Abstract
Myotonic dystrophy (DM) is a dominantly inherited, multisystemic disease caused by expanded CTG (type 1, DM1) or CCTG (type 2, DM2) repeats in untranslated regions of the mutated genes. Pathogenesis results from expression of RNAs from the mutated alleles that are toxic because of the expanded CUG or CCUG repeats. Increased understanding of the repeat-containing RNA (C/CUGexp RNA)-induced toxicity has led to the development of multiple strategies targeting the toxic RNA. Among these approaches, antisense oligonucleotides (ASOs) have demonstrated high potency in reversing the RNA toxicity in both cultured DM1 cells and DM1 animal models, thus offering great promise for the potential treatment of DM1. ASO targeting approaches will also provide avenues for the treatment of other repeat RNA-mediated diseases.
Introduction
RNA-mediated pathogenesis is emerging as an important pathological mechanism in a wide range of diseases, particularly in microsatellite expansion disorders (Wheeler and Thornton, 2007; Cooper et al., 2009). A growing number of autosomal dominant diseases, such as myotonic dystrophy (dystrophia myotonica; DM) (Brook et al., 1992; Harley et al., 1992; Mahadevan et al., 1992), spinocerebellar ataxia (types 8, 10, and 12) (Koob et al., 1999; Matsuura et al., 2000), Huntington's disease-like 2 (HDL2) (Stevanin et al., 2003; Rudnicki et al., 2007), fragile X-associated tremor ataxia syndrome (FXTAS) (Aziz et al., 2003; Hagerman et al., 2004), and frontotemporal dementia/amyotrophic lateral sclerosis (FTD/ALS) (DeJesus-Hernandez et al., 2011; Renton et al., 2011), have been found to be caused by mutations composed of expanded microsatellite repeats. Unlike the majority of inherited diseases caused by gene mutations in the protein-coding regions, however, several of the microsatellite expansion disorders are caused by expansion within noncoding regions, including introns, 5′ untranslated regions (UTRs), and 3′ UTRs (Ranum and Day, 2002; Cooper et al., 2009). How mutations that do not affect the protein-coding region of genes cause an autosomal dominant disease has been a conundrum. However, a large number of studies have revealed a critical role for the toxic gain-of-function of repeat-containing RNA, independent of the encoded protein, as a predominant pathogenic factor in several of these microsatellite expansion diseases.
The first disease demonstrated to be caused by RNA-mediated toxicity was DM (Wheeler and Thornton, 2007). DM is a dominantly inherited, multisystemic disorder caused by expanded CTG repeats in the 3′ UTR of the DMPK gene (type 1, DM1) or expanded CCTG repeats in the first intron of the CNBP gene (type 2, DM2) (Brook et al., 1992; Mahadevan et al., 1992; Liquori et al., 2001). DM is the most common adult-onset muscular dystrophy and the second most common cause of muscular dystrophy after Duchenne muscular dystrophy (DMD), affecting approximately 1 of 8000 people worldwide (Mahadevan et al., 1992; Day and Ranum, 2005). DM is characterized by progressive skeletal muscle weakness, myotonia, insulin resistance, cardiac arrhythmia, smooth muscle dysfunction, and neurological abnormalities (Harper, 2001).
The two types of DM are caused by different microsatellite expansions within different genes but share common (as well as different) clinical features (Harper, 2001; Udd and Krahe, 2012). DM1 is generally the more severe form. Unaffected individuals contain CTG repeat lengths of <38 units in the DMPK gene, whereas individuals with DM1 contain repeat lengths of 50 to >2000 units. The length of repeats directly correlates with the age of onset and the severity of disease (Harper, 2001).
To date, there is no effective treatment for DM. However, increased understanding of the repeat RNA-induced toxicity in DM pathogenesis has led to the rapid development of therapeutic strategies aimed at neutralizing or degrading the toxic RNA. These therapeutic approaches include antisense oligonucleotides (ASOs), small interfering RNAs (siRNAs), small molecules, ribozymes, and engineered small nuclear RNAs (snRNAs) (Furling et al., 2003; Mulders et al., 2009; Pushechnikov et al., 2009; Warf et al., 2009; Wheeler et al., 2009; François et al., 2011; Childs-Disney et al., 2012). Among these approaches, ASOs have demonstrated particularly promising results in removing CUGexp RNA, reversing downstream toxic consequences and suppressing the expansion of CTG repeats (Mulders et al., 2009; Wheeler et al., 2009; Nakamori et al., 2011). Here we review advances in DM pathogenesis and therapeutic approaches with a major focus on the application of ASOs in DM1.
