Abstract
Recombinant adeno-associated viruses (rAAVs) have been tested in humans and other large mammals without adverse events. However, one study of mucopolysaccharidosis VII correction in mice showed repeated integration of rAAV in cells from hepatocellular carcinoma (HCC) in the Dlk1–Dio3 locus, suggesting possible insertional mutagenesis. In contrast, another study found no association of rAAV integration with HCC, raising questions about the generality of associations between liver transformation and integration at Dlk1–Dio3. Here we report that in rAAV-treated ornithine transcarbamylase (Otc)–deficient mice, four examples of integration sites in Dlk1–Dio3 could be detected in specimens from liver nodule/tumors, confirming previous studies of rAAV integration in the Dlk1–Dio3 locus in the setting of another murine model of metabolic disease. In one case, the integrated vector was verified to be present at about one copy per cell, consistent with clonal expansion. Another verified integration site in liver nodule/tumor tissue near the Tax1bp1 gene was also detected at about one copy per cell. The Dlk1–Dio3 region has also been implicated in human HCC and so warrants careful monitoring in ongoing human clinical trials with rAAV vectors.
Zhong and colleagues use deep sequencing to examine the distribution of integrated AAV genomes in nodules/tumors from the livers of ornithine transcarbamylace (Otc) deficient mice. Using this approach, they report four examples of integration sites in the Dlk1-Dio3 locus; this site has previously been implicated in human hepatocellular carcinoma and may warrant careful monitoring in human clinical trials using AAV vectors.
Introduction
Previous studies of hepatocellular carcinoma (HCC) in mice after exposure to recombinant adeno-associated virus (rAAV) vectors have reached different conclusions regarding possible insertional mutagenesis. Donsante et al. (2007), after rAAV transduction of normal mice, found four examples of integration sites in HCC tumors in the delta-like homologue 1–deiodinase type 3 (Dlk1–Dio3) region, which contains a large collection of genes encoding microRNAs and snoRNAs on chromosome 12 (Donsante et al., 2007). Quantification of vector copy number in transformed cells showed that these integration sites reached an abundance of up to 28% of diploid genomes. Because murine liver cells may have higher ploidy than typical diploid cells, and tumor cells may be mixed with untransformed cells, this was proposed to be consistent with insertional activation, in which every transformed cell is derived from a single clone expected to contain the activating integration event. However, a subsequent study of hemophilia gene therapy in mice showed no elevation in HCC rates after rAAV exposure, and genome-wide surveys by pyrosequencing showed no integration sites in the Dlk1–Dio3 locus in tumor samples (Li et al., 2011). Recently, a study in which rAAV was deliberately targeted to the Dlk1–Dio3 locus resulted in HCC (Wang et al., 2012). Exposure to AAV or rAAV has not been associated with transformation in large animal models or humans, leaving the possible role of rAAV in transformation uncertain. rAAV-based vectors are presently in use in human clinical trials to treat cystic fibrosis, α-1 anti-trypsin deficiency, Parkinson's disease, Batten's disease, muscular dystrophy, Leber's congenital amaurosis, and hemophilia B (Flotte et al., 2004; Mandel et al., 2006; Kaplitt et al., 2007; Maguire et al., 2008; Worgall et al., 2008; Nathwani et al., 2011), highlighting the importance of further understanding possible genotoxicity by rAAV.
We therefore studied rAAV integration sites in liver nodules or tumors arising during a preclinical study of liver gene therapy for ornithine transcarbamylase (Otc) deficiency (OTCD) in mice using intraportally delivered rAAV serotype 2, 7, and 8 vectors (Bell et al., 2006). As in many other studies of insertional activation of oncogenes in human and mice (Hacein-Bey-Abina et al., 2003; Kohn et al., 2003; Donsante et al., 2007), we reasoned that recurrent vector integration events in a single locus in tumor tissue would indicate a potential contribution of insertional activation to transformation. The study in OTCD mice attempted to characterize the unexpected observation of liver nodules in long-term surviving vector-treated mice. The article of Bell et al. (2006) described the incidence and histological analysis of nodules and in a subset of tissues, also vector abundance. The strain of mice used in these studies has a high background of spontaneous nodules and HCC, making it difficult to ascribe a vector to tumor formation. The key findings were a statistically significant increase and nodules and HCC of vector-treated animals over untreated animals only when the vector expressed lacZ. There was no increase in nodules/HCC when the vector expressed OTC; in fact, a metabolic correction with gene therapy may have diminished background tumors, consistent with the recent observation that OTCD is associated with an increased risk of HCC (Wilson et al., 2012). Thus, we sought to investigate possible insertional activation by characterizing the distributions of integrated rAAV genomes in nodules/tumors and adjacent normal tissues from the study of Bell et al. (2006), using deep sequencing of junctions between rAAV and flanking host DNA, with particular focus on the Dlk1–Dio3 locus.
