Abstract
Down syndrome is the leading cause of prenatal chromosome abnormalities, accounting for 53% of all reported chromosome conditions. Testing strategies, guidelines, and screening options have expanded from their conception in the 1970s, and now include such options as anatomical ultrasound, maternal serum screening, and noninvasive prenatal testing. This review summarizes all currently available noninvasive diagnostic techniques for the detection of Down syndrome. By understanding fully each technology and the possible alternatives, the physician will be able to provide their patients with all the information necessary to make an informed decision regarding their medical management.
Keywords: Down syndrome, noninvasive screening, diagnostic techniques
Introduction
It has been roughly half a century since Lejeune et al1 first described Down syndrome in 1959. While the technology employed for prenatal detection of Down syndrome has expanded in leaps and bounds, the primary focus of prenatal care remains the same: to offer women the most thorough risk assessment with the least invasive procedure possible. Down syndrome is the leading cause of prenatal chromosome abnormalities, accounting for 53% of all reported chromosome conditions.2
Testing strategies, guidelines, and screening options have expanded from their conception in the 1970s. At that time, any woman aged 35 years or older was considered to be of advanced maternal age, and this was the sole criteria used by the American Congress of Obstetricians and Gynecologists to define pregnancies that should be offered amniocentesis or chorionic villus sampling. As of 2007, the American Congress of Obstetricians and Gynecologists has defined a pregnancy as “high-risk” when any of the following criteria are met: family history of aneuploidy, advanced maternal age, abnormal serum screen, or abnormal ultrasound findings.3,4 While multiple screening options are currently available, the only diagnostic tests offered prenatally for Down syndrome are amniocentesis and chorionic villus sampling. This review compares the risks, benefits, and limitations of all currently available prenatal screening methods for detection of Down syndrome.
Established techniques
Anatomical ultrasound
Anatomical ultrasound has been used since the 1980s to provide health care practitioners and expectant mothers with information regarding a pregnancy.5 With advances in technology, prenatal sonography has expanded from focusing on detection of major structural abnormalities (ie, cardiac defects, hydrops, duodenal atresia, or cystic hygroma) to include detection of soft markers to assist in identifying pregnancies that are at risk of various chromosomal conditions.
Bricker et al6 have defined soft markers as “structural changes detected at ultrasound scan which may be transient and in themselves have little or no pathological significance, but are thought to be more commonly found in fetuses with congenital abnormalities, particularly karyotypic abnormalities”.4–6 Soft markers that have been linked to Down syndrome include nuchal thickening, echogenic intracardiac focus, echogenic bowel, renal pelvic dilation, shortened long bones, absence of the nasal bone, pyelectasis, ventriculomegaly, clinodactyly, and sandal gap toe.5,7,8 In the absence of soft markers, the sensitivity of anatomical ultrasound to detect Down syndrome is relatively low at 50%. However, the presence of one soft marker is associated with an increased risk of Down syndrome, ie, one soft marker increases the risk by two-fold and three or more soft markers increases the risk by 100-fold.7 Many scoring indices have been created to help maximize sensitivity, while decreasing false-positive rates.7,9,10 These indices incorporate the presence of structural abnormalities and/or soft markers and maternal age to provide physicians with a guideline as to who should be offered more invasive procedures. While these guidelines have assisted in determining the criteria for a positive finding, they remain limited by the quality of the ultrasound and the expertise of the sonographer.
Maternal serum screening
Beginning in 1984, multiple marker screening provided physicians with a means of offering an individualized risk for Down syndrome without the inherent risk imposed by chorionic villus sampling or amniocentesis (Table 1).11,12 This second trimester screening, performed at 15–20 weeks gestation, is often referred to as the quad screen because it incorporates maternal age-related risk and four maternal serum biomarkers, ie, alpha-fetoprotein, free beta human chorionic gonadotropin, unconjugated estriol, and dimeric inhibin A levels.11 By combining maternal age with the quad screen, the detection rate is roughly 75% for Down syndrome in women younger than 35 years and >80% in women 35 years and older (with a positive screening rate of 5%).12
Table 1.
