC6 (MIM# 609413), PGBD3, SLC18A3 (MIM# 600336), CHAT, and OGDHL (Fig. 1). Some of these 11 genes are known to contribute to human disease. Recessive mutations in ERCC6 have been reported in patients with Cockayne syndrome type B (CSB, MIM# 133540) [Falik-Zaccai et al., 2008; Ghai et al., 2011]. Interestingly, two genes, CHAT, encoding choline acetyltransferase and the single exon SLC18A3 that maps to the first intron of CHAT and encodes vesicular acetylcholine transporter, are involved in cholinergic neurotransmission [Harold et al., 2003]. Point mutations in CHAT have been found in patients with autosomal recessive myasthenic syndrome with episodic apnea (CMS-EA; MIM# 254210) [Ohno et al., 2001]. Of note, a few of our patients manifest some features typical for CMS-EA such as hypotonia, sleep apnea, dysphagia, and ptosis. Usually, haploinsufficiency for enzymes is not deleterious; however, exceptions have been described [Bademci et al., 2010]. We suggest that the above features as well as chronic constipation, gastroesophageal and vesicoureteral refluxes, epilepsy, ataxia, dysphagia, and nystagmus present in some of our patients with heterozygous deletions at 10q11.2, could have been exacerbated by haploinsufficiency of SLC18A3 (cis-genetics effect) and may be mitigated with acetylcholinesterase inhibitors. In support of this notion, a 50–100% increase of the choline transporter 1 (Slc5a7) mRNA was found in the heterozygous Chat+/– mice and was proposed to compensate for the reduced Chat activity and restore the acetylocholine amount [Brandon et al., 2004]. Alternatively, the abnormal phenotype in patients with a heterozygous deletion of a gene responsible for an autosomal recessive trait can result from unmasking of a recessive mutation or functional polymorphism of the remaining allele [Kurotaki et al., 2005]. However, we did not identify any change in 221 amplicons sequenced in the second allele of CHAT in 13 patients with heterozygous deletions of 10q11.2.
Notably, none of the deletions presented here harbor the RET proto-oncogene that maps 2.5 Mb centromeric to the proximal LCR10q11.2 LCR cluster. Mutations in RET have been reported in patients with multiple endocrine neoplasia, type IIA (MEN2A; MIM# 171400), MEN, type IIB (MEN2B; MIM# 162300), Hirschsprung disease (HSCR; aganglionic megacolon; MIM# 142623), and medullary thyroid carcinoma (MTC; MIM# 155240). Interestingly, Puliti et al. [1993] described a patient with a cytogenetically visible deletion 10q11.2q21.2 and a variant of HSCR.
As expected from the NAHR mechanism, we also identified patients with the reciprocal microduplications. None of them were referred for microarray testing for any of the specific clinical features that we suggest may be due to haploinsufficiency of the CHAT and SLC18A3 genes, although detailed clinical descriptions were not available to us.
We report novel deletions and duplications at 10q11.21q11.23 that are likely caused by NAHR between LCRs. Our findings challenge the traditionally used paradigm in the diagnostic setting that aberrations inherited from a phenotypically normal parent are usually without clinical consequences. Large copy number alterations such as those described in this report are associated with unpredictable and variable phenotypic outcomes and pose diagnostic and counseling difficulties. Careful consideration of additional factors that may influence variable phenotype should be considered. Larger studies are needed to obtain a better understanding of this complex genomic region and its associated pathology. Further analysis of the 10q11.21q11.23 deletion in a well-phenotyped family might reveal that the deletion has recognizable phenotypic consequences, although the effect in some individuals may be more subtle depending on genetic and/or nongenetic modifiers.
Supplementary Material
Acknowledgments
We thank Ian M. Campbell for helpful discussion. We thank Erin Dodge and Aaron Theisen (Signature Genomics) for their editorial assistance and assistance with figure preparation. Lisa G. Shaffer, Jill A. Rosenfeld, and Blake C. Ballif are employees of Signature Genomic Laboratories, a subsidiary of PerkinElmer.
Contract grant sponsor: Polish Ministry of Science and Higher Education (R13-0005-04/2008 to P.S.).
