Abstract
Systemic autoimmune diseases are characterized by the development of autoantibodies directed against a limited subset of nuclear antigens, including DNA. DNA-specific B cells take up mammalian DNA through their B cell receptor, and this DNA is subsequently transported to an endosomal compartment where it can potentially engage TLR9. We have previously shown that ssDNA-specific B cells preferentially bind particular DNA sequences, and antibody specificity for short synthetic oligodeoxynucleotides (ODNs) has been shown. Since CpG-rich DNA, the ligand for TLR9 is found in low abundance in mammalian DNA, we sought to determine whether antibodies derived from DNA-reactive B cells showed binding preference for CpG-rich native dsDNA, and thereby select immunostimulatory DNA for delivery to TLR9. We examined a panel of anti-DNA antibodies for binding to CpG-rich and CpG-poor DNA fragments. We show that a number of anti-DNA antibodies do show preference for binding to certain native dsDNA fragments of differing sequence, but this does not correlate directly with the presence of CpG dinucleotides. An antibody with preference for binding to a fragment containing optimal CpG motifs was able to promote B cell proliferation to this fragment at 10-fold lower antibody concentrations than an antibody that did not selectively bind to this fragment, indicating that antibody binding preference can influence autoreactive B cell responses.
Keywords: Autoantibody, Systemic lupus erythematosus, Anti-DNA, TLR9, B-cell, CpG-rich DNA
1. Introduction
Systemic lupus erythematosus (SLE) and other systemic autoimmune diseases are characterized by the development of autoantibodies directed against a limited subset of self-antigens. A high percentage of the autoantigens targeted in SLE are normally found as either DNA-associated or RNA-associated macromolecules. DNA-related antigens include, single-stranded DNA, dsDNA, histones, and other DNA-binding proteins. RNA-related antigens include U-rich RNA, SmD, and other splicesome-associated proteins. In addition, a substantial number of autoreactive B cells recognize autologous IgG and these rheumatoid factors (RF) can potentially bind IgG immune complexes, which incorporate DNA or RNA-containing particles. These antigens have in common the presence of bound nucleic acids [1, 2].
We have previously shown that these bound nucleic acids are able to provide an adjuvant effect by activating either Toll-like receptor 9 (TLR9) or TLR7 after being taken up by the B cell receptor (BCR) on B cells or by Fcγ receptors on dendritic cells [3, 4]. AM14 BCR transgenic (Tg) mice express a prototypical autoimmune RF BCR, which binds IgG2a with low affinity [5]. When stimulated with IgG2a antibodies specific for DNA or RNA-associated antigens, AM14 B cells proliferate in a TLR9 or TLR7-dependent manner, respectively. This is dependent on the presence of mammalian DNA or RNA in the culture supernatant [3, 6]. Similarly, 3H9 dsDNA specific B cells, and 3H9/Vκ8 ssDNA specific B cells proliferate directly in response to DNA present in the culture supernatant [7, 8].
The TLR family is one of the major families of innate immune receptors. Ligands include a diverse array of pathogen-derived molecules, as well as some endogenous ligands hypothesized to serve as danger signals. TLR engagement on antigen presenting cells (APC) leads to upregulation of costimulatory molecules, cytokine production, and type I IFN production. Engagement of TLRs on B cells leads to proliferation, antibody production, and cytokine secretion. While most TLRs are expressed on the cell surface, the subset that recognize nucleic acids is localized intracellularly, where they serve to detect nucleic acids derived from viruses and bacteria. Included in this group are TLR3, which recognizes dsRNA, and TLR7 and TLR8, which recognize ssRNA [9]. The TLR9 signaling cascade is preferentially engaged by unmethylated CpG motifs, found at a higher frequency in microbial than mammalian DNA [10]. Optimal CpG motifs for activating mouse TLR9, as defined with synthetic oligonucleotides, have the base context PuPuCGPyPy, with the best motif being GACGTT [11]. Mammalian DNA is thought to be a relatively poor TLR9 ligand due to its low CG-content, CpG depletion, and CpG methylation [12, 13]. Therefore, how mammalian DNA is able to engage TLR9 in autoreactive B cells is unknown.
We have previously shown that immune complexes (IC) incorporating dsDNA fragments derived from CG-rich mammalian DNA can activate AM14 B cells better than ICs incorporating CG-poor mammalian DNA fragments. Thus TLR9 can distinguish CG-rich and CG-poor mammalian DNA. We also found that 3H9/Vκ8 ssDNA-specific antibody preferentially binds certain CG-rich DNA fragments over others, and that these fragments induced a stronger proliferative response [8]. These observations are consistent with the premise that the activation of DNA-reactive B cells requires a receptor that binds CG-rich DNA. Studies from a number of groups have found that sequence-specific antibodies can be generated. For example, immunization of mice with the DNA-binding domain of the human papillomavirus E2 protein bound to its target DNA sequence lead to the generation of antibodies specific for the target DNA sequence [14]. Another study using systemic evolution of ligands by exponential enrichment (SELEX) to examine the binding of ssDNA-reactive antibodies to DNA found that these antibodies bound to stem-loop structures, and that some of the antibodies were specific for certain sequences [15]. The anti-DNA antibody H241 has also been reported to bind to a (dG-dC)3 core in the context of a short single-stranded ODN [16], and 3H9R single-chain Fv has also been reported to have a binding preference for poly(dG)·poly(dC) over poly(dA)·poly(dT) [17]. Importantly, all of these studies examined binding to short synthetic ODN substrates. Whether antibody preference for sequence applies to long native dsDNA fragments, which are hypothesized to be the in vivo ligand in autoimmune disease is unknown.