Serum screenings
| Screening test | Sensitivity | False positive |
|---|---|---|
| FTS | 85%–90% | 5% |
| MMS | 81% | 5% |
| Integrated | 95% | 5% |
| Step-wise | 94%–96% | 5% |
| Contingent | 94%–95% | 5% |
It was not until the late 1990s that first trimester screening was introduced as an earlier screening option for the detection of Down syndrome. First trimester screening incorporates maternal age, nuchal translucency ultrasonography, and measurement of maternal serum free beta human chorionic gonadotropin and pregnancy-associated plasma protein A.3,4,12,13 Collection of blood for biochemical analysis and ultrasound assessment for nuchal translucency is typically performed between 11 and 13 6/7 weeks gestation. Increased nuchal translucency, reduction in pregnancy-associated plasma protein A levels, and an increase in beta human chorionic gonadotropin can be an indication of Down syndrome, and assist practitioners in identifying pregnancies at risk for the syndrome. A nuchal translucency measurement by itself has a detection rate for Down syndrome of about 70% with a 5% false-positive rate, but when combined with pregnancy-associated plasma protein A and beta human chorionic gonadotropin measurements, detection rates increase to 79%–90%, with a 5% false-positive screen rate.11,14 Various studies have been conducted to determine the optimal time for performing the first trimester screening, with the goal of providing a maximum detection rate while still maintaining a low false-positive rate.15–18 These studies suggest that earlier pregnancy-associated plasma protein A and beta human chorionic gonadotropin measurements taken at 9–10 weeks gestation, with nuchal translucency measurement taken at 12 weeks gestation, can increase the detection rate to 90%–93%, with a 3%–5% false-positive rate.15,17,18 A detection rate of 92%–95% with a 3%–5% false-positive rate can be achieved when pregnancy-associated plasma protein A measurements are done at 9–10 weeks gestation, with beta human chorionic gonadotropin and nuchal translucency measurements taken at 12 weeks gestation.15 First trimester screening gives women who receive prenatal care prior to 14 weeks gestation the ability to have information sooner than with second trimester screening. If the results reveal an increased risk of fetal aneuploidy, the woman can be offered genetic counseling with the option to choose either first-trimester chorionic villus sampling or second-trimester amniocentesis.
Independent sequential screening is defined as independently performed first-trimester and second-trimester screenings, with separate individualized risk assessments given.11,19 Although independent sequential screening increases the detection rate from 88%–91% to 94%, it also increases the false-positive rate from 5% to 11%.11,19 Thus, it is recommended that women who undergo first-trimester screening for aneuploidy should not also have second-trimester serum screening in the same pregnancy. If a higher detection rate is preferred, an integrated or sequential screening test which combines both first-trimester and second-trimester screening is suggested.
Integrated screening is defined as the process by which a patient’s individualized risk is calculated based on the combination of both the first-trimester and second-trimester screenings.11,19 Due to the nature of this screening method, a patient’s first-trimester screening results are not disclosed until second-trimester screening is performed and a combined risk based on both screenings can be calculated.11,19 There has been ethical debate regarding integrated screening because the patient’s first-trimester screening results are not disclosed until the conclusion of the second-trimester screening. This precludes patients who are at high risk based on first-trimester screening from being offered chorionic villus sampling and having more options available to them.