DM Pathogenesis
Several hypotheses were proposed to explain the complex clinical features of DM1, including the following: (1) myotonic dystrophy protein kinase (dystrophia myotonica-protein kinase; DMPK) haploinsufficiency (Fu et al., 1993; Hofmann-Radvanyi and Junien, 1993), (2) haploinsufficiency of neighboring genes (Klesert et al., 1997; Thornton et al., 1997), and (3) mutant DMPK RNA toxicity (Wang et al., 1995; Caskey et al., 1996). Early studies found reduced levels of DMPK mRNA and protein in DM1 patient samples, leading to the hypothesis that DMPK haploinsufficiency accounted for DM1 (Fu et al., 1993; Hofmann-Radvanyi and Junien, 1993; Novelli et al., 1993). In addition, because the CTG repeat expansion overlaps with the promoter region of the downstream SIX5 gene and increased CTG repeat length correlated with reduced levels of Six5, SIX5 haploinsufficiency was also proposed to cause DM1 (Klesert et al., 1997; Thornton et al., 1997). However, genetic inactivation of DMPK or SIX5 in mice failed to produce prominent features of DM1 (Jansen et al., 1996; Klesert et al., 2000; Sarkar et al., 2000), suggesting that altered expression of DMPK or SIX5 does not contribute to the predominant aspects of the disease. Accumulating evidence has established a role for RNA-induced toxicity in DM1 pathogenesis. First, the similarity between the disease features of DM1 (CTGexp) and DM2 (CCTGexp) is consistent with a predominant role for an RNA gain-of-function in DM pathogenesis. DM1 and DM2 repeat expansion mutations occur at two distinct loci with no apparent functional links, suggesting that genes harboring these mutations are unlikely to be directly responsible for the disease. Second, transgenic mice (called HSALR) expressing 250 repeats within the 3′ UTR of an unrelated gene (human skeletal α-actin, HSA) recapitulate critical features of DM1 in skeletal muscle, including myotonia and myopathy (Mankodi et al., 2000). Mice expressing the same transgenic RNA but with only five CUG repeats exhibited no myopathy. Third, induced expression of CUGexp RNA in skeletal muscle and heart reproduces muscle weakness and heart conduction abnormalities (Mahadevan et al., 2006; Wang et al., 2007; Orengo et al., 2008), establishing a prominent role for RNA-induced toxicity in DM1 pathogenesis. Further studies demonstrated that the expanded repeats (CUGexp or CCUGexp) cause the transcribed RNA to aggregate into nuclear foci readily detectable by in situ hybridization (Davis et al., 1997). Results from several laboratories have shown that CUGexp or CCUGexp RNA induces cellular toxicity by disrupting the function of RNA-binding proteins, activating downstream signaling pathways or other mechanisms, thereby playing a trans-dominant role in DM pathogenesis (Ranum and Day, 2004; Udd and Krahe, 2012).
CUGexp RNA-induced toxicity
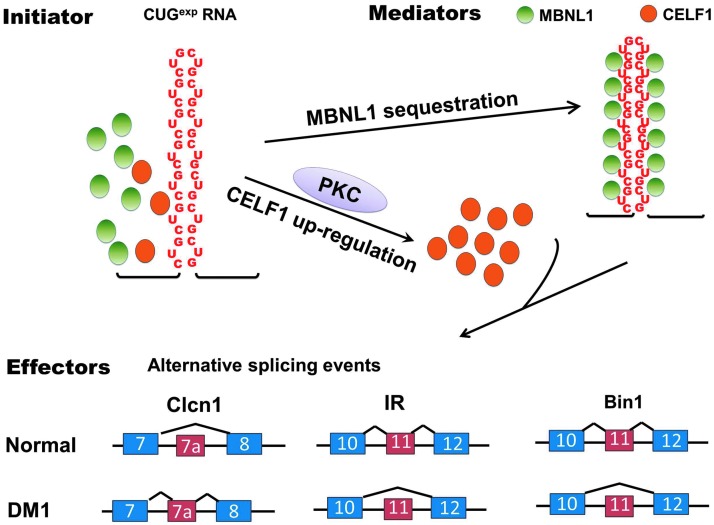
The RNA gain-of-function in DM has been characterized predominantly for CUGexp RNA in DM1. At least two mechanisms account for the repeat RNA-induced trans-dominant toxic effects. First, the CUGexp RNA forms a double-stranded hairpin structure that directly binds to the muscleblind-like (MBNL) family of proteins with high affinity, resulting in MBNL sequestration and loss-of-function (Miller et al., 2000; Mankodi et al., 2001). Second, CUGexp RNA activates protein kinase C (PKC) and upregulates CELF1 (CUGBP1 and ETR-3-like factor) protein levels, resulting in its gain-of-function (Kuyumcu-Martinez et al., 2007; Wang et al., 2007). MBNL and CELF proteins are antagonistic splicing factors that regulate alternative splicing events during development (Ho et al., 2004; Lin et al., 2006; Kalsotra et al., 2008). Disruption of their functions by CUGexp RNA leads to widespread splicing dysregulation, in which embryonic splicing patterns are expressed; several of these misregulated splicing events contribute to disease features (Fig. 1). CELF and MBNL proteins also regulate RNA processing in addition to splicing that is likely to be relevant to DM pathogenesis (Dasgupta and Ladd, 2012; Wang et al., 2012).
FIG. 1.
RNA-initiated toxicity in myotonic dystrophy type 1 (DM1) pathogenesis. The expanded CUG repeats in the myotonic dystrophy protein kinase (dystrophia myotonica-protein kinase; DMPK) 3′ untranslated region (UTR) form a hairpin structure that binds to the splicing regulator muscleblind-like 1 (MBNL1) with high affinity, resulting in MBNL1 sequestration and loss-of-function. The CUG expansion (CUGexp) RNA also upregulates the levels of a second splicing factor, CUGBP Elav-like family member 1 (CELF1), by activation of protein kinase C (PKC). Disrupted function of MBNL1 and CELF1 leads to misregulation of a wide range of alternative splicing events including Clcn1, IR, and BIN1, directly contributing to clinical features of DM1.
Loss-of-function of MBNL
Humans and mice express three MBNL paralogues: MBNL1, -2, and -3. MBNL1 was first identified on the basis of its affinity for CUGexp RNA and colocalization with CUGexp RNA foci (Miller et al., 2000; Mankodi et al., 2001). Expression of CUGexp RNA results in sequestration and loss of MBNL1 function (Lin et al., 2006). Genetic ablation of MBNL1 in mice results in extensive splicing defects in multiple tissues and reproduces critical features of DM1, including myotonia and cataracts (Kanadia et al., 2003; Lin et al., 2006; Du et al., 2010; Suenaga et al., 2012; Wang et al., 2012). The majority of splicing defects observed in patients with DM1 were also detected in MBNL1 knockout mice (Kanadia et al., 2003). Moreover, overexpressing MBNL1 in skeletal muscles of HSALR mice expressing CUGexp RNA alleviates myotonia and reverses aberrant splicing (Kanadia et al., 2006), establishing a critical role for MBNL1 loss-of-function in DM1.