Since the ligand for TLR9 is in low abundance in mammalian DNA, and DNA-specific B cells can show binding preference, this raised the hypothesis that DNA-reactive B cells might preferentially bind to CpG-rich DNA among a pool of genomic DNA, and thereby select immunostimulatory DNA for delivery to TLR9. To examine this possibility, we generated a panel of anti-DNA antibodies and examined their ability to bind to CpG-rich and CpG-poor substrates. We find that a number of anti-DNA antibodies do show preference for binding to certain DNA fragments, but this does not correlate directly with the presence of CpG dinucleotides. However, an antibody with a binding preference for an immunostimulatory DNA fragment is able to promote a proliferative response to the fragment at lower doses than antibodies that do not display this binding preference, indicating that antibody specificity does contribute to autoreactive B cell responses.
2. Materials and Methods
2.1 B cell proliferation
AM14 RF+ mice were obtained from crosses between MRL AM14 H chain transgenic (Tg) and BALB/c Vκ8 L chain Tg mice [5]. B cells were positively selected from spleen cell suspensions using anti-B220 microbeads (Miltenyi Biotech) and cultured as described previously [8, 18]. Proliferation was measured with a 6 h pulse of 3H-thymidine 24 h post-stimulation.
2.2 Antibodies
Anti-TNP antibody Hy1.2 has been described previously [3]. The IgG2a monoclonal anti-DNA antibodies PA4 and H241 were kindly provided by Dr. Mark Monestier [19] and Dr. David Stollar [16]. 6–120, 8D8, 3A5, 10G10, 11E8, 16F8, F2.2.G5, B5.E12, and E8.F1 were obtained from the fusion of MRL-lpr or MRL-gld spleen cells to the mouse myeloma fusion partner SP2 or NSO-bcl-2 [20]. These mAbs were initially identified as DNA-reactive by ELISA. 6–120 and 8D8 were subsequently found to give a homogeneous nuclear staining pattern in a HEp2 immunofluorescent screen and to stain crithidia kinetoplasts. They were therefore considered reactive to dsDNA. All IgG2a antibodies were purified on protein G sepharose.
2.3 DNA fragments
Mouse DNA was prepared from spleen, and E.coli DNA from DH5α using Qiagen DNeasy® Blood & Tissue Kit, and digested with DdeI to yield fragments ranging from 0.2–2 kb. The dsDNA fragments CGneg, CG50, Sumo, Senp1, and clone 11 and have been previously described [7, 8]. These fragments as well as unselected mouse DNA and E. coli DNA were biotinylated by filling-in 5’ overhangs from restriction digestion with Klenow(exo-) in the presence of biotin-16-2'-deoxy-uridine-5'-triphosphate [7]. Primers and enzymes were removed from all DNAs using the DNA Clean & Concentrator-25™ kit (Zymoresearch). All DNAs contained less than 0.1 EU/ml endotoxin when tested at 5× concentration.
2.4 ELISAs
MAbs were initially screened for their capacity to directly bind biotinylated soluble DNA fragments. 96-well plates were coated with 2 µg/ml goat anti-mouse IgG2a, blocked with 1%BSA/PBS, and anti-DNA antibodies were added at 2 µg/ml for 2 hr at RT. Biotin labeled DNA was then added at 300 ng/ml for 2 hr at RT. For competition ELISA, plates were coated directly with the mAbs and blocked with 1%BSA/PBS. Increasing concentrations of unlabeled competitor DNA fragment were then added to the wells in the presence of a fixed concentration of a biotinylated fragment. ELISAs were developed with Streptavidin-horseradish peroxidase (Southern Biotech) and TMB substrate (Sigma).
2.5 ANA and CrithiDNA
Purified antibodies were assayed on Hep-2 or CrithiDNA® slides (Antibodies, Inc.) at 4 µg/ml and detected with goat anti-mouse Ig(H+L)-FITC (Southern Biotech) at 10 µg/ml. CrithiDNA® slides were also costained with DAPI to confirm localization of DNA.
2.6 DNA selection
50 µl of packed protein G sepharose was incubated with 100 µg PA4 or 8D8 for 5 min in a final volume of 500 µl and washed 3× to remove unbound antibody. 2 µg of plasmid DNA was digested with restriction enzymes. LITMUS-CG50 was digested with EcoRI, BamHI, and DdeI. pCpG-CG50 and pCpG-clone 11 were prepared by digesting pCpG-mcs (Invivogen) with EcoRI and AvrII and mixing with CG50 or clone 11 fragment [8]. DNA was labeled by incubating digest with Klenow(exo-) Fragment in the presence of 33 µM dTTP, dCTP, and dGTP, and 10 µCi of [α32P]-dATP for 30 min at 37°C. Unincorporated nucleotides were removed with a ProbeQuant™ G-50 Micro column (GE Healthcare). Sepharose was then incubated with DNA in 500 µl of buffer C (50 mM Tris-HCl pH 8.0, 0.1 M NaCl, 1 mM dithiothreitol, 10 µg/ml gelatin) [21] for 1 hr at RT with rotation. Sepharose was then centrifuged and the unbound DNA was collected, followed by 500 µl washes with buffer C supplemented with the indicated concentrations of NaCl. Radioactivity in the fractions was counted and salt was removed from selected fractions using the DNA Clean & Concentrator-25™ kit (Zymoresearch). 10,000 cpm were loaded onto a 4.5% non-denaturing polyacrylamide gel and visualized using a phosphoimager.