In contrast, patients undergoing stepwise sequential screening are provided with their individual risk once the results from the first-trimester screen are available.11,19 Patients who screen positive during first-trimester screening are offered genetic counseling and are given the information regarding chorionic villus sampling and amniocentesis. Patients who screen negative are offered second-trimester screening and are provided with an adjusted risk number that incorporates both the first-trimester and second-trimester results. Incorporation of both the first-trimester and second-trimester screenings in this stepwise manner increases the detection rate to 94%–96% while still maintaining a low false-positive rate of 5%–6%.11,19
The final type of sequential screening is known as the contingent screening method. This method is similar to stepwise sequential screening in that scores are calculated based on results from both the first-trimester and second-trimester screening.11,19 However, the contingent screening method uses the first-trimester results to classify patients into three subgroups, ie, screen-positive, screen-negative, and borderline.11,19 Second-trimester screening is only offered to patients who fall into either the screen-negative or the borderline group. The detection rate for this method is 94%–95%, with a false-positive rate of about 5%.11,19
Noninvasive prenatal testing
It has been known since the mid 1950s that fetal cells are present in the maternal circulation.20,21 However, low yield (1 fetal cell/mL of maternal blood), inability to develop an efficient enrichment process, and the tendency for fetal DNA to disintegrate during chromosome extraction have inhibited utilization of these fetal cells for the development of a noninvasive prenatal test.20,22–24 Use of noninvasive prenatal testing targeted to fetal cells was finally abandoned when Bianchi et al25 demonstrated that fetal cells could remain in the maternal circulation for decades, thus making it impossible to distinguish new fetal cells from those of previous pregnancies.
It was not until 1997 when Lo et al26 demonstrated the existence of cell-free fetal DNA in the maternal circulation that the scientific community was presented with a new possible target for noninvasive prenatal testing. Fetal cells, theorized to be derived from the placenta, enter the maternal circulation where they undergo apoptosis. In the process, fetal DNA is cleaved into small 150–200 base pairs fragments, which are then released into the maternal bloodstream.20 These cell-free fetal DNA fragments can be detected as early as the fourth week of gestation and reliably after the seventh week of gestation.12,20,23 Unlike the longevity seen with fetal cells, cell-free fetal DNA has a half-life of only 16 minutes and is cleared from the maternal circulation within 2 hours of delivery.27 The low false-positive rate as well as the prevalence of cell-free fetal DNA in the maternal circulation (accounting for >10% of all cell-free DNA or 16–80 fetal genomes/mL of maternal blood), made cell-free fetal DNA a desirable target for noninvasive prenatal testing.12,20,23,28,29
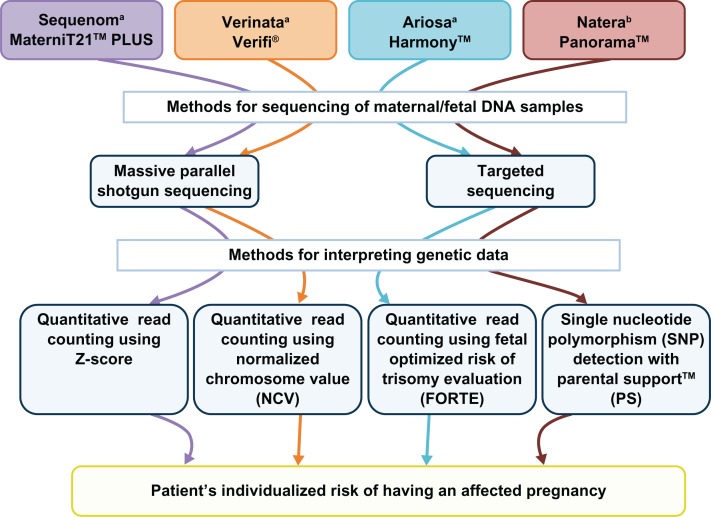
Over the next 15 years, scientists would work on developing and refining methods of detecting pregnancies at risk for aneuploidy. In October, 2011, Sequenom Inc (San Diego, CA, USA) was the first company to make a noninvasive pre-natal test commercially available for the detection of Down syndrome. To date, three companies (Verinata Health [Redwood, CA, USA], Ariosa Diagnostics [San Jose, CA, USA], and Sequenom Inc) offer a noninvasive prenatal test for the detection of trisomies 13 and 18 and Down syndrome, with a fourth company (Natera, San Carlo, CA, USA) expected to have a commercially available noninvasive prenatal test within the next few months (Figure 1).
Figure 1.
Currently used NIPT methodology.