Individuals affected by DM1 have variable levels of hypersomnia and cognitive and behavioral abnormalities (Harper, 2001; Weber et al., 2010). MBNL1 knockout mice recapitulated important pathological features and aberrant splicing events of DM in skeletal muscle but exhibited only minor effects in brain (Kanadia et al., 2003). Studies show that knockout of MBNL2, the more abundant MBNL paralogue in the nervous system, leads to extensive molecular and phenotypic defects in the mouse brain (Charizanis et al., 2012). Notably, MBNL2 knockout mice develop hypersomnia associated with an increased propensity for rapid eye movement (REM) sleep, reminiscent of the DM1-related symptom. In addition, these mice exhibit decreased synaptic NMDA receptor activity, defective long-term potentiation, and impaired learning and memory (Charizanis et al., 2012). Whether the widespread splicing changes directly contribute to brain abnormalities in mice and patients with DM1 remains to be determined. It is likely that CUGexp RNA-induced combinatorial loss-of-function of both MBNL1 and MBNL2 in different tissues extensively disrupts alternative splicing events implicated in DM1 pathogenesis.
Gain-of-function of CELF1
In addition to sequestration of MBNL1, CUGexp RNA also activates protein kinase C (PKC), leading to hyperphosphorylation of CELF1 protein, resulting in increased CELF1 stability and steady state levels (Kuyumcu-Martinez et al., 2007; Wang et al., 2007). Increased CELF1 protein levels were observed in DM1 skeletal muscle, heart, and myoblast cultures (Roberts et al., 1997; Timchenko et al., 2001; Dansithong et al., 2005; Jones et al., 2011). Elevated expression of CELF1 in skeletal muscle of transgenic mice produced muscle weakness and prominent central nuclei, a histological hallmark in DM1 (Ward et al., 2010). Increased expression of CELF1 in mouse hearts resulted in left ventricular systolic dysfunction and dilation, as well as PR interval prolongation (Koshelev et al., 2010), reproducing cardiac features of DM1. Induced expression of CELF1 in heart and muscle reproduced altered splicing events observed in DM1. Furthermore, CELF1 and MBNL1 play an antagonistic role in coregulating a subset of splicing, suggesting that the combined effects of MBNL1 loss-of-function and CELF1 gain-of-function contribute to DM pathogenesis (Ho et al., 2004; Dansithong et al., 2005).
Misregulation of alternative splicing
Misregulated splicing is a molecular hallmark of DM1 (Philips et al., 1998; Savkur et al., 2001; Charlet et al., 2002; Mankodi et al., 2003), with multiple aberrant splicing events caused by MBNL1 loss-of-function. For example, MBNL1 regulates the alternative splicing of a muscle-specific, voltage-dependent chloride channel, chloride channel-1 (CLCN1) (Kanadia et al., 2003). In patients with DM, HSALR mice, and MBNL1 knockout mice, aberrant inclusion of Clcn1 exon 7a introduces a premature termination codon (Charlet et al., 2002; Mankodi et al., 2002; Kanadia et al., 2003), resulting in diminished expression of Clc1 protein and myotonia (Lueck et al., 2007). MBNL1 also promotes the inclusion of exon 11 of the insulin receptor (IR) (Ho et al., 2004; Dansithong et al., 2005). Inclusion of exon 11 produces an IR isoform (IR-B) with high signaling activity, whereas exclusion of exon 11 generates an isoform (IR-A) with increased affinity for insulin but low signaling activity. In patients with DM1, MBNL1 sequestration results in aberrant skipping of exon 11 in IR that is directly associated with insulin resistance (Savkur et al., 2001). In addition, studies have shown that MBNL1 regulates alternative splicing of BIN1, a gene involved in T-tubule biosynthesis (Fugier et al., 2011). Missplicing of BIN1 in patients with DM1 leads to expression of an inactive form of BIN1 that is unable to bind phosphatidylinositol 5-phosphate and to tubulate membrane, resulting in abnormal T tubule structures. Correction of abnormal BIN1 splicing restores the sarcomere membrane structure and improves muscle contraction strength, implicating a role for BIN1 missplicing in muscle weakness (Fugier et al., 2011). However, MBNL1 knockout mice do not display overt muscle weakness or wasting, suggesting that additional mechanisms may be involved in the progressive muscle weakness in DM. One study shows that splicing of exon 29 in the CaV1.1 calcium channel, regulated by both MBNL1 and CELF1, is misregulated and may be associated with muscle weakness in DM (Tang et al., 2012). Additional abnormal splicing events including cardiac troponin T (cTNT), sarcoplasmic/endoplasmic reticulum calcium ATPase-1 (SERCA1), Lim binding domain-3 (LDB3), and myomesin (MYOMI1) have been observed in patients with DM and in mouse models (Philips et al., 1998; Lin et al., 2006; Hino et al., 2007; Koebis et al., 2011; Ohsawa et al., 2011). However, the contribution of these misspliced events in DM pathogenesis is unclear.
