Abstract
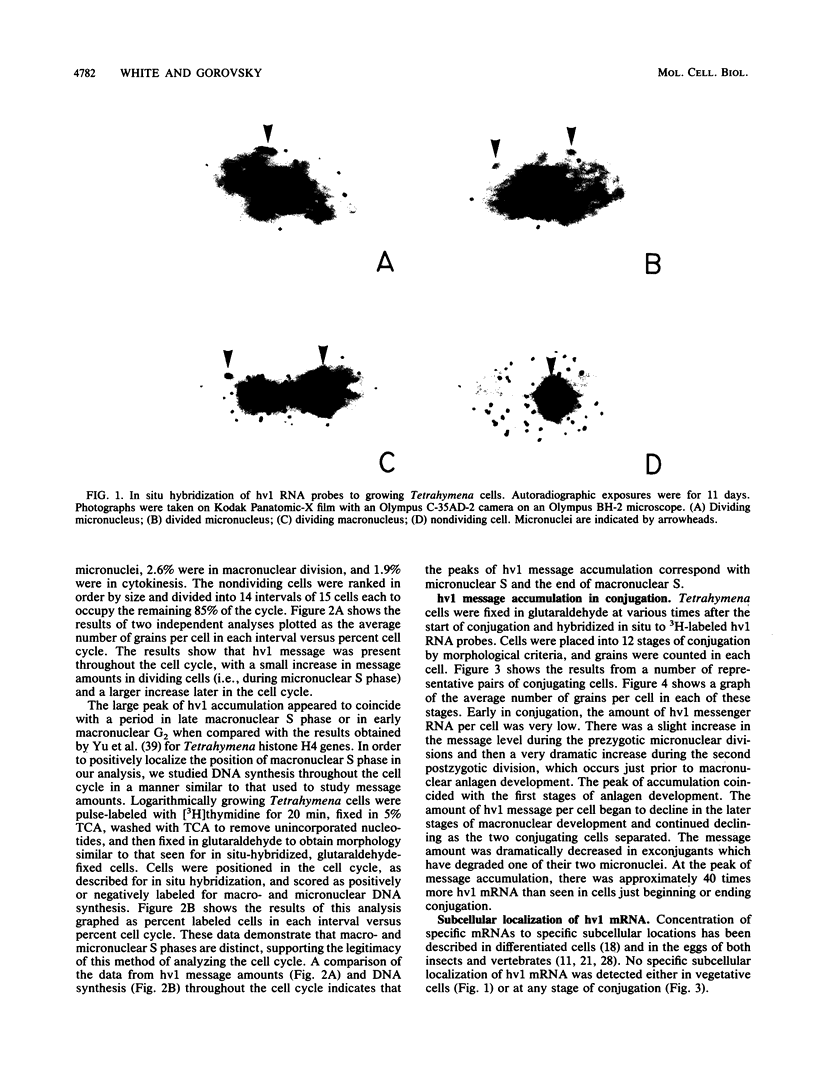
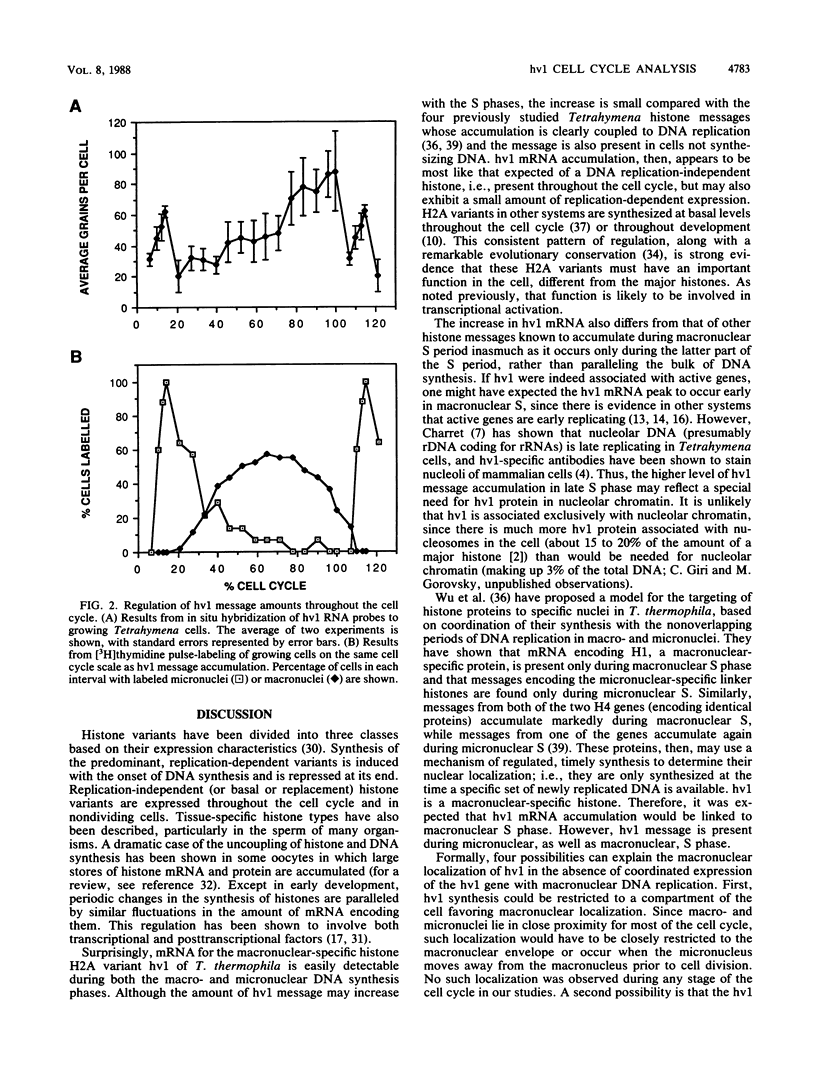
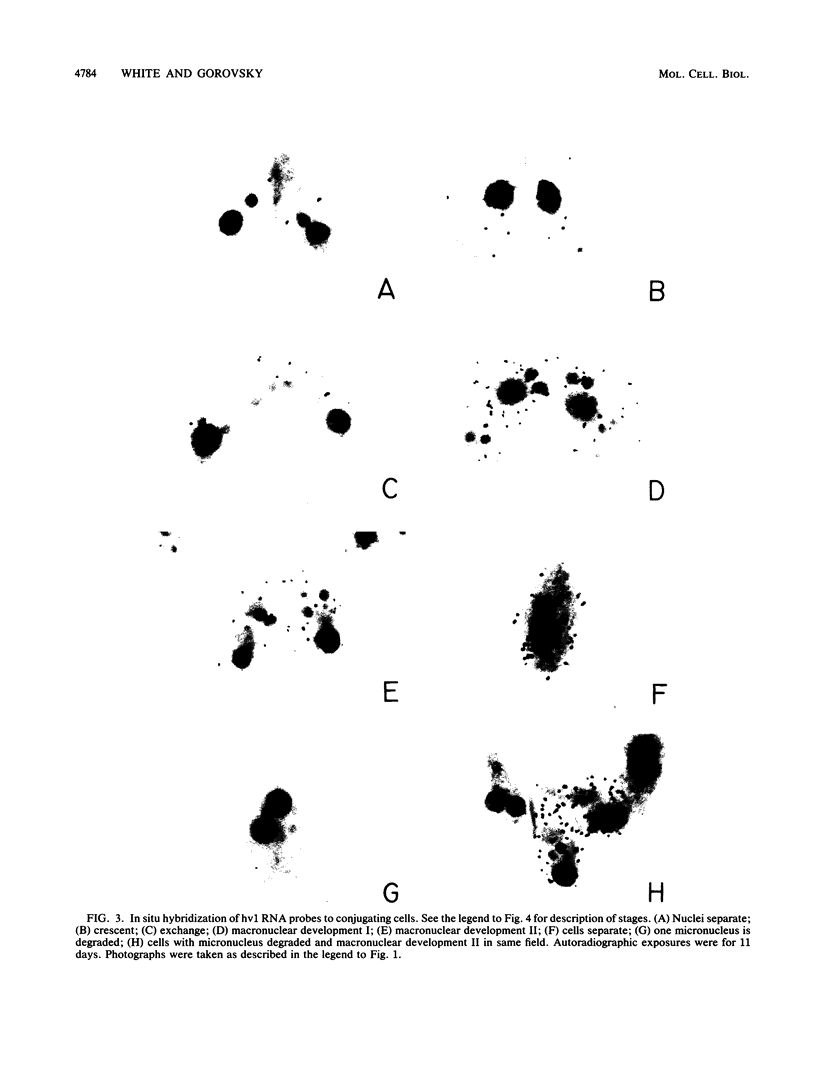
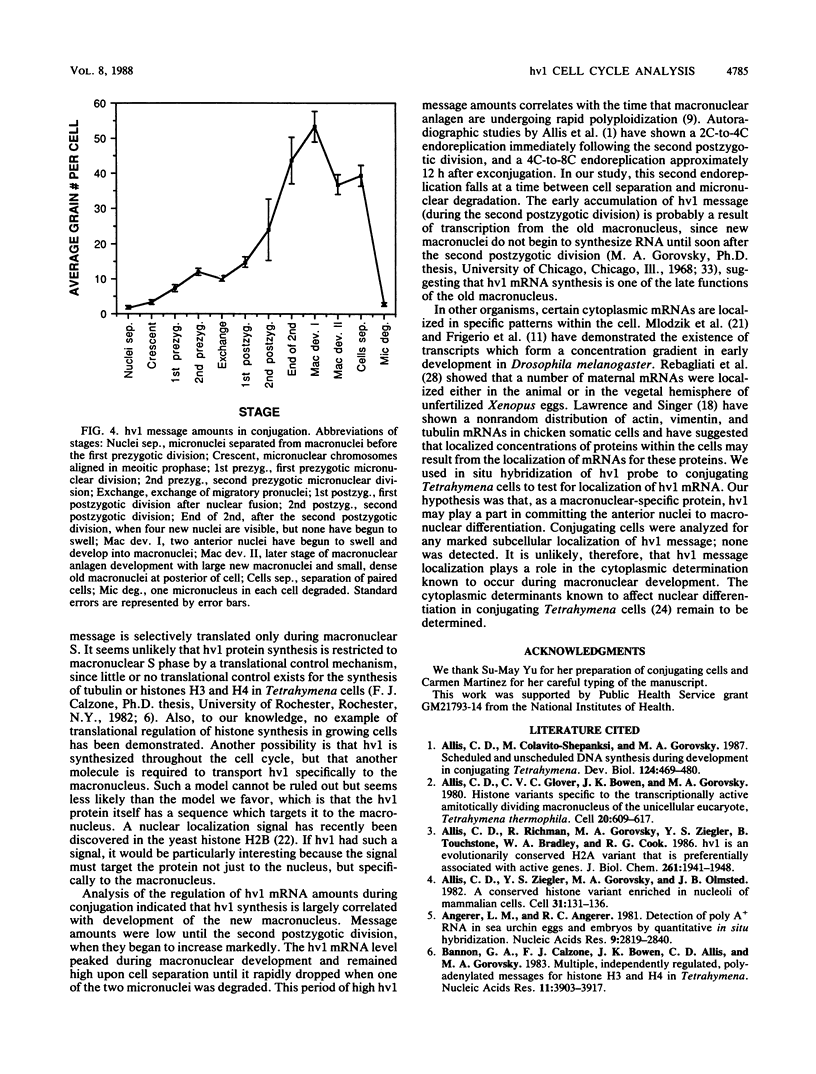
hv1 is a histone H2A variant found in the transcriptionally active Tetrahymena macronucleus but not in the transcriptionally inert micronucleus. This, along with a number of other lines of evidence, suggests that hv1 is associated with active genes. We have used a cDNA clone as a probe to study hv1 mRNA accumulation throughout the cell cycle and during conjugation. In situ hybridization to glutaraldehyde-fixed growing cells, whose position in the cell cycle was determined by size and morphology, showed that hv1 message is present throughout the cell cycle. The message was uniformly distributed in these vegetative cells. Compared with four other Tetrahymena histone genes studied to date (S. -M. Yu, S. Horowitz, and M. A. Gorovsky, Genes Dev., 1:683, 1987; M. Wu, C. D. Allis, and M. A. Gorovsky, Proc. Natl. Acad. Sci. USA 85:2205, 1988), hv1 mRNA is the only one that does not show a pattern of accumulation during the cell cycle that could explain the nuclear localization of its encoded protein. Thus, either hv1 or some molecule with which it associates contains a macronuclear-specific targeting sequence or there exists a cell cycle-regulated event that restricts its translation to the macronuclear S phase. In situ hybridization to conjugating cells revealed that hv1 message amounts increase just prior to macronuclear development and decline precipitously after the cells separate. The hv1 message showed no marked subcellular localization and is, therefore, unlikely to play a role in the cytoplasmic determination known to occur during macronuclear development.