Abstract
Although malignant cells can be recognized and controlled by the immune system, in patients with clinically apparent cancer immunosurveillance has failed. To better understand local immunoregulatory processes that impact on cancer progression, we correlated intratumoral immunological profiles with the survival of patients affected by primary clear cell renal cell carcinoma (ccRCC). A retrospective analysis of 54 primary ccRCC samples for 31 different immune response-related transcripts, revealed a negative correlation of CD68 (a marker of tumor-associated macrophages, TAMs) and FOXP3 (a marker of regulatory T cells, Tregs) with survival. The subsequent analysis of 12 TAM-related transcripts revealed an association between the genes coding for CD163, interferon regulatory factor 4 (IRF4) and fibronectin 1 (FN1), all of which have been linked to the M2 TAM phenotype, with reduced survival and increased tumor stage, whereas the opposite was the case for the M1-associated gene coding for inducible nitric oxide synthetase (iNOS). The M2 signature of (CD68+) TAMs was found to correlate with CD163 expression, as determined in prospectively collected fresh ccRCC tissue samples. Upon co-culture with autologous tumor cells, CD11b+ cells isolated from paired blood samples expressed CD163 and other M2-associated proteins, suggesting that the malignant cells promote the accumulation of M2 TAMs. Furthermore, the tumor-associated milieu as well as isolated TAMs induced the skewing of autologous, blood-derived CD4+ T cells toward a more immunosuppressive phenotype, as shown by decreased production of effector cytokines, increased production of interleukin-10 (IL-10) and enhanced expression of the co-inhibitory molecules programmed death 1 (PD-1) and T-cell immunoglobulin mucin 3 (TIM-3). Taken together, our data suggest that ccRCC progressively attracts macrophages and induces their skewing into M2 TAMs, in turn subverting tumor-infiltrating T cells such that immunoregulatory functions are increased at the expense of effector functions.
Keywords: T-cell response, clear cell renal cell carcinoma, immunoregulation, tumor immunology, tumor-associated macrophages
Introduction
It is well established that the immune system recognizes and can destroy tumor cells, and a significant association between immune infiltration and disease outcome has been described in many different cancers.1 However, the interactions between immune cells and tumor cells are complex and we are only beginning to understand them. According to current models, malignant cells and immune cells mutually influence each other, ultimately resulting in the escape of the former from immunosurveillance, such as in patients with clinically manifest cancer.2 Besides tumor cell-intrinsic escape mechanisms, including the downregulation of MHC Class I and II molecules and/or tumor-associated antigens (TAAs), neoplastic cells can create a microenvironment that interferes with immune effector mechanisms. Thus, cancer cells can secrete immunosuppressive cytokines such as interleukin (IL)-10 and transforming growth factor β (TGFβ)3,4 and promote the recruitment of immunoregulatory cell types such as regulatory T cells (Tregs), specific subsets of tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs).5-7 CD68 is expressed by macrophages and preclinical data indicate that TAMs most often exert tumor-promoting functions by stimulating angiogenesis, tumor-cell proliferation and metastasis, as well as by contributing to the subversion of adaptive immune responses.8 TAMs show a remarkable degree of plasticity and it has recently been shown that the conversion of pro-inflammatory M1 macrophages to so-called “alternatively-activated” M2 macrophages9 and/or the preferential accumulation10 of the latter within neoplastic lesions exert critical tumor-promoting effects. M1 macrophages express the transcription factor interferon regulatory factor (IRF)511 and are thought to bear antitumor activities because of an IL-12highIL-10low phenotype5 coupled to elevated expression levels of the inducible nitric oxide (NO) synthetase (iNOS),12 resulting in increased production of NO. M2 macrophages express the transcription factor IRF413 and are characterized by an IL12lowIL10high phenotype,5 the expression of the scavenger receptor CD163,14 the mannose receptor (MR)15 and increased levels of fibronectin 1 (FN1).16
The clinical outcome of renal cell cancer (RCC) patients varies considerably, especially among individuals presenting without metastases. The prognosis for localized disease is good with a five-year survival rate of more than 90% upon tumor removal via radical or partial nephrectomy.17 However, due to the lack of major symptoms, about one-third of patients bears metastatic disease at time of diagnosis, and 25–50% of patients treated for local disease will develop metastasis.18 The survival of metastatic RCC is dramatically low, with a five-year survival rate of less than 15%. In this setting, treatment options are often limited by radio- and chemotherapy resistance.19
RCCs are largely considered as immunogenic tumors and are frequently infiltrated by immune cells.20 However, in contrast to breast carcinoma, bladder carcinoma and other cancers (reviewed in ref. 1), an elevated number of tumor-infiltrating leukocytes (TILs) is associated with poor prognosis in RCC.21 To better understand the local mechanisms that preclude the control of clinically apparent RCC by the immune system, we correlated intratumoral immunological profiles with survival and tumor stage (pT) in patients affected by primary clear cell renal cell carcinoma (ccRCC), which is the most frequent subtype of RCC. Furthermore, we investigated the impact of the tumor microenvironment on T-cell phenotype and function.