Other mechanisms
In addition to MBNL1 and CELF1, RNA-binding proteins including heterogeneous nuclear ribonucleoprotein (hnRNP) H and Staufen have also been implicated in DM pathogenesis (Kim et al., 2005; Paul et al., 2006; Mahadevan, 2012; Ravel-Chapuis et al., 2012). Furthermore, antisense transcripts generated from the expanded repeats have been observed in DM1. For example, an antisense transcript adjacent to the SIX5 regulatory region is produced and may be involved in local chromatin remodeling (Cho et al., 2005). More recently, it has been shown that CUGexp RNA can also induce repeat-associated non-ATG-initiated (RAN) translation that may be involved in DM1 pathogenesis (Zu et al., 2011). There are likely to be multiple mechanisms for DM pathogenesis; however, expression of CUGexp RNA is likely to be a predominant factor, making the RNA the major target of current therapeutic approaches.
Therapeutic Strategies in DM1
Other than supportive care, no effective treatment is available for DM1. Several strategies have been developed to reduce the effects of DM1 pathogenesis, based on addressing the effects on CELF or MBNL. For example, overexpression of MBNL1 in HSALR mice reversed the myotonia and missplicing events (Kanadia et al., 2006; Chamberlain and Ranum, 2012). Inhibition of PKC activity in a heart-specific DM1 model alleviated the CUGexp RNA-induced cardiac conduction defects and partially reversed CELF1- regulated missplicing events (Wang et al., 2009). In an approach targeting misregulated splicing of a specific gene product, restoration of normal Clc1 splicing in HSALR mice directly reversed the myotonia (Wheeler et al., 2007). Although these approaches exhibited clear efficacy, they target specific consequences of the toxic RNA, leaving other pathogenic aspects intact. The most effective approach to eliminate the RNA-induced toxicity is to remove the toxic RNA. In support of this idea, reducing the expression of a tetracycline-inducible transgene encoding the human DMPK 3′ UTR with five CTG repeats efficiently reversed the myotonia and cardiac defects caused by overexpression of the transgene (Mahadevan et al., 2006), clearly demonstrating that removal of the pathogenic RNA can be beneficial to DM1.
To directly neutralize or remove the toxic RNA, various approaches including RNA interference (RNAi), ribozyme, small molecules, peptides, and ASOs have been developed (Langlois et al., 2003, 2005; Mulders et al., 2009; Warf et al., 2009; Wheeler et al., 2009, 2012; Garcia-Lopez et al., 2011; Childs-Disney et al., 2012; Lee et al., 2012; Parkesh et al., 2012). Among these approaches, ASOs have demonstrated particularly promising results in reversing the RNA toxicity, including restoration of normal activities of the RNA-binding proteins affected by CUGexp RNA, correction of missplicing events, and alleviation of myotonia. Here we focus on advances applying ASOs to target the toxic RNA in both DM1 patient cells and animal models.
A brief overview of ASO technology
ASOs target the RNA through base pairing to complementary nucleotide sequences. Natural nucleotides are not suitable for therapeutic application because of their sensitivity to cellular nucleases. Various chemical modifications have significantly improved the stability and binding affinity of ASOs such as phosphorothioate (PS), locked nucleic acid (LNA), 2′-O-methoxyethyl (MOE), 2′-O-methyl (OMe), peptide nucleic acid (PNA), and phosphorodiamidate morpholino (PMO) (Kole et al., 2012). Of note, among these modifications, only the PS backbone modification is compatible with endogenous RNase H activity that specifically cleaves the RNA moiety in a DNA–RNA duplex (Lima et al., 2004).
ASOs function via two general mechanisms: steric blocking or targeting enzymatic cleavage by RNase H. Steric-blocking ASOs are typically uniformly modified (MOE, OMe, LNA, or PMO) ASOs that bind to the RNA and prevent the binding of factors (proteins or RNAs) without inducing RNA degradation (Bennett and Swayze, 2010). For example, ASOs specifically designed to block splice sites at intron–exon junctions or cis-acting splicing regulatory elements have been used to redirect the splicing of dystrophin in DMD and SMN2 in spinal muscular atrophy (SMA) and are currently being used in clinical trials (van Deutekom et al., 2007; Kinali et al., 2009; Hua et al., 2010, 2011; Muntoni and Wood, 2011). In contrast, ASOs competent for RNase H-mediated degradation are typically designed as “gapmers” containing 6–10 central nucleotides with RNase H-competent PS modifications flanked by 3 or 4 nucleotides at the 5′ and 3′ ends that contain modifications that further stabilize the ASO but are not RNase H competent (Bennett and Swayze, 2010). On binding to the target RNA, the “gap” in the gapmer–RNA duplex allows RNase H-mediated cleavage, resulting in specific degradation of RNA. Two gapmers are currently in clinical trials: (1) mipomersen, a gapmer targeting the apolipoprotein B100 mRNA, is used to reduce the level of low-density lipoprotein cholesterol in patients with familial hypercholesterolemia (Raal et al., 2010); and (2) OGX-11, a gapmer targeting the clusterin mRNA, is used to induce cancer cell apoptosis in patients with metastatic prostate cancer (Chi et al., 2010).
Application of ASOs to target toxic CUGexp RNA in DM1
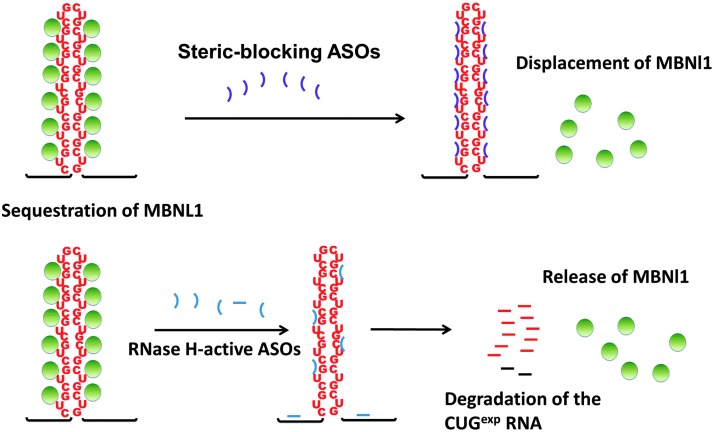
ASOs delivered to cells go to the nucleus (Leonetti et al., 1991), making them attractive reagents in targeting nuclear transcripts such as CUGexp RNA. Both steric-blocking ASOs and gapmers have been tested in DM1 patient cells and mouse models to target the CUGexp RNA with high specificity and efficacy, demonstrating great potential for future DM1 treatment (Fig. 2).