Materials and Methods
Isolation of total cellular DNAs from liver tumor and adjacent normal tissues
The adjacent normal liver and tumor/nodule DNA samples were isolated from samples according to Bell et al. (2006). Briefly, the tissue samples were collected from the Otc-deficient male mice at 12 months after intraportal injection with rAAV2, 7, and 8 vectors expressing either the mOTC or the LacZ gene driven by the human thyroid hormone-binding globulin (TBG) promoter. The genetic background of those mice was B6C3F1, which was bred by crossing C57BL/6 with C3H mice. Frozen liver and tumor tissue was homogenized in a lysis buffer using TissueLyser II (Qiagen, Valencia, CA) and total cellular DNA was extracted by the Qiagen DNEasy kit (Qiagen).
DNA sequencing
rAAV inverted terminal repeat (ITR)–host genomic DNA (gDNA) junction libraries were prepared using standard methods, which are described in the Supplementary Methods section (Supplementary Data are available online at www.liebertonline.com/hum). The purified libraries of rAAV ITR–host gDNA junction fragments were sequenced by a Genome Sequencer FLX pyrosequencer (Roche/454 Life Sciences, Branford, CT) at the University of Pennsylvania DNA Sequencing Facility.
Genome-wide analysis of rAAV integration sites
Pyrosequencing reads (stored and manipulated in a MySQL database) were first decoded using DNA barcodes, which separated the sequence reads by mouse and tissue of origin. Reads were then aligned against the linker and rAAV ITR primers using the Crossmatch program and subsequently trimmed. Reads passing the trimming criteria were aligned against vector sequences using BLAT (stepSize=5; tileSize=10). BLAT hits were processed as described by Li et al. (2011) to distinguish concatemers from authentic integration events. Sequences carrying authentic integration site junctions were aligned to the mouse genome (UCSC freeze mm8) using BLAT. To call a read as an authentic rAAV integration site, 15 bases of ITR DNA were required to be present between the primer site and the flanking mouse DNA to exclude reads resulting from mispriming on the mouse genome, the match to the mouse genome was required to start within three bases of the ITR end, and the match to the mouse genome was required to be 70% of sequence size with 98% identity. Integration sites in the mouse genome were analyzed as described in previous publications (Berry et al., 2006; Li et al., 2011). A detailed explanation of genomic heatmaps can be found in Ocwieja et al.'s (2011) supplementary text “Guide to Interpreting Genomic Heat Maps Summarizing Integration Site Distributions.” Cancer-related genes were identified with a collection of cancer-related gene lists from seven different sources. Integration site data sets and oligonucleotides used for analysis are listed in Supplementary Tables S1 and S2. DNA sequences have been deposited at Genebank under the accession numbers JS187093 - JS564611, library accession LIBGSS_038841.
Browsing rAAV integration sites on the mouse genome
The integration sites studied here can be viewed using the UCSC genome browser.
Results and Discussion
We chose to study the five mice from the Bell et al.'s (2006) report with the highest numbers of vector copies in tumors based on polymerase chain reaction (PCR) analyses, because these were more likely to be caused by insertional activation and harbor monoclonal integration sites (Fig. 1). Ligation-mediated PCR and pyrosequencing of rAAV integration sites (Li et al., 2011) yielded a total of 999,442 sequence reads. Dereplication yielded 608 unique integration sites from the tumor/nodule samples and 972 unique integration sites from the adjacent normal liver controls (Fig. 1; Supplementary Methods and Tables S1 and S2).