Results
Correlation between the expression of immune response-related transcripts and survival in ccRCC
To better understand the local immunosuppressive mechanisms that preclude the immune system to control clinically manifest RCC, we correlated intratumoral immunological profiles with survival in 54 patients affected by primary ccRCC. We retrospectively analyzed the expression of 31 immune response-related transcripts (Table S1A) by quantitative reverse transcription real-time PCR (qRT-PCR) using a collection of formalin-fixed paraffin-embedded (FFPE) tumor tissues from ccRCC patients, for which information on tumor size and disease course was available. The samples of patients who died of tumor-unrelated causes were excluded from the analysis (Table S2). Using the Cox proportional hazard model, we found no significant correlation between the degree of leukocyte infiltration (Ct values of CD45 normalized to those of the 18S rRNA) and survival (Fig. 1A).
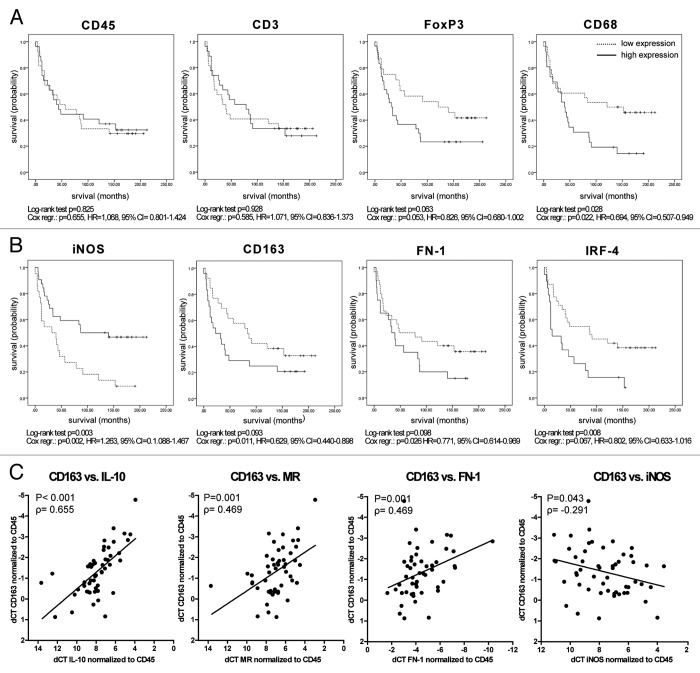
Figure 1.FOXP3 and CD68 and genes associated to M2 tumor-associated macrophages inversely correlate with survival in clear cell renal cell carcinoma patients. (A–C) Fifty-four clear cell renal cell carcinoma (ccRCC) formalin-fixed paraffin-embedded (FFPE) tumor samples were subjected to a retrospective qRT-PCR analysis for different immune response-related genes. ∆Ct levels of CD45 were calculated by normalization to the endogenous control (18S rRNA), ∆Ct levels of all other genes were calculated by normalization to CD45. Survival analysis was performed using the Cox proportional hazard model and, after dichotomizing the data based on the mean-expression level, the log-rank test of the Kaplan-Meier estimator. The results of both statistical tests are displayed for selected genes. Patients that were still alive at time of analysis are marked with a tick. Kaplan-Meier survival curves show the relationship between gene expression and survival for CD45, CD3, FOXP3 and CD68 (A) and iNOS, CD163, FN1 and IRF4 (B). Multivariate Cox regression analysis revealed that the correlation of target gene expression and survival is independent of tumor grade for CD68: p = 0.02, hazard ratio (HR) = 0.704, 95% CI 0.520–0.954; iNOS: p = 0.011, HR = 1.261, 95% CI 1.055–1.506 and CD163 p = 0.015, HR = 0.645, 95% CI 0.453–0.919, as well as independent of patient age for CD68: p = 0.04, HR = 0.720, 95% CI 0.524–0.989; iNOS: p = 0.012, HR = 1.299, 95% CI 1.060–1.592 and CD163: p = 0.013, HR = 0.636, 95% CI 0.445–0.909. (C) ∆Ct levels of CD163 plotted against ∆Ct values of IL-10-, MR-, FN1- and iNOS-coding transcripts. Results of Spearman Rho correlation analysis are depicted. Each dot represents an individual patient.