3. Results
3.1 Antibodies reactive with dsDNA appear to preferentially bind the CpG-rich dsDNA fragment CG50 and E.coli DNA
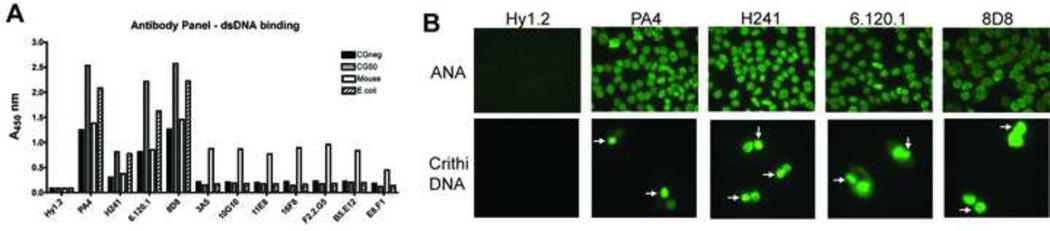
In order to examine antibody preference for sequence, we assembled a panel of 11 IgG2a anti-DNA antibodies from autoimmune-prone MRL-lpr and MRL-gld mice. PA4 and H241 had previously been shown to bind dsDNA. They were compared to 7 additional antibodies, also isolated from autoimmune mice and initially selected for their ability to bind either mouse or E.coli DNA. The anti-TNP antibody Hy1.2 was included as a negative control. Antibodies were first examined for their ability to bind directly to representative biotin-labeled CpG-poor and CpG-rich dsDNA fragments. These included CGneg, a 629 bp fragment with no CpG dinucleotides, CG50, a 607 bp fragment containing 50 optimal CpG motifs (Table 1), and biotin-labeled mouse and E.coli DNA. Seven antibodies showed weak reactivity only to mouse DNA. The other four antibodies (PA4, H241, 6.120.1, and 8D8) bound all 4 DNA preparations and showed preference for binding to the CpG-rich dsDNA fragments CG50 and E.coli DNA (Fig 1a). These 4 antibodies also gave a homogeneous nuclear staining pattern on ANA slides, and positive staining of the kinetoplast on CrithiDNA slides, thereby confirming reactivity with dsDNA (Fig 1b).
Table I.
| CpG-poor | Size (bp) | CpG | Optimal CpG |
|---|---|---|---|
| CGneg | 629 | 0 | 0 |
| Sumo | 619 | 1 | 0 |
| Senp1 | 557 | 4 | 0 |
| Mouse | 100–1000 | Depleted | Depleted |
| CpG-rich | Size (bp) | CpG |
Optimal CpG |
| Clone 11 | 573 | 42 | 2 |
| CG50 | 607 | 52 | 52 |
| E.coli | 200–2000 | Enriched | Enriched |
Fig. 1. Anti-DNA antibodies show binding preference for CpG-rich DNA fragments.
(A) Plates were coated with anti-IgG2a, incubated with IgG2a antibodies derived from hybridomas, and the indicated biotinylated dsDNA was added at 300 ng/ml. Binding of DNA to antibody was detected with SA-HRP. Results are representative of 3 experiments. (B) Hep-2 or CrithiDNA slides were incubated with 2 µg/ml of the indicated IgG2a antibodies. Binding was detected with anti-IgG-FITC. Kinetoplast is indicated by white arrow.
3.2 Anti-DNA antibody PA4 preferentially binds to the CpG-rich dsDNA fragment CG50
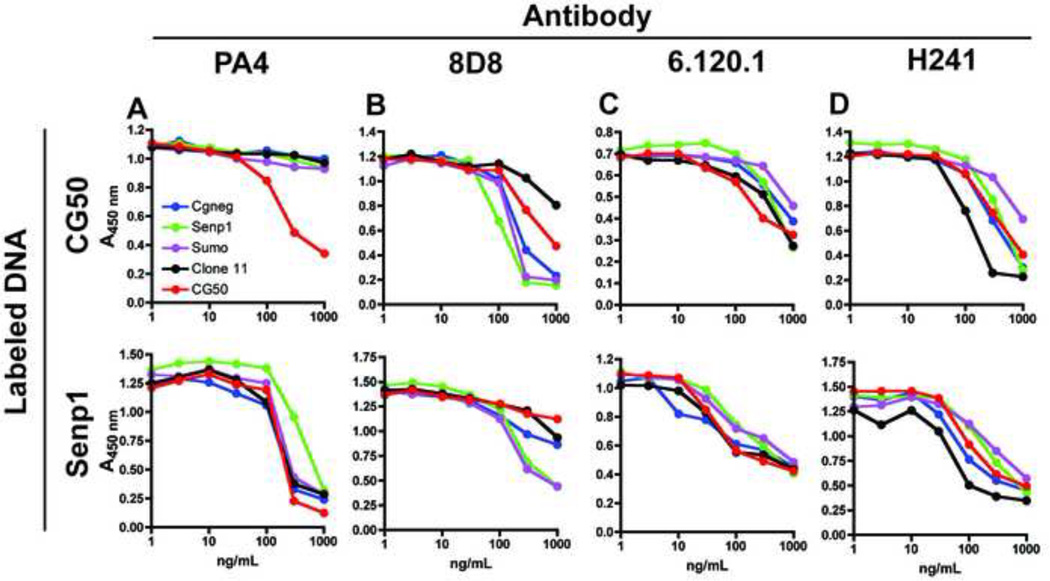
Interpretation of direct binding assays can be confounded by effects of the coupling reaction on DNA structure or by uneven labeling of the DNA ligands. To further evaluate the sequence specificity of the 4 DNA-reactive antibodies, we extended our analysis to competition assays in which it was possible to directly compare the ability of CpG-rich and CpG poor fragments to compete for binding to a labeled fragment. We first examined the binding of an anti-DNA antibody, PA4 [19], isolated from a drug-induced model of lupus, to CpG-rich and CpG-poor dsDNA fragments. Consistent with previous reports, we found that the relative ability of unlableled fragments to compete for binding to a labeled fragment was dependent on the particular fragment used as ligand [7]. Conditions were optimized to measure the capacity of plate-bound PA4 antibody to bind to the biotin-labeled fragments CG50 and Senp1, a 557 bp fragment containing only 4 CpG dinucleotides and no optimal motifs. The ability of PA4 to bind these fragments in the presence of increasing concentrations of additional dsDNA fragments was then evaluated. The competitor fragments included CG-rich (CG50 and clone 11) and CG-poor (CGneg, Sumo, Senp1) DNA sequences. When Senp1 was used as ligand, all the fragments competed comparably. However, if CG50 was used as ligand, unlabeled CG50 was able to compete for binding of biotin-labeled CG50 much more effectively than the other fragments (Fig. 2a), indicating that PA4 preferentially bound to the CG50 fragment. However, this enhanced binding activity did not extend to clone 11, a 573 bp fragment containing 42 CpG-dinucleotides derived from a mouse CpG island, that was enriched for CG dinucleotides.