Notes:Figure 1 depicts a flowchart of the various methodologies employed by the four commercially developed NIPTs. Figure based on information obtained from references 31–42. aDenotes currently commercially offered NIPT; bdenotes soon to be released NIPT.
Figure 1 describes the various strategies employed by the four companies to obtain and analyze maternal and/or fetal DNA from the maternal serum sample. Two companies, Sequenom Inc and Verinata Health, utilize massively parallel shotgun sequencing, commonly referred to as shotgun sequencing,30–31 while the other two companies, Natera and Ariosa Diagnostics, utilize targeted sequence analysis.33–36 The main difference between these two technologies is that shotgun sequencing results in amplification of all genetic information, while targeted sequencing results in amplification of only the genetic information of interest, ie, chromosome(s). Targeted sequencing enables companies such as Natera and Ariosa Diagnostics to perform noninvasive prenatal testing on smaller sample sizes compared with Sequenom Inc and Verinata Health. Massively parallel shotgun sequencing technology requires more “reads” (or DNA fragments) than those required by targeted sequencing to ensure that there are enough fragments from the chromosomes of interest to provide accurate results. A major disadvantage of targeted sequencing is the inability to rule out other chromosomal conditions, such as microduplications and deletions, because they have not been selected for. While this does not seem to be a major limiting factor at this time, due to the fact that companies are not currently reporting nonvalidated findings, it may become a shortcoming in the future as companies aim to expand noninvasive prenatal testing to include other chromosomal abnormalities, such as microdeletions and microduplications.
All four companies currently use variations of two methodologies for analyzing genetic data, ie, quantitative read counting or single nucleotide polymorphism detection. Three of the companies (Sequenom Inc, Verinata Health, and Ariosa Diagnostics) use quantitative read counting, while Natera uses bioinformatic algorithms to analyze single nucleotide polymorphism data obtained by next-generation aneuploidy testing using single nucleotide polymorphisms.
Quantitative read counting analyzes the number of chromosome fragments present from each chromosome or chromosomes of interest. Because the amount of genetic material from each chromosome is directly proportional to the chromosome size, companies can use a known euploid reference sample to calculate the expected proportion of genetic information from each chromosome given a euploid pregnancy.31,32,37–39 For example, chromosome 21 generally accounts for 1.5% of the human genome.31,37–39 Any change in the actual proportions of genetic information from chromosome 21 that is 2.5–3.1 standard deviations from the mean is determined to be an aneuploidy.30,31,40 The benefit of this technology is that it does not require any differentiation between maternal and fetal genetic information. However, this is also a limitation resulting from the fact that, in a fetus with Down syndrome, the contribution of genetic information from chromosome 21 increases from 1.5% to 2.25%, for an overall change of 0.75%. When taking into account the fact that fetal DNA only represents 10% of the total DNA in the sample, the overall change in the amount of chromosome 21 cell-free-DNA would only increase from 1.5% to 1.575%. When the fetal fraction is below 10%, it can result in inconclusive results, or a no-call. One company using quantitative read counting, Ariosa Diagnostics, has aimed to address this issue by creating the FORTE (fraction optimized risk of trisomy evaluation) assay.33,34 This assay utilizes polymorphic and nonpolymorphic regions that are known to differ between fetal and maternal DNA to determine the fetal fraction as well as the overall proportional representation of chromosomal fragments.