FIG. 2.
Application of antisense oligonucleotides (ASOs) to eliminate RNA toxicity in myotonic dystrophy type 1 (DM1). Steric blocking ASOs bind to CUGexp RNA with high affinity and disrupt the RNA–MBNL1 (muscleblind-like 1) interactions, thereby displacing MBNL1 from the toxic RNA and reversing some features of DM1. In contrast, RNase H-active ASOs bind to the CUGexp RNA and directly induce the degradation of the toxic RNA, resulting in release of MBNL1, restoration of its normal function, and reversal of disease features.
In 2009, two separate studies showed that steric-blocking ASOs can disrupt the CUGexp RNA–MBNL1 interactions and reverse RNA-induced toxicity in mouse models and DM1 myoblasts. Using a 25-nucleotide CAG morpholino (CAG25) that binds to CUGexp RNA with high affinity, Wheeler and colleagues (2009) demonstrated that CAG25 effectively prevented the formation of the RNA–MBNL1 complex and displaced MBNL1 from the preformed RNA–protein complex in vitro (Wheeler et al., 2009). When applied in vivo to the skeletal muscle of HSALR transgenic mice, a single intramuscular injection of CAG25 dissolved the RNA ribonuclear foci, resulting in the release of MBNL1, reversal of myotonia, and restoration of normal splicing transitions for at least 14 weeks (Wheeler et al., 2009).
In an independent study, Mulders and colleagues screened 2′OMe-phosphorothioate ASOs that annealed along the DMPK mRNA including the CUG repeat region and found that PS58, a (CAG)7 oligonucleotide complementary to the CUG repeats, could effectively induce degradation of CUGexp RNA in DM1 myoblasts and mouse models (Mulders et al., 2009). Although wild-type DMPK mRNA contains CUG repeats, both CAG25 and PS58 appeared to specifically target the mutant CUGexp RNA, possibly due to the high binding capacities of the expanded repeats. Intriguingly, ASOs used in both studies significantly reduced levels of CUGexp RNA (50% for CAG25 PMO, 80% for PS58) even though the ASOs were not designed to induce enzymatic cleavage of the target RNA (Mulders et al., 2009; Wheeler et al., 2009). The breakdown of the RNA–MBNL1 complex by ASOs facilitated the export of RNA to the cytoplasm (Wheeler et al., 2009), suggesting the RNA is made available for degradation machinery in the nucleus or cytoplasm. However, the released CUGexp RNA from the ribonuclear foci also raise the possibility that CUGexp RNA, free of MBNL1 binding, can induce cellular toxicity through MBNL1-independent mechanisms (Cooper, 2009; Mahadevan, 2012).
The most attractive approach to eliminate RNA toxicity is to directly degrade the toxic RNA. RNase H activity removes the RNA primer for DNA synthesis on Okazaki fragments during DNA replication. The nuclear localization and essentially ubiquitous expression of RNase H (Suzuki et al., 2010) make RNase H-active ASOs an ideal tool with which to target nuclear-retained CUGexp RNA in the cell types affected in DM1. Two studies have demonstrated the high efficacy and potency of using RNase H-active ASOs in targeting the CUGexp RNA in both DM1 cell culture and mouse models. Using gapmers that contain CAG sequences complementary to the CUG repeats, Lee and colleagues showed that LNA- or MOE-flanked gapmers can effectively reduce the levels of toxic RNA and disrupt RNA foci in a cell culture model of DM1 (Lee et al., 2012). These effects were highly specific to expanded CUG transcripts, as repeats of normal length were not significantly affected, suggesting selective gapmer targeting of the mutant allele. When administered to the skeletal muscle of a DM1 mouse model by direct injection, gapmers targeting the CUG repeats also showed efficacy in removing the CUGexp RNA. However, injected ASOs induced histological abnormalities in skeletal muscle (Lee et al., 2012). It is possible that the toxicity is due to the combination of the ASO and an inflammatory response induced by intramuscular injection. If this is the case, systemic administration is expected to produce less toxicity.
Combined administration of gapmers (RNase H-active) and the CAG25 morpholino (RNase H-inactive) was used to test whether disruption of the RNA foci with CAG25 would increase availability of CUGexp RNA to degradation by the CAG gapmer. In both DM1 cell culture and skeletal muscle of DM1 mouse models, combined administration produced synergistic effects, suggesting that combined application of ASOs through different pathways may help to maximize therapeutic efficacy. These studies showed limited effects, however, because of competition between gapmer and morpholino ASOs both targeting the CUG repeats. An improved strategy would use CAG25 to disrupt RNA–protein interactions and gapmer ASOs to target another region of the DMPK mRNA.