Footnotes
Web Resources
Blast2 (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
Database of Genomic Variants (http://projects.tcag.ca/variation/)
Human Genome Segmental Duplication Database (http:// projects.tcag.ca/humandup/)
Miropeats (http://genome.wustl.edu/software/miropeats)
Online Mendelian Inheritance in Man (http://www.omim.org)
Repeatmasker (http://www.repeatmasker.org)
UCSC genome browser (http://genome.ucsc.edu)
References
- Bademci G, Edwards TL, Torres AL, Scott WK, Zuchner S, Martin ER, Vance JM, Wang L. A rare novel deletion of the tyrosine hydroxylase gene in Parkinson disease. Hum Mutat. 2010;31:E1767–E1771. doi: 10.1002/humu.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, Asamoah A, Brock PL, Gowans GC, Conway RL, Graham JM, Jr., Medne L, Zackai EH, Shaikh TH, Geoghegan J, Selzer RR, Eis PS, Bejjani BA, Shaffer LG. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, McDonald M, Campbell L, Ming JE, Zackai EH, Bejjani BA, Shaffer LG. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008a;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, McDonald-McGinn DM, Zackai EH, Hersh JH, Bejjani BA, Shaffer LG. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet. 2008b;74:469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- Barber JC, Maloney VK, Huang S, Bunyan DJ, Cresswell L, Kinning E, Benson A, Cheetham T, Wyllie J, Lynch SA, Zwolinski S, Prescott L, Crow Y, Morgan R, Hobson E. 8p23.1 duplication syndrome; a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet. 2008;16:18–27. doi: 10.1038/sj.ejhg.5201932. [DOI] [PubMed] [Google Scholar]
- Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, Fong CT, Salamone J, Freedenberg D, Hannig VL, Prock LA, Miller DT, Raffalli P, Harris DJ, Erickson RP, Cunniff C, Clark GD, Blazo MA, Peiffer DA, Gunderson KL, Sahoo T, Patel A, Lupski JR, Beaudet AL, Cheung SW. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- Bisgaard AM, Kirchhoff M, Nielsen JE, Brandt C, Hove H, Jepsen B, Jensen T, Ullmann R, Skovby F. Transmitted cytogenetic abnormalities in patients with mental retardation: pathogenic or normal variants? Eur J Med Genet. 2007;50:243–255. doi: 10.1016/j.ejmg.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Brandon EP, Mellott T, Pizzo DP, Coufal N, D'Amour KA, Gobeske K, Lortie M, López-Coviella I, Berse B, Thal LJ, Gage FH, Blusztajn JK. Choline transporter 1 maintains cholinergic function in choline acetyltransferase haploinsufficiency. J Neurosci. 2004;24:5459–5466. doi: 10.1523/JNEUROSCI.1106-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, Lupski JR, Chinault AC, Beaudet AL. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Wellcome Trust Case Control Consortium. Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Earthrowl ME, Grafham DV, Rubenfield M, French L, Steward CA, Sims SK, Jones MC, Searle S, Scott C, Howe K, Hunt SE, Andrews TD, Gilbert JG, Swarbreck D, Ashurst JL, Taylor A, Battles J, Bird CP, Ainscough R, Almeida JP, Ashwell RI, Ambrose KD, Babbage AK, Bagguley CL, Bailey J, Banerjee R, Bates K, Beasley H, Bray-Allen S, Brown AJ, Brown JY, Burford DC, Burrill W, Burton J, Cahill P, Camire D, Carter NP, Chapman JC, Clark SY, Clarke G, Clee CM, Clegg S, Corby N, Coulson A, Dhami P, Dutta I, Dunn M, Faulkner L, Frankish A, Frankland JA, Garner P, Garnett J, Gribble S, Griffiths C, Grocock R, Gustafson E, Hammond S, Harley JL, Hart E, Heath PD, Ho TP, Hopkins B, Horne J, Howden PJ, Huckle E, Hynds C, Johnson C, Johnson D, Kana A, Kay M, Kimberley AM, Kershaw JK, Kokkinaki M, Laird GK, Lawlor S, Lee HM, Leongamornlert DA, Laird G, Lloyd C, Lloyd DM, Loveland J, Lovell J, McLaren S, McLay KE, McMurray A, Mashreghi-Mohammadi M, Matthews L, Milne S, Nickerson T, Nguyen M, Overton-Larty E, Palmer SA, Pearce AV, Peck AI, Pelan S, Phillimore B, Porter K, Rice CM, Rogosin A, Ross MT, Sarafidou T, Sehra HK, Shownkeen R, Skuce CD, Smith M, Standring L, Sycamore N, Tester J, Thorpe A, Torcasso W, Tracey A, Tromans A, Tsolas J, Wall M, Walsh J, Wang H, Weinstock K, West AP, Willey DL, Whitehead SL, Wilming L, Wray PW, Young L, Chen Y, Lovering RC, Moschonas NK, Siebert R, Fechtel K, Bentley D, Durbin R, Hubbard T, Doucette-Stamm L, Beck S, Smith DR, Rogers J. The DNA sequence and comparative analysis of human chromosome 10. Nature. 2004;429:375–381. doi: 10.1038/nature02462. [DOI] [PubMed] [Google Scholar]
- Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, Rosenfeld JA, Lamb AN, Sahoo T. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:119–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, Smith WE, Simon-Fayard E, Alexander AA, Kulharya AS, Ketterling RP, Clark RD, Jalal SM. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik-Zaccai TC, Laskar M, Kfir N, Nasser W, Slor H, Khayat M. Cockayne syndrome type II in a Druze isolate in Northern Israel in association with an insertion mutation in ERCC6. Am J Med Genet A. 2008;146A:1423–1429. doi: 10.1002/ajmg.a.32309. [DOI] [PubMed] [Google Scholar]
- Fewtrell MS, Tam PK, Thomson AH, Fitchett M, Currie J, Huson SM, Mulligan LM. Hirschsprung's disease associated with a deletion of chromosome 10 (q11.2q21.2): a further link with the neurocristopathies? J Med Genet. 1994;31:325–327. doi: 10.1136/jmg.31.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns JP, Bulcke J, Verdu P, Carton H, Kleczkowska A, Van den Berghe H. Apparent late-onset Cockayne syndrome and interstitial deletion of the long arm of chromosome 10 (del(10)(q11.23q21.2)). Am J Med Genet. 1991;40:343–344. doi: 10.1002/ajmg.1320400320. [DOI] [PubMed] [Google Scholar]
- Ghai SJ, Shago M, Shroff M, Yoon G. Cockayne syndrome caused by paternally inherited 5 Mb deletion of 10q11.2 and a frameshift mutation of ERCC6. Eur J Med Genet. 2011;54:272–276. doi: 10.1016/j.ejmg.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, deRavel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels A, Stewart HH, Hennekam RC, Cooper GM, Regan R, Knight SJ, Eichler EE, Vermeesch JR. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Peirce T, Moskvina V, Myers A, Jones S, Hollingworth P, Moore P, Lovestone S, Powell J, Foy C, Archer N, Walter S, Edmonson A, McIlroy S, Craig D, Passmore PA, Goate A, Hardy J, O'Donovan M, Williams J, Liddell M, Owen MJ, Jones L. Sequence variation in the CHAT locus shows no association with late-onset Alzheimer's disease. Hum Genet. 2003;113:258–267. doi: 10.1007/s00439-003-0960-2. [DOI] [PubMed] [Google Scholar]
- Holden JJ, MacDonald EA. Brief clinical report: interstitial deletion of the long arm of chromosome 10: del(10)(q11.2q21). Am J Med Genet. 1985;20:245–248. doi: 10.1002/ajmg.1320200206. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M, Gerdes T, Brunebjerg S, Bryndorf T. Investigation of patients with mental retardation and dysmorphic features using comparative genomic hybridization and subtelomeric multiplex ligation dependent probe amplification. Am J Med Genet A. 2005;139:231–233. doi: 10.1002/ajmg.a.31019. [DOI] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O'Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Schulze H, Strauss G, Ott C-E, Hall J, Trotier F, Fleischhauer S, Greenhalgh L, Newbury-Ecob RA, Neumann LM, Habenicht R, Konig R, Seemanova E, Megarbane A, Ropers H-H, Ullmann R, Horn D, Mundlos S. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, Schinzel A, Baumer A, Anderlid BM, Schoumans J, Knoers NV, van Kessel AG, Sistermans EA, Veltman JA, Brunner HG, de Vries BB. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Shen JJ, Touyama M, Kondoh T, Visser R, Ozaki T, Nishimoto J, Shiihara T, Uetake K, Makita Y, Harada N, Raskin S, Brown CW, Hoglund P, Okamoto N, Lupski JR. Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet Med. 2005;7:479–483. doi: 10.1097/01.gim.0000177419.43309.37. [DOI] [PubMed] [Google Scholar]