Unlike quantitative read counting, the next-generation aneuploidy testing using single nucleotide polymorphisms system created by Natera utilizes the Parental Support™ statistical algorithm to analyze the sequenced fetal cell-free DNA together with maternal genetic information, paternal genetic information, and HapMap technology.35,36 This algorithm provides a series of possible “hypotheses” of fetal genotypes (eg, monosomy, disomy, trisomy) based on known common crossover points on the chromosome(s) of interest, parental information, fetal fraction, and fetal chromosome copy number. The sample-specific confidence interval for each hypothesis is calculated, and a “hypothesis” is considered confirmed when the confidence interval is above 98%.36
While the results seen from these four companies are promising (Table 2), it is important to remember that noninvasive prenatal testing is for screening and is not a diagnostic tool.30,32,35,36,41 Noninvasive prenatal test results group patients into three possible categories, ie, low risk of aneuploidy, high risk of aneuploidy, or no-call (undeterminable). The clinical implications for low-risk and high-risk populations are the same for noninvasive prenatal testing as they are for first-trimester screening and multiple marker screening. Individuals who are determined to be in the low-risk category would not require any further diagnostic evaluation, while those in the high-risk category are recommended to undergo diagnostic testing (ie, chorionic villus sampling or amniocentesis). Individuals who fall into the third category of “no-call” would require a repeat sample to be drawn. For these reasons, noninvasive prenatal testing is not intended to serve as a replacement for amniocentesis or chorionic villus sampling, but rather a methodology that would enable fewer unnecessary diagnostic procedures by detecting patients who are at high risk for aneuploidy more accurately, when compared with the detection rates for first-trimester screening and multiple marker screening. Physicians should ensure that their patients understand that these tests are not designed to circumvent an unwanted diagnostic procedure, and in actuality may result in the recommendation of such a diagnostic procedure should test results reveal a high risk for aneuploidy.
Table 2.
Accuracy of commercially available NIPT
| Sequenom MaterniT21™ PLUS | Verinata Verifi® | Ariosa Harmony™ | Natera Panorama™ | |
|---|---|---|---|---|
| Sensitivity | 98.6%–99% (209/212) | 100% (89/89) | 100% (81/81) | 100% (19/19) |
| Specificity | 99.80% (1468/1471) | 100% (404/404) | 99.97% (2887/2888) | 100% (362/362) |
| False positive | 0.2% (3/1471) | 0 | 0.03% (1/2888) | 0 |
| No call rate | 3.4 | 5.8 | 4.7%–5.7% | 12.6%* |
Notes:Table 2 depicts the outcome from the validation studies and current no call rates from the four commercially developed NIPTs.
denotes a no call rate that incorporates all aneuploidies analyzed by Natera (Trisomy 13, 18, 21, and sex chromosome abnormalities).
While noninvasive prenatal tests do have an increased specificity and sensitivity compared with first-trimester screening or multiple marker screening, limitations do exist. False-positive results may be present. Possible causes for false-positive results could be placental mosaicism, vanishing twin syndrome, or an unidentified maternal condition, such as mosaicism or cancer. Secondly, the majority of noninvasive prenatal tests are currently only offered to women with singleton pregnancies who are at high risk for Down syndrome because of family history of aneuploidy, advanced maternal age, an abnormal serum screen, or abnormal ultrasound findings. With the exception of Ariosa Diagnostics, which has conducted validation studies for Harmony® in the low-risk population, it is important to note that these tests have not been validated in low-risk or multiple gestation populations, and their accuracy is unknown at this time.42
Future directions
It has long been a goal of prenatal genetic diagnosis to develop a noninvasive test that would allow for detection of aneuploidy and eliminate the need for invasive testing, such as chorionic villus sampling or amniocentesis. While this is still not currently possible, developments in noninvasive prenatal testing are the initial steps in this direction. While the scope of this review does not include other aneuploidy conditions, all of the companies discussed do offer screening for trisomy 13 and 18, with varying degrees of success. The noninvasive prenatal tests marketed by Verinata Health and Natera also include a screen for sex chromosome abnormalities, such as Turner syndrome, Kleinfelter syndrome, and 47, XYY syndrome. Validation studies are currently underway for noninvasive prenatal testing in the detection of fetal aneuploidies in the general population as well as multiple gestations. Furthermore, the future of noninvasive prenatal testing may expand beyond the common aneuploidies to include other chromosomal conditions, such as microdeletions and microduplications. With the ever-expanding testing options that are now available to the expectant mother, it is imperative that physicians remain up to date on these technologies and have a clear understanding of the risks, benefits, and limitations of these technologies. By fully understanding each technology and the possible alternatives, physicians can provide their patients with all the information necessary to make an informed decision regarding medical management.