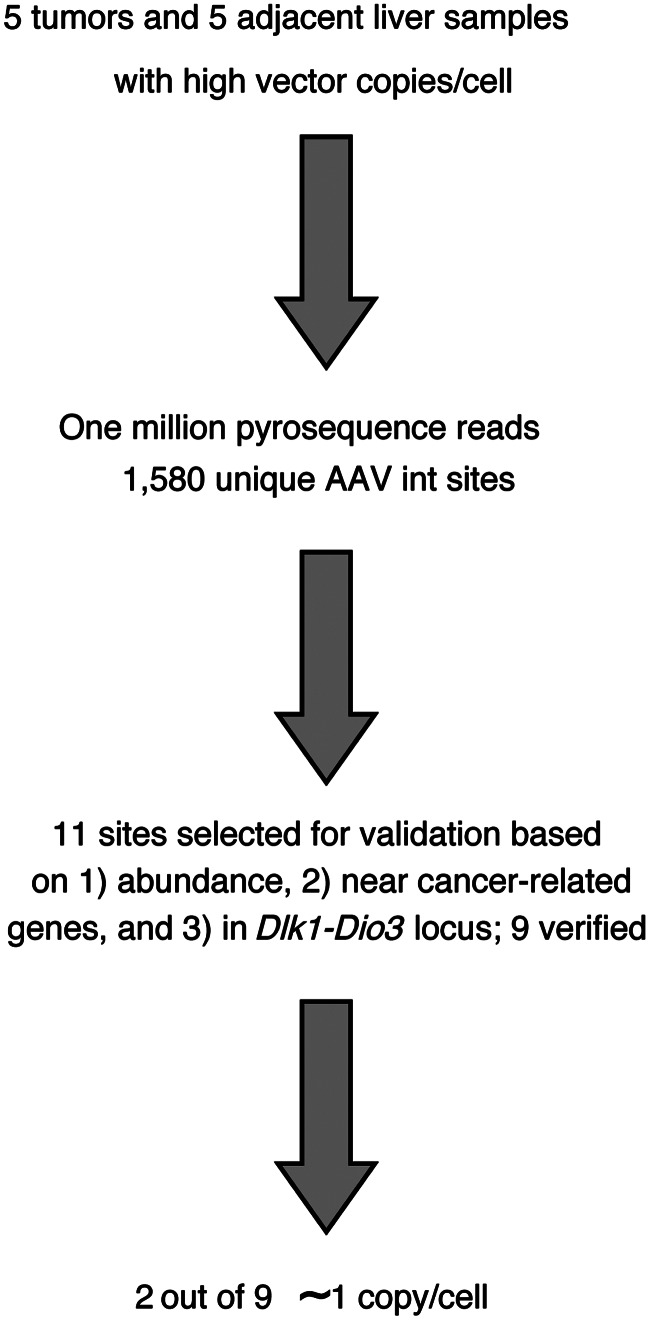
FIG. 1.
Flow chart summarizing the analysis of rAAV vector integration site distributions. Initially, 10 samples were selected from the study of Bell et al. (2006), consisting of paired tumor and normal liver DNA specimens from five mice containing liver nodules or tumors after exposure to rAAV. These were used to generate 10 integration site libraries, one per specimen, that were sequenced using the 454/Roche pyrosequencing method, yielding 1,580 unique integration site positions. Of these, 11 were selected for verification because of possible involvement in transformation based on high relative abundance (n=8), proximity to cancer-related genes (n=3), or location in the Dlk1–Dio3 locus (n=5), which was previously suggested to be associated with rAAV integration in hepatocellular carcinoma. Note that the sum of the numbers above is greater than 11 because some sites were chosen based on more than one criteria (see Table 1). Of the 11 integration sites, 9 could be validated by PCR amplification, cloning in bacterial plasmids, and DNA sequencing. Of these, two out of nine were present at ∼1 copy per cell (Table 1). rAAV, recombinant adeno-associated virus; PCR, polymerase chain reaction.