Fig. 2. Anti-DNA antibodies show distinct binding preferences.
Plates were coated with 1–2 µg/ml antibody and the indicated concentration of unlabeled CGneg or CG50 was added in the presence of 20–30 ng/ml biotinylated CG50 (top) or Senp1 (bottom). Bound DNA was detected with SA-HRP. Results are representative of 3 experiments.
3.3 Other anti-dsDNA mAbs show variable binding to DNA fragments by competition ELISA
A similar format was used to examine the other 3 dsDNA-binding antibodies (Fig 2b, c, d). Again, two different DNA ligands were used, but did not seem to have a major effect on the experimental outcome. Each antibody seemed to have a relatively unique binding preference. Unexpectedly, 8D8 seemed to bind the CG-poor fragments the most avidly. 6.120.1 bound slightly better to CG50, and H241 seemed to bind best to clone 11.
3.4 Anti-DNA antibody PA4 preferentially binds to the CpG-rich dsDNA fragment CG50, but not another CpG-rich dsDNA fragment
To more directly assess the relative binding of PA4 and 8D8 to specific DNA fragments, antibodies were immobilized on protein G sepharose and incubated with a mix of 32P-labeled DNA fragments derived from restriction digestion of plasmid DNA. Sepharose was then washed with low salt buffer (0.1 M NaCl) to remove unbound DNA, and DNA was eluted with increasing concentrations of NaCl (0.3 M to 1 M). DNA content of the fractions was then analyzed on non-denaturing polyacrylamide gels. Specific binding of DNA to PA4/sepharose, but not Hy1.2/sepharose was detected (Fig S1a). Binding of DNA to PA4/sepharose was not saturated at the concentration of DNA used (Fig S1b), and antibody was not eluted from protein G sepharose with high salt washes (Fig S1c).
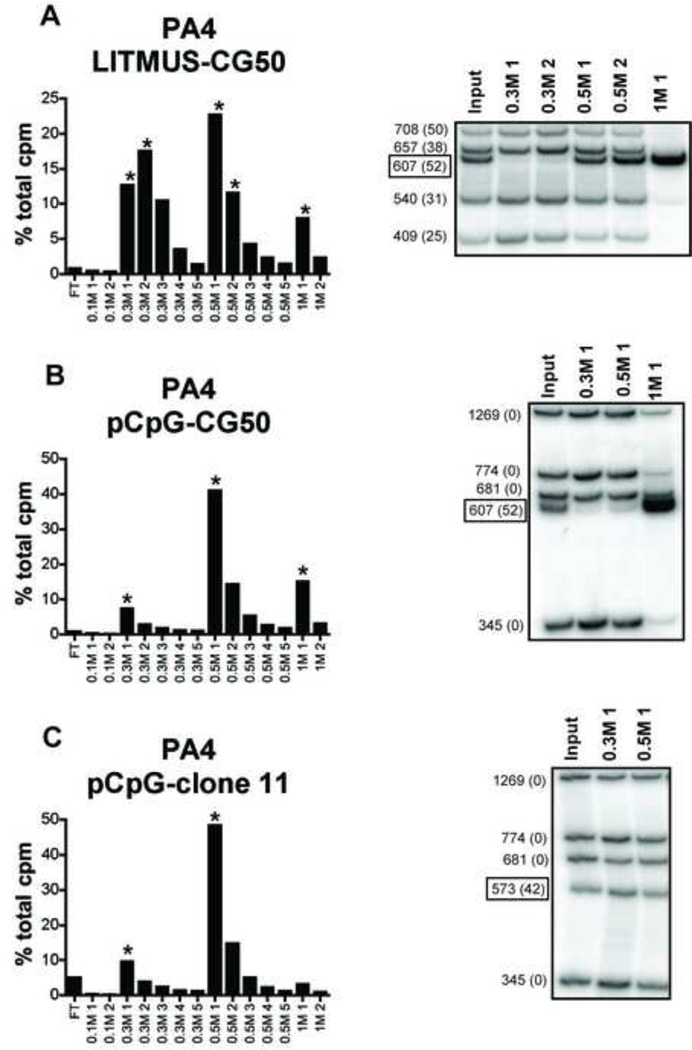
To confirm the apparent preferential binding of PA4 to CG50, we incubated PA4/sepharose with a restriction digest of LITMUS-CG50, made up of 5 electrophoretically distinct fragments, one of which was CG50. The bound DNA was eluted in distinct peaks at 0.3 M, 0.5 M, and 1 M salt. The 1 M eluate contained almost exclusively the CG50 fragment (Fig 3a). By contrast, the 540 bp and 409 bp fragments were enriched in the lower salt fractions. This confirmed that PA4 bound the CG50 fragment with high affinity, consistent with the ELISA data. However, the other DNA fragments in this mixture also contained CpG dinucleotides (number indicated in parenthesis), due to their plasmid origin, and therefore PA4 does not simply have a general preference for DNA enriched in CpG dinucleotides.