In the experiments described previously, ASOs were delivered locally to a single muscle by intramuscular injection. Whether systemic application of ASOs is useful in removing the toxic CUGexp RNA was unclear, because of the poor intracellular uptake of ASOs in some tissues, particularly those most affected in DM1: skeletal muscle, heart, and brain (Muntoni and Wood, 2011). Notably, a robust reduction of toxic RNA in skeletal muscles was observed after subcutaneous injection of gapmers that target the transgenic HSA mRNA containing 250 CUG repeats in HSALR mice (Wheeler et al., 2012). The reduced level of CUGexp RNA was accompanied by loss of nuclear RNA foci, release of MBNL1, correction of myotonia, and restoration of misregulated splicing, with no apparent off-target effects (Wheeler et al., 2012). The most astonishing effect was that the silencing of the CUGexp RNA was sustained for 1 year after the treatment was terminated. The persistent effects appear to be specific to nuclear-retained RNAs, as ASOs targeting Pten mRNA that is rapidly transported to the cytoplasm were ineffective (Wheeler et al., 2012). RNase H-active ASOs thus provide clear advantages to effectively target the nuclear-trapped CUGexp RNA for potential therapeutic applications.
Perspective and Conclusions
ASOs are a promising approach to eliminate RNA toxicity; however, challenges remain for large-scale therapeutic application in DM1. First, DM1 is a multisystemic disease and therefore requires efficient delivery of ASOs to multiple tissues. Although systemic delivery effectively removed CUGexp RNA, reversed the splicing events, and alleviated myotonia in skeletal muscle, efficient gapmer delivery to heart and brain is more challenging (Bennett and Swayze, 2010; Muntoni and Wood, 2011). Second, with the high potency of gapmers comes the potential for off-target effects (Bennett and Swayze, 2010). Although no apparent off-target effects were observed in animal experiments with the ASOs targeting human ACAT1 in the HSALR transcript (Wheeler et al., 2012), a therapeutic ASO will target human DMPK. Because the entire DMPK mRNA is the target, there are likely to be multiple ASOs that will induce efficient mRNA degradation that can then be screened for minimal off-target effects. In addition to targeting the “wrong” mRNA, ASOs may also have aptameric-like properties by interacting with certain proteins (Marques and Williams, 2005). The long-term pharmacokinetics and potential for toxicity of gapmers need to be fully explored before their large-scale clinical application. Third, there are concerns regarding the potential for reduced expression of DMPK protein. The nuclear localization of both RNase H and the toxic CUGexp RNA leads to preferential degradation of RNA from the expanded allele. However, there is likely to be some level of degradation of mRNA from the normal allele. Given that RNA from the expanded allele is sequestered, the mutation produces haploinsufficiency of DMPK. Degradation of RNA from the mutant allele will not add to this deficiency and will be beneficial by removing the toxic RNA. However, degradation of mRNA from the normal allele will further decrease DMPK protein expression. In DMPK knockout mice, there is little effect of hemizygosity but complete loss of DMPK function produced cardiac abnormalities in older animals (Jansen et al., 1996). Therefore, the effects on the wild-type allele will need to be carefully monitored.
In parallel with the rapid development of ASOs, other strategies including screening of small chemicals and peptides that specifically bind to CUGexp RNA have also made promising progress (Warf et al., 2009; Garcia-Lopez et al., 2011; Childs-Disney et al., 2012). The strategy for most small-molecule approaches is to alter RNA–protein interactions potentially making CUGexp RNA more soluble. Combinatorial approaches using ASOs with small molecules could lead to effective CUGexp RNA degradation with reduced doses of both reagents. Chemicals such as the peptide ABP1 and rationally designed small molecules that bind to CUGexp RNA with high affinity would be good candidates for further pharmaceutical and therapeutic development (Garcia-Lopez et al., 2011; Childs-Disney et al., 2012). In addition, linking ASOs with peptides or nanoparticles may significantly improve drug delivery to DM-affected tissues such as heart (Yin et al., 2011; Betts et al., 2012).
The understanding of pathogenic mechanisms of DM has progressed rapidly in the 20 years since the identification of the DM1 disease gene in 1992. Although many details of the pathogenic mechanism remain to be delineated, the identification of the crucial role of the toxic RNA has led to the rapid development of therapeutic strategies in DM1. The efficacy, potency, and high selectivity of ASOs in targeting the pathogenic RNA offer the promise for potential treatment in DM1. Successful development of RNA-targeting therapeutic strategy in DM1 will also offer new opportunities for treatment of other RNA-dominant diseases caused by microsatellite expansion.
Acknowledgments
Z.G. is supported by a fellowship from the Myotonic Dystrophy Foundation. T.A.C. is supported by National Institutes of Health grants R01AR45653, R01HL045565, and R01AR060733 and by the Muscular Dystrophy Association.
Author Disclosure Statement
The authors have declared that no conflict of interest exists.