For each of the five mice studied, integration sites were compared between the liver nodules and flanking normal tissue (Supplementary Table S2). The relative abundance of reads provided a first indication of the abundance of cells hosting vectors at these locations, though this estimate is considerably complicated by biases in the recovery process (discussed further below). The positions of all integration sites on the mouse genome can be viewed along with user-configurable annotation as described in the Materials and Methods section. The distribution of integration sites was then analyzed in detail (Fig. 2). Figure 2 shows that integration sites are distributed along the length of each chromosome, without obvious clustering in hotspots. Global distributions of rAAV integration sites were analyzed relative to genomic features, and were generally as in previous studies. Supplementary Figure S1 presents a statistical analysis for integration site distributions relative to many types of genomic features. Integration sites usually showed a slight enrichment in gene-dense regions and near gene 5′ ends compared with computational random distributions. Features associated with gene 5′ ends such as CpG islands were also enriched, though with a considerable sample-to-sample variation. Analysis of the integration site distributions relative to the AAV serotypes suggested some possible differences, though the power was limited because of the modest number of mice studied (Supplementary Fig. S1).
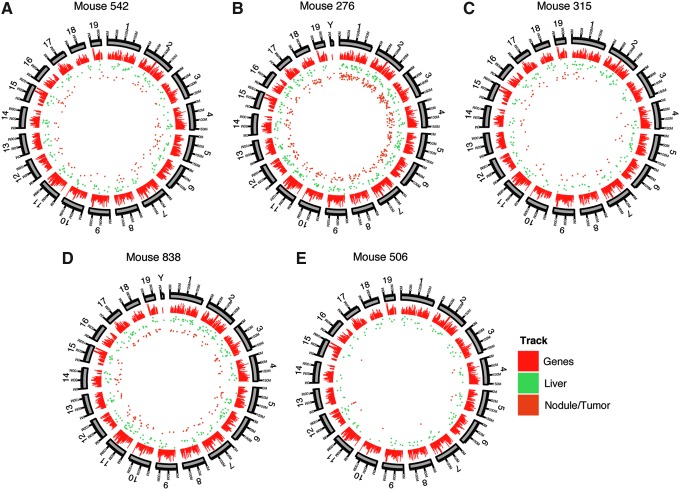
FIG. 2.
Distribution of rAAV integration sites in liver nodule/tumor and flanking normal tissue for the five mice studied. For each mouse, the chromosomes are shown numbered sequentially in circular ideograms (outermost ring). Progressing inward, relative gene density is shown by the red bars, positions of integration sites in normal liver is shown by the green dots, and integration sites in nodule/tumor samples is shown by the brown dots.
We next sought to quantify the absolute abundance of integration sites that were candidates for involvement in insertional activation as an indication of expansion of vector-marked cell clones. Recovery of rAAV–host DNA junctions is complicated by the palindromic structure of the ITR sequences at each end of the rAAV genome, because the DNA secondary structure can interfere with PCR amplification and DNA sequencing (Supplementary Fig. S2). Thus, the population of integration sites studied may be incomplete, and quantifying vector abundance by counting sequence reads provides only an initial estimate.
Integration sites were selected for verification based on three criteria: relative abundance of sequence reads, proximity to cancer-associated genes, and proximity to the Dlk1–Dio3 locus (Table 1; note that some sites were chosen based on more than one criteria). Eleven integration sites that met these criteria were selected from 1,580 unique rAAV integration sites revealed by pyrosequencing, 10 from tumor/nodule (i.e., 276TM, 315TM, and 506TM, respectively), and 1 Dlk1–Dio3 site from flanking normal tissue (i.e., 276LV) (Table 1). These four samples had sufficient DNA for follow-up analysis. We first attempted to reamplify the integration sites using targeted PCR primers, followed by cloning products into bacterial plasmids and sequencing by the Sanger method (Supplementary Methods). Extensive efforts were made for each site, involving amplification attempts with multiple primer pairs. A total of 9 sites in 3 samples (i.e., 276TM, 276LV, and 506TM) were verified out of the 11 attempted (Table 1). A total of five sites in the Dlk1–Dio3 region could be verified, one from normal tissue and four from tumor-nodule tissue, thus showing a trend toward enrichment in the tumor-nodule samples (p=0.076; Fisher's exact test).
Table 1.