Fig. 3. Anti-DNA antibody PA4 preferentially binds CG50 fragment, but not CpG island clone 11 fragment.
The anti-DNA antibody PA4 was immobilized on Protein G sepharose and incubated with 32P-labeled DNA fragments resulting from restriction enzyme digestion of LITMUS-CG50 (A), pCpG-mcs-CG50 (B), or pCpG-mcs-clone 11 (C). Sequential salt washes (shown on the x-axis) were used to elute bound DNA from anti-DNA antibodies. Selected fractions (*) were analyzed on a non-denaturing gel for DNA content. Boxed DNA fragment indicated is inserted (non-plasmid) DNA (CG50 or clone 11). DNA fragment size is indicated at the left and number of CpG dinucleotides is shown in parenthesis.
To further test the ability of PA4 to distinguish CpG-rich DNA fragments from non-CpG-rich fragments, we used the CpG-negative plasmid pCpG-mcs. A mixture of pCpG-mcs fragments mixed with CG50 was added to PA4/sepharose. The CG50 fragment was again selectively bound by PA4 relative to the CpG-negative fragments (Fig 3b). To test the ability of PA4 to selectively bind to an additional CpG-rich DNA fragment, we also used the CpG island derived fragment, clone 11 [8]. In contrast to the binding preference displayed for CG50, PA4 failed to selectively bind to CpG island clone 11 (Fig 3c). Overall these data suggest that PA4 displays a high preference for binding to the CG50 fragment, but this does not directly correlate with the presence of CpG motifs or CpG dinucleotides.
3.5 Anti-DNA antibody 8D8 does not preferentially bind to the CpG-rich dsDNA fragment CG50, but does bind other CpG-poor dsDNA fragments
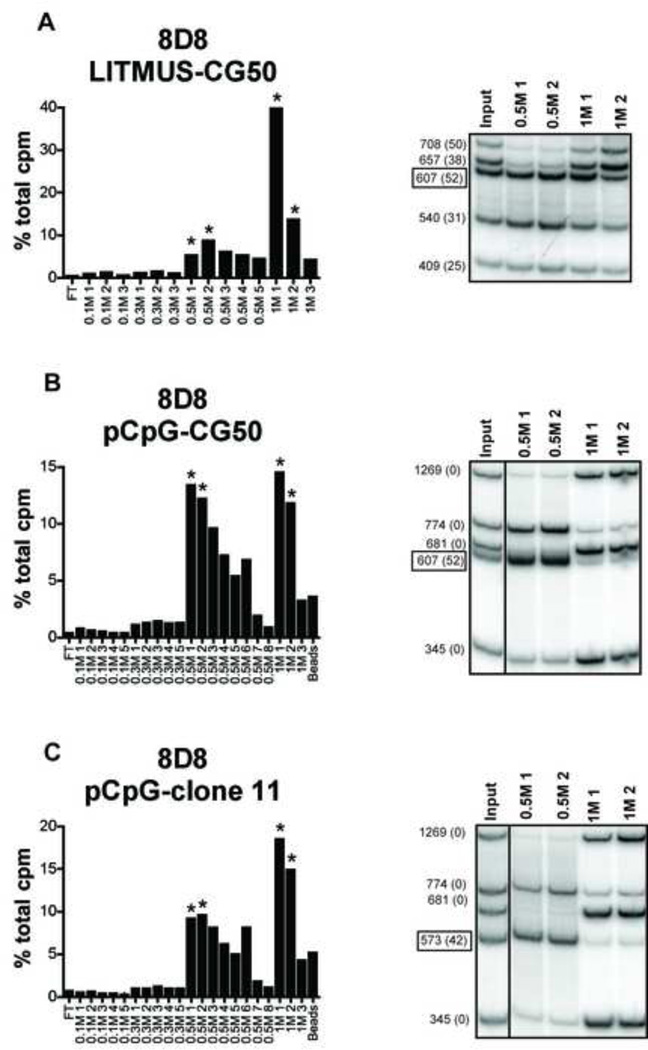
8D8 was also tested for binding to digested LITMUS-CG50 DNA. Higher salt concentrations were needed to elute the DNA fragments from 8D8 than PA4, suggesting that 8D8 had a higher affinity for DNA. DNA fragments were differentially eluted from 8D8 at 0.5 M and 1 M salt, however in this case the CG50 fragment eluted at low salt, while the 708 bp and 657 bp fragments were selectively eluted in high salt (Fig 4a). 8D8 was further evaluated for binding to the mixture of pCpG-mcs fragments with CG50 or CpG island clone 11. DNA was again eluted in 2 peaks at 0.5 M and 1 M. CG50 and a 774 bp plasmid fragment were eluted at 0.5 M, while three other plasmid fragments were eluted at 1 M (Fig 4b). Similar results were obtained for clone 11, where clone 11 and the 774 bp plasmid fragment eluted at 0.5 M and the three other plasmid fragments again selectively eluted at 1 M (Fig 4c). These data confirm that like PA4, 8D8 also selectively binds to certain DNA fragments over others. However, binding did not correlate with the presence of CpG motifs. These data again confirmed the ELISA results and clearly showed that in contrast to PA4, 8D8 preferentially bound CG-poor DNA over clone 11.