References
- Aziz M. Stathopulu E. Callias M., et al. Clinical features of boys with fragile X premutations and intermediate alleles. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;121B:119–127. doi: 10.1002/ajmg.b.20030. [DOI] [PubMed] [Google Scholar]
- Bennett C.F. Swayze E.E. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Betts C.A. Hammond S.M. Yin H.F. Wood M.J. Optimizing tissue-specific antisense oligonucleotide-peptide conjugates. Methods Mol. Biol. 2012;867:415–435. doi: 10.1007/978-1-61779-767-5_27. [DOI] [PubMed] [Google Scholar]
- Brook J.D. McCurrach M.E. Harley H.G., et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- Caskey C.T. Swanson M.S. Timchenko L.T. Myotonic dystrophy: Discussion of molecular mechanism. Cold Spring Harbor Symp. Quant. Biol. 1996;61:607–614. [PubMed] [Google Scholar]
- Chamberlain C.M. Ranum L.P. Mouse model of muscleblind-like 1 overexpression: Skeletal muscle effects and therapeutic promise. Hum. Mol. Genet. 2012;21:4645–4654. doi: 10.1093/hmg/dds306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K. Lee K.Y. Batra R., et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet B.N. Savkur R.S. Singh G., et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- Chi K.N. Hotte S.J. Yu E.Y., et al. Randomized phase II study of docetaxel and prednisone with or without OGX-011 in patients with metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2010;28:4247–4254. doi: 10.1200/JCO.2009.26.8771. [DOI] [PubMed] [Google Scholar]
- Childs-Disney J.L. Hoskins J. Rzuczek S.G., et al. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem. Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D.H. Thienes C.P. Mahoney S.E., et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Cooper T.A. Molecular biology: Neutralizing toxic RNA. Science. 2009;325:272–273. doi: 10.1126/science.1177452. [DOI] [PubMed] [Google Scholar]
- Cooper T.A. Wan L. Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansithong W. Paul S. Comai L. Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- Dasgupta T. Ladd A.N. The importance of CELF control: Molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins [Wiley interdisciplinary reviews] RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.M. Mccurrach M.E. Taneja K.L., et al. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J.W. Ranum L.P. Genetics and molecular pathogenesis of the myotonic dystrophies. Curr. Neurol. Neurosci. Rep. 2005;5:55–59. doi: 10.1007/s11910-005-0024-1. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M. Mackenzie I.R. Boeve B.F., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H. Cline M.S. Osborne R.J., et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François V. Klein A.F. Beley C., et al. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat. Struct. Mol. Biol. 2011;18:85–87. doi: 10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- Fu Y.H. Friedman D.L. Richards S., et al. Decreased expression of myotonin-protein kinase messenger RNA and protein in adult form of myotonic dystrophy. Science. 1993;260:235–238. doi: 10.1126/science.8469976. [DOI] [PubMed] [Google Scholar]
- Fugier C. Klein A.F. Hammer C., et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- Furling D. Doucet G. Langlois M.A., et al. Viral vector producing antisense RNA restores myotonic dystrophy myoblast functions. Gene Ther. 2003;10:795–802. doi: 10.1038/sj.gt.3301955. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez A. Llamusi B. Orzaez M., et al. In vivo discovery of a peptide that prevents CUG–RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman R.J. Leavitt B.R. Farzin F., et al. Fragile-X-associated tremor/ataxia syndrome (FXTAS) in females with the FMR1 premutation. Am. J. Hum. Genet. 2004;74:1051–1056. doi: 10.1086/420700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley H.G. Brook J.D. Rundle S.A., et al. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- Harper P.S. Myotonic Dystrophy. W.B. Saunders; London: 2001. [Google Scholar]
- Hino S. Kondo S. Sekiya H., et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- Ho T.H. Charlet B.N. Poulos M.G., et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Radvanyi H. Junien C. Myotonic dystrophy: Over-expression or/and under-expression? A critical review on a controversial point. Neuromuscul. Disord. 1993;3:497–501. doi: 10.1016/0960-8966(93)90104-r. [DOI] [PubMed] [Google Scholar]
- Hua Y. Sahashi K. Hung G., et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y. Sahashi K. Rigo F., et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G. Groenen P.J. Bachner D., et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 1996;13:316–324. doi: 10.1038/ng0796-316. [DOI] [PubMed] [Google Scholar]
- Jones K. Jin B. Iakova P., et al. RNA foci, CUGBP1, and ZNF9 are the primary targets of the mutant CUG and CCUG repeats expanded in myotonic dystrophies type 1 and type 2. Am. J. Pathol. 2011;179:2475–2489. doi: 10.1016/j.ajpath.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A. Xiao X. Ward A.J., et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia R.N. Johnstone K.A. Mankodi A., et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia R.N. Shin J. Yuan Y., et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H. Langlois M.A. Lee K.B., et al. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinali M. Arechavala-Gomeza V. Feng L., et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: A single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesert T.R. Otten A.D. Bird T.D. Tapscott S.J. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat. Genet. 1997;16:402–406. doi: 10.1038/ng0897-402. [DOI] [PubMed] [Google Scholar]
- Klesert T.R. Cho D.H. Clark J.I., et al. Mice deficient in Six5 develop cataracts: Implications for myotonic dystrophy. Nat. Genet. 2000;25:105–109. doi: 10.1038/75490. [DOI] [PubMed] [Google Scholar]
- Koebis M. Ohsawa N. Kino Y., et al. Alternative splicing of myomesin 1 gene is aberrantly regulated in myotonic dystrophy type 1. Genes Cells. 2011;16:961–972. doi: 10.1111/j.1365-2443.2011.01542.x. [DOI] [PubMed] [Google Scholar]
- Kole R. Krainer A.R. Altman S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob M.D. Moseley M.L. Schut L.J., et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat. Genet. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- Koshelev M. Sarma S. Price R.E., et al. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez N.M. Wang G.S. Cooper T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois M.A. Lee N.S. Rossi J.J. Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol. Ther. 2003;7:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Langlois M.A. Boniface C. Wang G., et al. Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J. Biol. Chem. 2005;280:16949–16954. doi: 10.1074/jbc.M501591200. [DOI] [PubMed] [Google Scholar]