Verification of Adeno-Associated Virus Integration Sites
| Samplea | Vector | Tissue | Mouse age at injection (genotype) | Mouse age at necropsy | Chromosome | Strand | Position | Nearest gene (Dlk-Dio locus?) | Pyrosequence reads | Criteria for selection for verificationb | Succeeded in verifying? | Attempted to quantify? | Copies/cell in sample |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 276LV | OTC | Healthy liver | chr12 | − | 110061934 | Rian* (Dlk-Dio) | 25 | DD | Y | Y | 0.003 | ||
| 276TM | OTC | Nodule/tumor | chr12 | + | 110059316 | Mir341, Rian*(Dlk-Dio) | 8728 | DD, A | Y | Y | 0.007 | ||
| 276TM | OTC | Day 157 (spf ash) | Day 530 | chr4 | − | 154474234 | Cdk11b* | 1499 | C, A | Y | Y | 0.0002 | |
| 276TM | OTC | chr6 | + | 52675302 | Tax1bp1* | 815 | C, A | Y | Y | 0.8 | |||
| 276TM | OTC | chr9 | − | 9199504 | Arhgap42* | 90 | A | Y | Y | 0.001 | |||
| 276TM | OTC | chr9 | + | 9199549 | Arhgap42* | 4 | A | Y | Y | 0.002 | |||
| 315TM | LACZ | Nodule/tumor | Day 126 (spf ash) | Day 489 | chr10 | + | 126713917 | Mars∼* | 845 | C, A | N | N | |
| 315TM | LACZ | chr14 | + | 56882944 | Efha1* | 16 | A | N | N | ||||
| 506TM | OTC | Nodule/tumor | Day 93 (spf) | Day 451 | chr12 | − | 110086679 | Rian*(Dlk-Dio) | 2489 | DD, A | Y | Y | 0.0001 |
| 506TM | OTC | chr12 | − | 110046356 | Rtl1 (Dlk-Dio) | 73 | DD | Y | Y | 1.2 | |||
| 506TM | OTC | chr12 | − | 110086616 | Rian* (Dlk-Dio) | 8 | DD | Y | Y | 0.003 |
Integration sites from 542TM and 838TM were not analyzed because of a lack of sufficient genomic DNA.
Criteria for selecting sites to verify: A, high abundance (up to top four in the sample depending on the amount of DNA available); C, near cancer-associated gene; DD, in the Dlk–Dio locus.
Indicates within the gene; ∼, oncogene.
OTC, ornithine transcarbamylase.
Quantitative PCR was then conducted for these three samples, each of which has 2–3 copies of vector genome per cell as detected by a vector-specific primer/probe set, to quantify the number of integrants per cell in tumor and normal tissues at the confirmed loci (Supplementary Table S2 and Supplementary Methods). A total of 48 different amplicons were tested over the 9 sites for efficiency of amplification. Because of the secondary structure, several primers were tested near the edge of the ITR for each site. This allowed two integration sites to be documented as present at ∼1 copy per cell in the tumor samples by quantitative PCR, as is required in the insertional activation model (Supplementary Table S3 and Supplementary Fig. S3), though we note that potential polyploidy in liver cells complicates this picture. It is unknown whether the lower numbers for the remainder of the sites were because of authentic low-level abundance of these sequences or unsolved problems in amplification through the ITR.
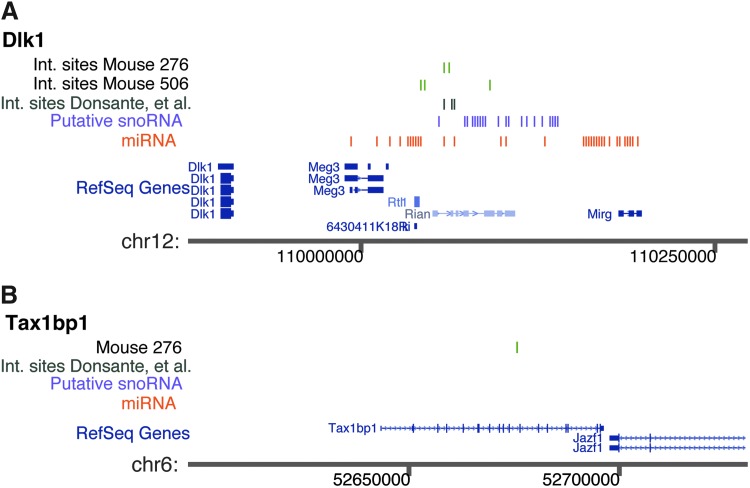
One of the sites that were confirmed at ∼1 copy per cell resided in the Dlk1–Dio3 locus in the Rtl1 gene. A total of five integration sites were detected in the Dlk1–Dio3 locus, four from the nodule/tumor samples and one in the normal liver controls (Fig. 3 and Table 1). The rAAV ITR sequences were partially rearranged in two out of six integration sites at Dlk1–Dio3, as was often the case for rAAV integration sites. The other integration site with a copy number near 1 per cell was in the Tax1-binding protein 1 (Tax1bp1) gene, which hosted two integration sites over the full data set (Fig. 3). The Tax1bp1 gene has been implicated in suppressing inflammation by negatively regulating NFκB.