Fig. 4. Anti-DNA antibody 8D8 preferentially binds plasmid fragments, but not CG50 of CpG island clone 11 fragments.
The anti-DNA antibody 8D8 was immobilized on Protein G sepharose and incubated with 32P-labeled DNA fragments resulting from restriction enzyme digestion of LITMUS-CG50 (A), pCpG-mcs-CG50 (B), or pCpG-mcs-clone 11 (C). Sequential salt washes (shown on the x-axis) were used to elute bound DNA from anti-DNA antibodies. Selected fractions (*) were analyzed on a non-denaturing gel for DNA content. Boxed DNA fragment indicated is inserted (non-plasmid) DNA (CG50 or clone 11). DNA fragment size is indicated at the left and number of CpG dinucleotides is shown in parenthesis.
3.6 Anti-DNA antibody PA4 induces B cell proliferation to CG50 at 10-fold lower doses than 8D8
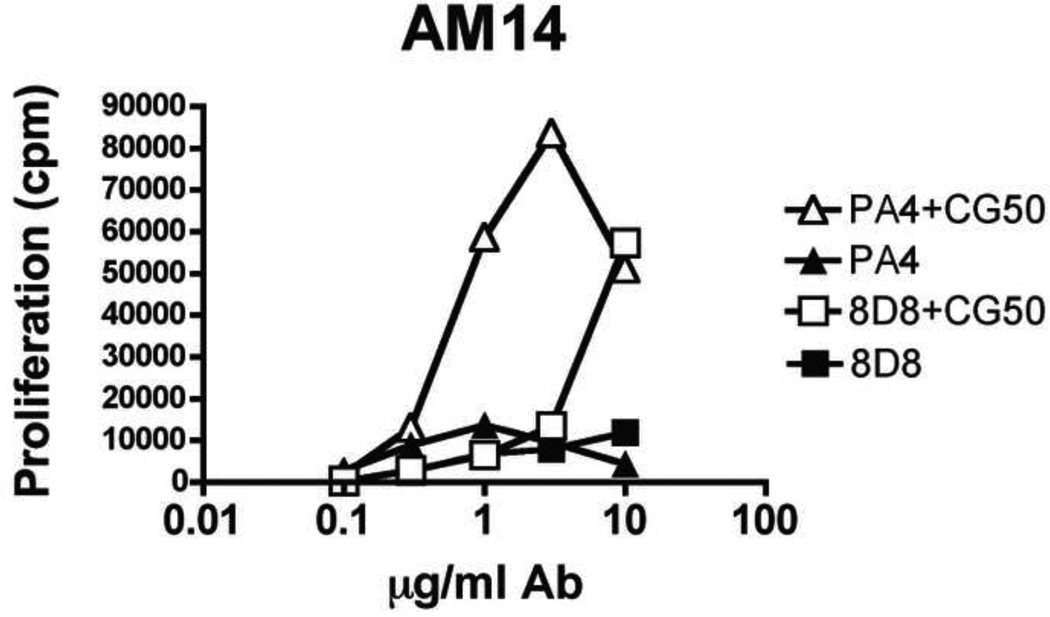
In order to determine if the preference of PA4 for CG50 was functionally relevant to autoreactive B cell responses, we stimulated AM14 B cells with increasing concentrations of either PA4 or 8D8 in complex with a fixed amount of CG50 fragment. Despite 8D8’s apparent higher affinity for DNA, PA4 was able to induce high levels of proliferation at 1 µg/ml, while 8D8 induced equivalent levels of proliferation only when used at 10 µg/ml (Fig 5). Both responses were TLR9-dependent (data not shown). This indicates that antibodies with preference for immunostimulatory DNA are able to induce B cell proliferation at lower doses compared to antibodies that do not display this preference.
Fig. 5. Anti-DNA antibody PA4 induces proliferation to CG50 at lower antibody doses than 8D8.
Purified AM14 B cells were stimulated with the indicated concentration of PA4 or 8D8 in the presence or absence of 300 ng/ml CG50 and proliferation was measured. Results are representative of 2 experiments.
4. Discussion
Autoimmune diseases such as SLE are associated with autoantibodies specific for a number of nucleic acid containing autoantigens, including dsDNA. We previously demonstrated that delivery of CpG-containing, but not CpG-free, dsDNA immune complexes to TLR9 via the BCR activates proliferation of rheumatoid factor producing AM14 B cells [7, 8]. Because unselected total mammalian DNA fails to activate TLR9 [7], presumably due to its low CpG content, it is likely that there must be a mechanism(s) that selectively provides CpG-containing DNA to TLR9 in SLE. One possibility is that DNA reactive autoantibodies preferentially bind CpG containing DNA. A number of previous studies have demonstrated that DNA-reactive autoantibodies can selectively bind to specific base sequences such as poly(dA-dT) or poly(dG-dC) or to DNA conformations such as Z-DNA [16, 22, 23]. Moreover, DNAs purified from the Ig fraction of serum from SLE patients were enriched in CpG dinucleotides supporting the idea that DNA reactive autoantibodies may select for immunostimulatory DNAs [24].