- Lobo S, Cervenka J, London A, Pierpont ME. Interstitial deletion of 10q: clinical features and literature review. Am J Med Genet. 1992;43:701–703. doi: 10.1002/ajmg.1320430410. [DOI] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Räber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJ, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BB, Vermeesch JR, Barber JC, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K, Tsujino A, Brengman JM, Harper CM, Bajzer Z, Udd B, Beyring R, Robb S, Kirkham FJ, Engel AG. Choline acetyltransferase mutations cause myasthenic syndrome associated with episodic apnea in humans. Proc Natl Acad Sci USA. 2001;98:2017–2022. doi: 10.1073/pnas.98.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JD. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995a;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Parsons JD. Improved tools for DNA comparison and clustering. Comput Appl Biosci. 1995b;11:603–613. doi: 10.1093/bioinformatics/11.6.603. [DOI] [PubMed] [Google Scholar]
- Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, Glaze D, Krull K, Lee JA, Lewis RA, Mendoza-Londono R, Robbins-Furman P, Shaw C, Shi X, Weissenberger G, Withers M, Yatsenko SA, Zackai EH, Stankiewicz P, Lupski JR. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Puliti A, Covone AE, Bicocchi MP, Bolino A, Lerone M, Martucciello G, Jasonni V, Romeo G. Deleted and normal chromosome 10 homologs from a patient with Hirschsprung disease isolated in two cell hybrids through enrichment by immunomagnetic selection. Cytogenet Cell Genet. 1993;63:102–106. doi: 10.1159/000133510. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzàlez JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Ledbetter DH, Lupski JR. Molecular cytogenetics of contiguous gene syndromes: mechanisms and consequences of gene dosage imbalance. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. Metabolic and molecular basis of inherited disease. McGraw Hill; New York: 2001. pp. 1291–1324. [Google Scholar]
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Shah PS, Murthy P, Skidmore D, Shaffer LG, Bejjani BA, Chitayat D. Williams syndrome in a preterm infant with phenotype of Alagille syndrome. Am J Med Genet A. 2008;146A:2407–2411. doi: 10.1002/ajmg.a.32356. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O'Hara R, Casalunovo T, Conlin LK, D'Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SD, Hansen KL, Pasztor LM, DiLiberti JH, Jorgenson RJ, Young RS, Moore CM. Deletions of the long arm of chromosome 10. Am J Med Genet. 1985;20:181–196. doi: 10.1002/ajmg.1320200122. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, Fitzpatrick CA, Segraves R, Richmond TA, Guiver C, Albertson DG, Pinkel D, Eis PS, Schwartz S, Knight SJ, Eichler EE. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Schroer RJ, Novara F, De Gregori M, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, Porter K, Prigmore E, Krepischi-Santos AC, Varela MC, Koiffmann CP, Lees AJ, Rosenberg C, Firth HV, de Silva R, Carter NP. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, Morris CA, Scherer SW, Osborne LR. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, Lupski JR. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, Banna L, Brereton AV, Hill A, Bisgaard AM, Muller I, Hultschig C, Erdogan F, Wieczorek G, Ropers HH. ArrayCGHidentifiesreciprocal16p13.1duplicationsanddeletionsthatpredispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, FitzPatrick DR, Maher E, Martin H, Parnau J, Pindar L, Ramsay J, Shaw-Smith C, Sistermans EA, Tettenborn M, Trump D, de Vries BB, Walker K, Raymond FL. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, Gallo N, Morrow BE, Shaffer LG, Babcock M, Chernos J, Bernier F, Sprysak K, Christiansen J, Haase S, Elyas B, Lilley M, Bamforth S, McDermid HE. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenger-Hain JL, Roberson J, Van Dyke DL, Weiss L. Interstitial deletion of chromosome 10, del(10) (q11.2q22.1) in a boy with developmental delay and multiple congenital anomalies. Am J Med Genet. 1993;46:438–440. doi: 10.1002/ajmg.1320460418. [DOI] [PubMed] [Google Scholar]
- Zogopoulos G, Ha KC, Naqib F, Moore S, Kim H, Montpetit A, Robidoux F, Laflamme P, Cotterchio M, Greenwood C, Scherer SW, Zanke B, Hudson TJ, Bader GD, Gallinger S. Germ-line DNA copy number variation frequencies in a large North American population. Hum Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.