FIG. 3.
Integration sites of rAAV in tumor/nodule samples at the (A) Dlk1–Dio3 and (B) Tax1bp1 loci. Integration sites are shown as bright green vertical lines, and the mouse of origin is shown to the left. The integration site positions reported by Donsante et al. (2007) are shown in (A) by the blue-green lines. The snoRNA annotation (blue lines) was derived from the annotation of the syntenic region of the human genome, and the miRNA annotation (red lines) was from experimental data in mice. All annotation was adapted from tables at http://genome.ucsc.edu/.
In murine cancer models, insertional activation is often used for discovery of cancer-associated genes—the finding of monoclonal insertion sites in independent tumors near a specific gene suggests direct involvement of that gene in transformation. Here we found an additional example of integration in the Dlk1–Dio3 region in a tumor/nodule sample, reaching ∼1 copy per cell, consistent with the idea that the insertion contributed to transformation. The level of marking at Dlk1–Dio3 in the tumor/nodule sample here was higher than that in the previous study (Donsante et al., 2007), strengthening the case for insertional activation. Three additional sites that are of unknown importance were also found in nodule/tumor tissue in the Dlk1–Dio3 locus, though for these we could not show high-level marking in tumors. We also identified another gene with a monoclonal integration event, Tax1bp1, which encodes a negative regulator of NFκB and inflammatory pathways, and is a potential negative regulator of cell growth (Verstrepen et al., 2011), though for this gene we have not observed multiple independent events suggestive of insertional activation. Complicating the picture, large numbers of additional integration sites were isolated from each of the tumors, and their origin is unclear. One explanation is that these represent admixture of untransformed cells. However, an alternative is that these are from transformed cells of different origins, which would imply that the cancers were probably of polyclonal origin, since it seems unlikely that so many different integrants would reside in the same cell. Unfortunately, tissue samples were not available that would allow for follow-up analysis, for example, by carrying out a detailed study of alterations in gene expression in genes near sites of rAAV integration.
Data linking Dlk1–Dio3 integration events with liver transformation must, nevertheless, be interpreted with some caution. The mouse study, which was the subject of these molecular analyses, was not designed to assess vector-mediated carcinogenesis. In fact, there was a statistically significant increase in tumors of animals treated with rAAV expressing lacZ over a fairly high background of tumors in non-vector-treated animals. However, treatment with rAAV-expressing OTC did not increase tumor formation and, in fact, may have diminished tumors. The paradox is that the tissues that harbored Dlk1–Dios3 integrations came from rAAV.OTC-treated animals (two nodules and one adjacent normal liver). The very nature of these kinds of studies makes it impossible to rule out that the rAAV integration events marked cells that progressed to nodules or tumors for reasons unrelated to integration. As reported in humans (Wilson et al., 2012), the OTC mice may be predisposed to HCC—chronic liver damage caused by accumulation of toxic metabolites and inappropriate regulation of nucleotide pools by elevated pyrimindine production, both of which are characteristic of OTC and associated with liver cancer. This in fact may explain why rAAV.OTC gene therapy could be protective. In addition, if integration events are involved in transformation, they are likely only one of multiple genetic insults.
Since the initial report of Donsante et al. (2007), considerable new data have accumulated on the function of the Dlk1–Dio3 region. The locus contains genes for ∼60 microRNAs and multiple snoRNAs and is imprinted, and correct expression is functionally associated with stem cell pluripotency (Stadtfeld et al., 2010). A recent study implicated a stem-like expression pattern in the syntenic human region at 14q32.2 as diagnostic for poor survival in human HCC (Luk et al., 2011). Furthermore, the Rian gene within the Dlk1–Dio3 locus was identified by transposon insertion screens as a common integration site in murine HCC (Akagi et al., 2004; Dupuy et al., 2009), and a targeted study showed that integration within this locus can be associated with HCC (Wang et al., 2012). Thus, the role of rAAV integration in causing tumors in the OTC model reported here is unclear, but adds to the picture of sensitivity of the Dlk1–Dio3 locus in combination with these other studies. These data, together with previous studies, emphasize that potential rAAV genotoxicity associated with integration in this locus warrants monitoring.