Acknowledgments
This work was supported by a grant from the National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development (1K23HD058043-01A1 to JV).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lejeune J, Gautier M, Turpin R. Study of somatic chromosomes from 9 mongoloid children. C R Hebd Seances Acad Sci. 1959;248:1721–1722. French. [PubMed] [Google Scholar]
- 2.Wellesley D, Dolk H, Boyd PA, et al. Rare chromosome abnormalities, prevalence and prenatal diagnosis rates from population-based congenital anomaly registers in Europe. Eur J Hum Genet. 2012;20:521–526. doi: 10.1038/ejhg.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ACOG Committee on Practice Bulletins ACOG Practice Bulletin No 77: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2007;109:217–227. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 4.ACOG Committee on Practice Bulletins ACOG Practice Bulletin No 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet Gynecol. 2007;110:1459–1467. doi: 10.1097/01.AOG.0000291570.63450.44. [DOI] [PubMed] [Google Scholar]
- 5.Getz L, Kirkengen AL. Ultrasound screening in pregnancy: advancing technology, soft markers for fetal chromosomal aberrations, and unacknowledged ethical dilemmas. Soc Sci Med. 2003;56:2045–2057. doi: 10.1016/s0277-9536(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 6.Bricker L, Garcia J, Henderson J. Ultrasound screening in pregnancy: a systematic review of the clinical effectiveness, cost-effectiveness and women’s views. Health Technol Assess. 2000;4:i–vi. 1–193. [PubMed] [Google Scholar]
- 7.Nyberg DA, Souter VL. Sonographic markers of fetal trisomies: second trimester. J Ultrasound Med. 2001;20:655–674. doi: 10.7863/jum.2001.20.6.655. [DOI] [PubMed] [Google Scholar]
- 8.Benacerraf BR. The history of the second-trimester sonographic markers for detecting fetal Down syndrome, and their current role in obstetric practice. Prenat Diagn. 2010;30:644–652. doi: 10.1002/pd.2531. [DOI] [PubMed] [Google Scholar]
- 9.Winter TC, Uhrich SB, Souter VL, Nyberg DA. The ‘genetic sonogram’: comparison of the index scoring system with the age-adjusted US risk assessment. Radiology. 2000;215:775–782. doi: 10.1148/radiology.215.3.r00ma36775. [DOI] [PubMed] [Google Scholar]
- 10.Bromley B, Lieberman E, Benacerraf BR. The incorporation of maternal age into the sonographic scoring index for the detection at 14– 20 weeks of fetuses with Down’s syndrome. Ultrasound Obstet Gynecol. 1997;10:321–324. doi: 10.1046/j.1469-0705.1997.10050321.x. [DOI] [PubMed] [Google Scholar]
- 11.Malone FD, Janick JA, Ball RH, et al. First-trimester or second-trimester screening, or both, for Down’s syndrome. N Engl J Med. 2005;353:2001–2011. doi: 10.1056/NEJMoa043693. [DOI] [PubMed] [Google Scholar]
- 12.Simpson JL. Invasive procedures for prenatal diagnosis: any future left? Best Pract Res Clin Obstet Gynaecol. 2012;26:625–638. doi: 10.1016/j.bpobgyn.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Driscoll DA, Gross SJ. Screening for fetal aneuploidy and neural tube defects. Genet Med. 2009;11:818–821. doi: 10.1097/GIM.0b013e3181bb267b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer K, Spencer CE, Power M, Dawson C, Nicolaides KH. Screening for chromosomal abnormalities in the first trimester using ultrasound and maternal serum biochemistry in a one-stop clinic: a review of three years prospective experience. BJOG. 2003;110:281–286. [PubMed] [Google Scholar]
- 15.Wright D, Spencer K, Kagan KK. First-trimester combined screening for trisomy 21 at 7–14 weeks’ gestation. Ultrasound Obstet Gynecol. 2010;36:404–411. doi: 10.1002/uog.7755. [DOI] [PubMed] [Google Scholar]
- 16.Tørring N. Performance of first-trimester screening between gestational weeks 7 and 13. Clin Chem. 2009;55:1564–1567. doi: 10.1373/clinchem.2009.125922. [DOI] [PubMed] [Google Scholar]
- 17.Kirkegaard I, Petersen OB, Uldbjerg N, Tørring N. Improved performance of first-trimester combined screening for trisomy 21 with the double test taken before a gestational age of 10 weeks. Prenat Diagn. 2008;28:839–844. doi: 10.1002/pd.2057. [DOI] [PubMed] [Google Scholar]