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Colavito-Shepanski M., Gorovsky M. A. Scheduled and unscheduled DNA synthesis during development in conjugating Tetrahymena. Dev Biol. 1987 Dec;124(2):469–480. doi: 10.1016/0012-1606(87)90500-8. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Richman R., Gorovsky M. A., Ziegler Y. S., Touchstone B., Bradley W. A., Cook R. G. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J Biol Chem. 1986 Feb 5;261(4):1941–1948. [PubMed] [Google Scholar]
- Allis C. D., Ziegler Y. S., Gorovsky M. A., Olmsted J. B. A conserved histone variant enriched in nucleoli of mammalian cells. Cell. 1982 Nov;31(1):131–136. doi: 10.1016/0092-8674(82)90412-3. [DOI] [PubMed] [Google Scholar]
- Angerer L. M., Angerer R. C. Detection of poly A+ RNA in sea urchin eggs and embryos by quantitative in situ hybridization. Nucleic Acids Res. 1981 Jun 25;9(12):2819–2840. doi: 10.1093/nar/9.12.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon G. A., Calzone F. J., Bowen J. K., Allis C. D., Gorovsky M. A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983 Jun 25;11(12):3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charret R. L'ADN nucléolaire chez Tetrahymena pyriformis: chronoloe de sa réplication. Exp Cell Res. 1969 Mar;54(3):353–361. doi: 10.1016/0014-4827(69)90214-6. [DOI] [PubMed] [Google Scholar]
- Cox K. H., DeLeon D. V., Angerer L. M., Angerer R. C. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984 Feb;101(2):485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- Doerder F. P., Debault L. E. Cytofluorimetric analysis of nuclear DNA during meiosis, fertilization and macronuclear development in the ciliate Tetrahymena pyriformis, syngen 1. J Cell Sci. 1975 May;17(3):471–493. doi: 10.1242/jcs.17.3.471. [DOI] [PubMed] [Google Scholar]