- Lee J.E. Bennett C.F. Cooper T.A. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti J.P. Mechti N. Degols G., et al. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima W.F. Nichols J.G. Wu H., et al. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- Lin X. Miller J.W. Mankodi A., et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Liquori C.L. Ricker K. Moseley M.L., et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Lueck J.D. Lungu C. Mankodi A., et al. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am. J. Physiol. Cell Physiol. 2007;292:C1291–C1297. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- Mahadevan M. Tsilfidis C. Sabourin L., et al. Myotonic dystrophy mutation: An unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Mahadevan M.S. Myotonic dystrophy: Is a narrow focus obscuring the rest of the field? Curr. Opin Neurol. 2012;25:609–613. doi: 10.1097/WCO.0b013e328357b0d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan M.S. Yadava R.S. Yu Q., et al. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A. Logigian E. Callahan L., et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi A. Urbinati C.R. Yuan Q.P., et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Mankodi A. Takahashi M.P. Jiang H., et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Mankodi A. Teng-Umnuay P. Krym M., et al. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann. Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- Marques J.T. Williams B.R. Activation of the mammalian immune system by siRNAs. Nat. Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- Matsuura T. Yamagata T. Burgess D.L., et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- Miller J.W. Urbinati C.R. Teng-Umnuay P., et al. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders S.A. van den Broek W.J. Wheeler T.M., et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F. Wood M.J. Targeting RNA to treat neuromuscular disease. Nat. Rev. Drug Discov. 2011;10:621–637. doi: 10.1038/nrd3459. [DOI] [PubMed] [Google Scholar]
- Nakamori M. Gourdon G. Thornton C.A. Stabilization of expanded (CTG)*(CAG) repeats by antisense oligonucleotides. Mol. Ther. 2011;19:2222–2227. doi: 10.1038/mt.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli G. Gennarelli M. Zelano G., et al. Failure in detecting mRNA transcripts from the mutated allele in myotonic dystrophy muscle. Biochem. Mol. Biol. Int. 1993;29:291–297. [PubMed] [Google Scholar]
- Ohsawa N. Koebis M. Suo S., et al. Alternative splicing of PDLIM3/ALP, for α-actinin-associated LIM protein 3, is aberrant in persons with myotonic dystrophy. Biochem. Biophys. Res. Commun. 2011;409:64–69. doi: 10.1016/j.bbrc.2011.04.106. [DOI] [PubMed] [Google Scholar]
- Orengo J.P. Chambon P. Metzger D., et al. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkesh R. Childs-Disney J.L. Nakamori M., et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J. Am. Chem. Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S. Dansithong W. Kim D., et al. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips A.V. Timchenko L.T. Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Pushechnikov A. Lee M.M. Childs-Disney J.L., et al. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: Application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raal F.J. Santos R.D. Blom D.J., et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- Ranum L.P. Day J.W. Dominantly inherited, non-coding microsatellite expansion disorders. Curr. Opin. Genet. Dev. 2002;12:266–271. doi: 10.1016/s0959-437x(02)00297-6. [DOI] [PubMed] [Google Scholar]
- Ranum L.P. Day J.W. Myotonic dystrophy: RNA pathogenesis comes into focus. Am. J. Hum. Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A. Belanger G. Yadava R.S., et al. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012;196:699–712. doi: 10.1083/jcb.201108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A.E. Majounie E. Waite A., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. Timchenko N.A. Miller J.W., et al. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki D.D. Holmes S.E. Lin M.W., et al. Huntington's disease-like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- Sarkar P.S. Appukuttan B. Han J., et al. Heterozygous loss of Six5 in mice is sufficient to cause ocular cataracts. Nat. Genet. 2000;25:110–114. doi: 10.1038/75500. [DOI] [PubMed] [Google Scholar]
- Savkur R.S. Philips A.V. Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Stevanin G. Fujigasaki H. Lebre A.S., et al. Huntington's disease-like phenotype due to trinucleotide repeat expansions in the TBP and JPH3 genes. Brain. 2003;126:1599–1603. doi: 10.1093/brain/awg155. [DOI] [PubMed] [Google Scholar]
- Suenaga K. Lee K.Y. Nakamori M., et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PLoS One. 2012;7:e33218. doi: 10.1371/journal.pone.0033218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. Holmes J.B. Cerritelli S.M., et al. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.Z. Yarotskyy V. Wei L., et al. Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of CaV1.1 calcium channel. Hum. Mol. Genet. 2012;21:1312–1324. doi: 10.1093/hmg/ddr568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C.A. Wymer J.P. Simmons Z., et al. Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat. Genet. 1997;16:407–409. doi: 10.1038/ng0897-407. [DOI] [PubMed] [Google Scholar]
- Timchenko N.A. Cai Z.J. Welm A.L., et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Udd B. Krahe R. The myotonic dystrophies: Molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012;11:891–905. doi: 10.1016/S1474-4422(12)70204-1. [DOI] [PubMed] [Google Scholar]
- van Deutekom J.C. Janson A.A. Ginjaar I.B., et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Wang E.T. Cody N.A. Jog S., et al. Transcriptome-wide regulation of Pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.S. Kearney D.L. de Biasi M., et al. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.S. Kuyumcu-Martinez M.N. Sarma S., et al. PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J. Clin. Invest. 2009;119:3797–3806. doi: 10.1172/JCI37976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Pegoraro E. Menegazzo E., et al. Myotonic dystrophy: Evidence for a possible dominant-negative RNA mutation. Hum. Mol. Genet. 1995;4:599–606. doi: 10.1093/hmg/4.4.599. [DOI] [PubMed] [Google Scholar]
- Ward A.J. Rimer M. Killian J.M., et al. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M.B. Nakamori M. Matthys C.M., et al. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber Y.G. Roebling R. Kassubek J., et al. Comparative analysis of brain structure, metabolism, and cognition in myotonic dystrophy 1 and 2. Neurology. 2010;74:1108–1117. doi: 10.1212/WNL.0b013e3181d8c35f. [DOI] [PubMed] [Google Scholar]
- Wheeler T.M. Thornton C.A. Myotonic dystrophy: RNA-mediated muscle disease. Curr. Opin. Neurol. 2007;20:572–6. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- Wheeler T.M. Lueck J.D. Swanson M.S., et al. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T.M. Sobczak K. Lueck J.D., et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T.M. Leger A.J. Pandey S.K., et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. Saleh A.F. Betts C., et al. Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice. Mol. Ther. 2011;19:1295–1303. doi: 10.1038/mt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T. Gibbens B. Doty N.S., et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]