Supplementary Material
Acknowledgments
This research was supported by Public Health Service Grant P01 HL59407-11 from the National Institutes of Health (to G.G.) and in part by a University of Massachusetts Medical School internal grant (to G.G.), as well as a research grant from the Fanconi Anemia Research Fund, Inc. (to L.Z.), R01 AI082020 (to F.D.B.), P01 HD057247 (to J.M.W.), P30 DK047757 (to J.M.W.), and P01 HL059407 (to J.M.W.). T.B. is a Special Fellow of the Leukemia and Lymphoma Society.
Author Disclosure Statement
J.M.W. is a consultant to ReGenX Holdings and is a founder of, holds equity in, and receives a grant from affiliates of ReGenX Holdings; in addition, he is an inventor on patents licensed to various biopharmaceutical companies, including affiliates of ReGenX Holdings. No competing financial interests exist for all other coauthors.
References
- Akagi K. Suzuki T. Stephens R.M. Jenkins N.A., et al. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P. Moscioni A.D. Mccarter R.J. Wu D., et al. Analysis of tumors arising in male B6C3F1 mice with and without AAV vector delivery to liver. Mol. Ther. 2006;14:34–44. doi: 10.1016/j.ymthe.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Berry C. Hannenhalli S. Leipzig J. Bushman F.D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donsante A. Miller D.G. Li Y. Vogler C., et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- Dupuy A.J. Rogers L.M. Kim J. Nannapaneni K., et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotte T.R. Brantly M.L. Spencer L.T. Byrne B.J., et al. Phase I trial of intramuscular injection of a recombinant adeno-associated virus alpha 1-antitrypsin (rAAV2-CB-hAAT) gene vector to AAT-deficient adults. Hum. Gene Ther. 2004;15:93–128. doi: 10.1089/10430340460732490. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. von Kalle C. Schmidt M. Mccormack M.P., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Kaplitt M.G. Feigin A. Tang C. Fitzsimons H.L., et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- Kohn D.B. Sadelain M. Dunbar C. Bodine D., et al. American Society of Gene Therapy (ASGT) ad hoc subcommittee on retroviral-mediated gene transfer to hematopoietic stem cells. Mol. Ther. 2003;8:180–187. doi: 10.1016/s1525-0016(03)00212-0. [DOI] [PubMed] [Google Scholar]
- Li H. Malani N. Hamilton S.R. Schlachterman A., et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood. 2011;117:3311–3319. doi: 10.1182/blood-2010-08-302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk J.M. Burchard J. Zhang C. Liu A.M., et al. DLK1-DIO3 Genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J. Biol. Chem. 2011;286:30706–30713. doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire A.M. Simonelli F. Pierce E.A. Pugh E.N., Jr., et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel R.J. Manfredsson F.P. Foust K.D. Rising A., et al. Recombinant adeno-associated viral vectors as therapeutic agents to treat neurological disorders. Mol. Ther. 2006;13:463–483. doi: 10.1016/j.ymthe.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Nathwani A.C. Tuddenham E.G. Rangarajan S. Rosales C., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocwieja K.E. Brady T.L. Ronen K. Huegel A., et al. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 2011;7:e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M. Apostolou E. Akutsu H. Fukuda A., et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen L. Verhelst K. Carpentier I. Beyaert R. TAX1BP1, a ubiquitin-binding adaptor protein in innate immunity and beyond. Trends Biochem. Sci. 2011;36:347–354. doi: 10.1016/j.tibs.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Wang P.R. Xu M. Toffanin S. Li Y., et al. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11264–11269. doi: 10.1073/pnas.1117032109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.M. Shchelochkov O.A. Gallagher R.C. Batshaw M.L. Hepatocellular carcinoma in a research subject with ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2012;105:263–265. doi: 10.1016/j.ymgme.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worgall S. Sondhi D. Hackett N.R. Kosofsky B., et al. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum. Gene Ther. 2008;19:463–474. doi: 10.1089/hum.2008.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.