In an effort to test this hypothesis, we examined the binding specificities of a number of randomly chosen anti-DNA antibodies to determine whether they preferentially bind CpG enriched DNA sequences. We found that the anti-DNA antibody PA4 had a strong preference for the experimentally constructed CG50 fragment, but not for the more physiologically relevant CpG-rich clone 11 fragment, indicating that PA4 does not specifically bind CpG dinucleotides. CG50 is highly repetitive, containing 50 repeats of the optimal mouse CpG motif GACGTT. The anti-DNA antibody H241 has been shown to make contacts with 6–8 bases within dsDNA [16], so it is possible that PA4 recognizes the repeated CpG motif in CG50, or another repeated sequence in CG50, as opposed to the simple CpG dinucleotide. Alternatively, PA4 could recognize a secondary structure specific to the CG50 fragment. Similar DNA conformations may account for the preferential binding of 8D8 to specific fragments encoded within the LITMUS and pCpG plasmids. Future experiments using a SELEX approach will be necessary to more precisely define the sequence(s) bound by these antibodies. In contrast to PA4, 8D8 was shown to preferentially bind 3 CpG-negative fragments, encoded by the CpG-deficient plasmid pCpG, more strongly than CG50. Importantly, this difference in binding specificity was shown to influence B cell activation. The combination of PA4 with the CG50 fragment was able to induce proliferation at 10-fold lower antibody concentrations compared to 8D8.
Our data show that anti-DNA antibodies could selectively bind nucleotide sequences in native dsDNAs that are hundreds of base pairs long, the hypothesized in vivo antigen in autoimmune disease. However, we found that not all anti-DNA antibodies are specific for immunostimulatory, CpG-containing dsDNAs. Therefore BCR specificity cannot account for the CpG-specific TLR9-dependent activation of DNA-reactive B cells in the context of systemic autoimmunity. An alternative possibility is that immunostimulatory (CG-rich) DNA becomes preferentially accessible to B cells as a result of certain forms of cell death and/or cell stress. For example, CpG-enriched regions of the mammalian genome such as the CpG islands found in the promoter regions of many genes might be selectively released during cell death. Other sources of CpG-rich DNA could include retrotransposable elements or mitochondrial DNA. In fact, cell injury can result in the rapid release of mitochondrial DNA to the circulation where it has been found to activate neutrophils through a TLR9-dependent mechanism [25, 26].
In the in vivo situation in autoimmune disease, where a complex mix of DNA ligands are likely to be present, a B cell that selectively binds to a sequence that is a strong TLR9 ligand will become activated more easily. However, from a disease perspective, it is important to keep in mind that certain factors, such as type I IFN or reduced expression of the inhibitory receptor FcγRIIB, enhance the response of autoreactive B cells to CpG-poor DNA through mechanisms that are still dependent on TLR9, and could therefore contribute to the activation of autoreactive B cells such as 8D8 [8, 27]. In addition, base modifications associated with oxidation or other forms of DNA damage that can occur in vivo may contribute to the detection of DNA by either the BCR or TLR9 [28]. Exactly how all these parameters contribute to the initial events that trigger the onset of systemic autoimmune disease remains to be determined.
Supplementary Material
(A) 10 µL of packed protein G sepharose was incubated with 20 µg PA4 or Hy1.2 antibody for 5 min. and washed 3× to remove unbound antibody. 100 ng of digested 32P-labeled LITMUS-CG50 plasmid DNA was incubated with sepharose for 15 min. at RT and cpms in flow-through, washes, and associated with sepharose were counted. DNA binds specifically to PA4 sepharose. (B) 25 µL of packed protein G sepharose was incubated with 50 µg PA4 antibody for 5 min. and washed to remove unbound antibody. The indicated amounts of 32P-labeled CG50 fragment was incubated with sepharose for 1 hr. at RT and cpms associated with sepharose were counted. DNA binding to antibody is not saturated at concentrations used. (C) 100 µL packed protein G sepharose was incubated with 200 µg PA4 antibody for 5 min, washed 3× to remove unbound antibody, and separated into 5 tubes. 250 µL of buffer C supplemented with the indicated amount of salt, or 0.1 M glycine pH 2.7, was incubated with sepharose for 5 min. at RT, spun, and supernatants were collected. Salt was removed from samples using a Centricon, and samples were run on an SDS-PAGE gel and stained with coomassie blue. PA4 and Hy1.2 are retained on protein G sepharose at the salt concentrations used to elute DNA.
Research Highlights.