- Ernst S. G., Miller H., Brenner C. A., Nocente-McGrath C., Francis S., McIsaac R. Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res. 1987 Jun 11;15(11):4629–4644. doi: 10.1093/nar/15.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio G., Burri M., Bopp D., Baumgartner S., Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986 Dec 5;47(5):735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Gabrielli F., Hancock R., Faber A. J. Characterisation of a chromatin fraction bearing pulse-labelled RNA. 2. Quantification of histones and high-mobility-group proteins. Eur J Biochem. 1981 Nov;120(2):363–369. doi: 10.1111/j.1432-1033.1981.tb05713.x. [DOI] [PubMed] [Google Scholar]
- Gilbert D. M. Temporal order of replication of Xenopus laevis 5S ribosomal RNA genes in somatic cells. Proc Natl Acad Sci U S A. 1986 May;83(9):2924–2928. doi: 10.1073/pnas.83.9.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M. A., Holmquist G. P., Gray M. C., Caston L. A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984 May 18;224(4650):686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Guinta D. R., Korn L. J. Differential order of replication of Xenopus laevis 5S RNA genes. Mol Cell Biol. 1986 Jul;6(7):2536–2542. doi: 10.1128/mcb.6.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986 May 9;45(3):407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Allis C. D., Bruns P. J. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res. 1982 Jul;140(1):227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Fjose A., Gehring W. J. Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 1985 Nov;4(11):2961–2969. doi: 10.1002/j.1460-2075.1985.tb04030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Langevin G. L., Singer R. H., Garcea R. L., Hereford L. M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987 Nov;7(11):4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newrock K. M., Cohen L. H., Hendricks M. B., Donnelly R. J., Weinberg E. S. Stage-specific mRNAs coding for subtypes of H2A and H2B histones in the sea urchin embryo. Cell. 1978 Jun;14(2):327–336. doi: 10.1016/0092-8674(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Rebagliati M. R., Weeks D. L., Harvey R. P., Melton D. A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985 Oct;42(3):769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- Ridsdale J. A., Davie J. R. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched in beta-globin gene sequences. Nucleic Acids Res. 1987 Feb 11;15(3):1081–1096. doi: 10.1093/nar/15.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D. Cell-cycle regulation of histone gene expression. Cell. 1986 May 23;45(4):471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkert D., Allis C. D. Timing of the appearance of macronuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J Cell Biol. 1984 Jun;98(6):2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard J., Kaneshiro E., Gorovsky M. A. Cytochemical studies on the problem of macronuclear subnuclei in tetrahymena. Genetics. 1972 Feb;70(2):251–260. doi: 10.1093/genetics/70.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Allis C. D., Gorovsky M. A. Cell-cycle regulation as a mechanism for targeting proteins to specific DNA sequences in Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2205–2209. doi: 10.1073/pnas.85.7.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Yu S. M., Gorovsky M. A. In situ dot blots: quantitation of mRNA in intact cells. Nucleic Acids Res. 1986 Oct 10;14(19):7597–7615. doi: 10.1093/nar/14.19.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]