- 18.Borrell A, Casals E, Fortuny A, et al. First-trimester screening for trisomy 21 combining biochemistry and ultrasound at individually optimal gestational ages. An interventional study. Prenat Diagn. 2004;24:541–545. doi: 10.1002/pd.949. [DOI] [PubMed] [Google Scholar]
- 19.Krantz DA, Hallahan TW, Macri VJ, Macri JN. Genetic sonography after first-trimester Down syndrome screening. Ultrasound Obstet Gynecol. 2007;29:666–670. doi: 10.1002/uog.4029. [DOI] [PubMed] [Google Scholar]
- 20.Wright CF, Burton H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum Reprod Update. 2009;15:139–151. doi: 10.1093/humupd/dmn047. [DOI] [PubMed] [Google Scholar]
- 21.Bianchi DW, Flint AF, Pizzimenti MF, Knoll JH, Latt SA. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci U S A. 1990;87:3279–3283. doi: 10.1073/pnas.87.9.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchi DW, Williams JM, Sullivan LM, Hanson F, Klinger KW, Shuber AP. PCR quantitation of fetal cells in maternal blood in normal and aneuploid pregnancies. Am J Hum Genet. 1997;61:822–829. doi: 10.1086/514885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sifakis S, Papantoniou N, Kappou D, Antsaklis A. Noninvasive prenatal diagnosis of Down syndrome: current knowledge and novel insights. J Perinat Med. 2012;40:319–327. doi: 10.1515/jpm-2011-0282. [DOI] [PubMed] [Google Scholar]
- 24.Sekizawa A, Purwosunu Y, Farina A, et al. Development of noninvasive fetal DNA diagnosis from nucleated erythrocytes circulating in maternal blood. Prenat Diagn. 2007;27:846–848. doi: 10.1002/pd.1792. [DOI] [PubMed] [Google Scholar]
- 25.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo Y, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 27.Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NA. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birch L, English CA, O’Donoghue K, Barigye O, Fisk NM, Keer JT. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clin Chem. 2005;51:312–320. doi: 10.1373/clinchem.2004.042713. [DOI] [PubMed] [Google Scholar]
- 29.Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palomaki GE, Kloza EM, Lambert-Messerlian GM. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 31.Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204:205. e1–e11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 32.Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 33.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:319. e1–e9. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 34.Sparks AB, Wang ET, Struble CA, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann B, Hill M, Gemelos G, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn. 2012;32:1233–1241. doi: 10.1002/pd.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage M. Merging into clinical practice: updates in non-invasive prenatal testing. Abstract 304 presented at the 31st National Society of Genetic Counselors Annual Education Conference; Boston, MA. October 24–27, 2012. [Google Scholar]
- 37.Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–20468. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:322. e1–e5. doi: 10.1016/j.ajog.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 39.Ashoor G, Syngelaki A, Poon L, Rezende J, Nicolaides K. Fetal fraction in maternal plasma cell-free DNA at 11–13 weeks’ gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. 2013;41:26–32. doi: 10.1002/uog.12331. [DOI] [PubMed] [Google Scholar]
- 40.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norton ME, Brah H, Weiss J, et al. Non-Invasive Chromosomal Evaluation (NICE) study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207:137. e1–e8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 42.Nicolaides H, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am J Obstet Gynecol. 2012;207:374. e1–e6. doi: 10.1016/j.ajog.2012.08.033. [DOI] [PubMed] [Google Scholar]