Anti-DNA mAbs derived from autoimmune prone mice preferentially bind specific DNA fragments
Antibody binding preference appears to be independent of CpG content
Antibodies that bind CpG-rich DNA most avidly form immune complexes that effectively activate RF B cells
Acknowledgements
The authors would like to thank Drs. Marko Radic and Marc Monestier for helpful discussions. This study was supported by NIH/NIAMS P01-AR050256 and NIH/NIAID T32 AI07309.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Plotz PH. The autoantibody repertoire: searching for order. Nat Rev Immunol. 2003;3:73–78. doi: 10.1038/nri976. [DOI] [PubMed] [Google Scholar]
- 2.Christensen SR, Shlomchik MJ. Regulation of lupus-related autoantibody production and clinical disease by Toll-like receptors. Semin Immunol. 2007;19:11–23. doi: 10.1016/j.smim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak- Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 4.Boule MW, Broughton C, Mackay F, Akira S, Marshak-Rothstein A, Rifkin IR. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatinimmunoglobulin G complexes. J Exp Med. 2004;199:1631–1640. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shlomchik MJ, Zharhary D, Saunders T, Camper SA, Weigert MG. A rheumatoid factor transgenic mouse model of autoantibody regulation. Int Immunol. 1993;5:1329–1341. doi: 10.1093/intimm/5.10.1329. [DOI] [PubMed] [Google Scholar]
- 6.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–1177. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viglianti GA, Lau CM, Hanley TM, Miko BA, Shlomchik MJ, Marshak-Rothstein A. Activation of autoreactive B cells by CpG dsDNA. Immunity. 2003;19:837–847. doi: 10.1016/s1074-7613(03)00323-6. [DOI] [PubMed] [Google Scholar]
- 8.Uccellini MB, Busconi L, Green NM, Busto P, Christensen SR, Shlomchik MJ, et al. Autoreactive B cells discriminate CpG-rich and CpG-poor DNA and this response is modulated by IFN-alpha. J Immunol. 2008;181:5875–5884. doi: 10.4049/jimmunol.181.9.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 11.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerutti ML, Centeno JM, Goldbaum FA, de Prat-Gay G. Generation of sequencespecific, high affinity anti-DNA antibodies. J Biol Chem. 2001;276:12769–12773. doi: 10.1074/jbc.M100260200. [DOI] [PubMed] [Google Scholar]
- 15.Stevens SY, Glick GD. Evidence for sequence-specific recognition of DNA by antisingle- stranded DNA autoantibodies. Biochemistry. 1999;38:560–568. doi: 10.1021/bi981899o. [DOI] [PubMed] [Google Scholar]
- 16.Stollar BD, Zon G, Pastor RW. A recognition site on synthetic helical oligonucleotides for monoclonal anti-native DNA autoantibody. Proc Natl Acad Sci U S A. 1986;83:4469–4473. doi: 10.1073/pnas.83.12.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radic MZ, Cocca BA, Seal SN. Initiation of systemic autoimmunity and sequence specific anti-DNA autoantibodies. Crit Rev Immunol. 1999;19:117–126. [PubMed] [Google Scholar]
- 18.Avalos AM, Latz E, Mousseau B, Christensen SR, Shlomchik MJ, Lund F, et al. Differential cytokine production and bystander activation of autoreactive B cells in response to CpG-A and CpG-B oligonucleotides. J Immunol. 2009;183:6262–6268. doi: 10.4049/jimmunol.0901941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monestier M, Novick KE, Losman MJ. D-penicillamine- and quinidine-induced antinuclear antibodies in A.SW (H-2s) mice: similarities with autoantibodies in spontaneous and heavy metal-induced autoimmunity. Eur J Immunol. 1994;24:723–730. doi: 10.1002/eji.1830240335. [DOI] [PubMed] [Google Scholar]
- 20.Ray S, Diamond B. Generation of a fusion partner to sample the repertoire of splenic B cells destined for apoptosis. Proc Natl Acad Sci U S A. 1994;91:5548–5551. doi: 10.1073/pnas.91.12.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekker SC, Young KE, von Kessler DP, Beachy PA. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. Embo J. 1991;10:1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stollar BD. Molecular analysis of anti-DNA antibodies. Faseb J. 1994;8:337–342. doi: 10.1096/fasebj.8.3.7511550. [DOI] [PubMed] [Google Scholar]
- 23.Thomas TJ, Meryhew NL, Messner RP. DNA sequence and conformation specificity of lupus autoantibodies. Preferential binding to the left-handed Z-DNA form of synthetic polynucleotides. Arthritis Rheum. 1988;31:367–377. doi: 10.1002/art.1780310308. [DOI] [PubMed] [Google Scholar]
- 24.Sano H, Takai O, Harata N, Yoshinaga K, Kodama-Kamada I, Sasaki T. Binding properties of human anti-DNA antibodies to cloned human DNA fragments. Scand J Immunol. 1989;30:51–63. doi: 10.1111/j.1365-3083.1989.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avalos AM, Uccellini MB, Lenert P, Viglianti GA, Marshak-Rothstein A. FcgammaRIIB regulation of BCR/TLR-dependent autoreactive B-cell responses. Eur J Immunol. 2010;40:2692–2698. doi: 10.1002/eji.200940184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajizadeh S, DeGroot J, TeKoppele JM, Tarkowski A, Collins LV. Extracellular mitochondrial DNA and oxidatively damaged DNA in synovial fluid of patients with rheumatoid arthritis. Arthritis Res Ther. 2003;5:R234–R240. doi: 10.1186/ar787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) 10 µL of packed protein G sepharose was incubated with 20 µg PA4 or Hy1.2 antibody for 5 min. and washed 3× to remove unbound antibody. 100 ng of digested 32P-labeled LITMUS-CG50 plasmid DNA was incubated with sepharose for 15 min. at RT and cpms in flow-through, washes, and associated with sepharose were counted. DNA binds specifically to PA4 sepharose. (B) 25 µL of packed protein G sepharose was incubated with 50 µg PA4 antibody for 5 min. and washed to remove unbound antibody. The indicated amounts of 32P-labeled CG50 fragment was incubated with sepharose for 1 hr. at RT and cpms associated with sepharose were counted. DNA binding to antibody is not saturated at concentrations used. (C) 100 µL packed protein G sepharose was incubated with 200 µg PA4 antibody for 5 min, washed 3× to remove unbound antibody, and separated into 5 tubes. 250 µL of buffer C supplemented with the indicated amount of salt, or 0.1 M glycine pH 2.7, was incubated with sepharose for 5 min. at RT, spun, and supernatants were collected. Salt was removed from samples using a Centricon, and samples were run on an SDS-PAGE gel and stained with coomassie blue. PA4 and Hy1.2 are retained on protein G sepharose at the salt concentrations used to elute DNA.