Subsequently, we quantified the transcripts of immune response-related genes upon normalization to the expression level of CD45, which allows for the qualitative analysis of the infiltrate independent of the degree of infiltration. We correlated each of the 31 transcripts with survival, using Cox regression analysis (Fig. 1A; Fig. S1A). We excluded the data for lymphotoxin α (LTα), arginase 1, B and T lymphocyte associated (BTLA), IL-2 and IL-17 from the analysis because of very low or undetectable expression. Univariate Cox regression analysis revealed a significant correlation of elevated FOXP3 (a marker of Tregs) and CD68 (a marker of macrophages) mRNA levels with reduced survival, whereas the abundance of CD3 transcripts (identifying T cells as a whole) did not correlate with patient survival (Fig. 1A). We correlated the abundance of CD68 transcripts with that of CD3, CD4 and CD8 transcripts, finding no correlation between CD68 and CD3 (correlation coefficient −0.082, two-tailed p = 0.556) or CD68 and CD8 (correlation coefficient −0.031, two-tailed p = 0.882), but a significant correlation between CD68 and CD4 (correlation coefficient 0.391, two-tailed p = 0.003). The latter correlation is positive, presumably reflecting the co-existence of macrophages and regulatory T cells. Furthermore, high expression levels of perforin and tumor necrosis factor α (TNFα) correlated with increased survival, while high expression levels of the lymphotoxin β receptor (LTβR) were associated with reduced survival (Fig. S1A). Since these associations turned out to be significant by means of only one out of two statistical tests employed, these genes were not considered to significantly correlate with survival. No signal was observed for cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and IL-10 in a few samples, and an estimated Ct value (40) was set for these samples (Fig. S1A). When such “no-signal” samples were excluded from the analysis, high expression values of CTLA-4 and IL-10 significantly correlated with reduced survival (Fig. S1B). Also other genes (marked with an asterisk in Fig. S1) failed to provide a signal in a few samples. However, statistical analyses did not significantly vary when “no-signal” samples were excluded (data not shown).
While the correlation of FOXP3 expression levels and survival just reached the level of statistical significance using univariate Cox regression analysis, the correlation between CD68 mRNA levels and reduced survival was independent of tumor size and patient age, as assessed by multivariate Cox regression analysis (Fig. 1). To validate qRT-PCR results, we quantified CD68 by immunohistochemistry on seven ccRCC tumor samples and we invariably found a clear correlation between the mRNA and protein levels of CD68 (Fig. S2).
The expression of M2-associated transcripts correlates with reduced survival in ccRCC
To further investigate the phenotype and impact of TAMs on ccRCC patient survival, we analyzed the same samples (Table S2) for the expression levels of additional 12 TAM-associated genes (Fig. 1B; Table S1B; Fig. S1C). We found a significant correlation between decreased survival and low levels of iNOS or high levels of CD163 transcripts (Fig. 1B). Multivariate Cox regression analysis revealed that both correlations are independent of tumor stage and patient age (Fig. 1). Along similar lines, a high expression of the M2-associated genes FN1 and IRF4 tended to correlate with reduced survival (Fig. 1B). Furthermore, we observed a positive correlation between the abundance of CD163 transcripts and that of mRNAs coding for the M2-associated proteins MR, IL-10 and FN1 as well as a negative correlation between the expression levels of CD163 and iNOS (an M1-associated factor) (Fig. 1C) suggesting that TAMs with an M2-like signature negatively influence the survival of ccRCC patients.
CD163high TAMs exhibit an M2-phenotype
We further characterized TAMs from 12 fresh primary ccRCC tumor samples by flow cytometry. Tumors from most patients contained a subpopulation of CD45+CD3−CD19−CD68+CD11b+CD163high cells (termed T2 population hereafter), which was absent in matched peripheral blood samples. In the peripheral blood as well as in tumors, we found populations that we termed P1 and T1 population, respectively, both of which were characterized by a CD45+CD3−CD19−CD68+CD11b+CD163low phenotype (Fig. 2A). Both T1 and T2 TAMs expressed higher levels of MHC Class II molecules than P1 cells, suggesting that TAMs exist in an activated state (Fig. 2B; Fig. S4). Based on the elevated expression of CD163 and MR, the T2 population classifies as M2 TAMs. Furthermore, the T2 but neither the T1 nor the P1 population exhibited increased expression of the programmed death ligand 1 (PD-L1) (Fig. 2B; Fig. S4). To further strengthen the presumption that CD163 associates with M2 TAMs, we analyzed the expression levels of different M1- and M2-related genes on sorted T2 TAMs by qRT-PCR (Table S1). When we compared T2 and P1 cells for the expression of M2-associated genes, the former exhibited a strong elevation of FN1- and IL-10-coding transcripts that was accompanied by a slight increase in the expression of IRF-4 and of the transcription factor c-MYC,22 whereas the expression of the M1-associated genes IRF5, iNOS and IL-12 was low (Fig. 2C). Except for the elevated expression of CD163 and MR, no clear differences between the expression of M1- and M2-assocaited genes were observed in the T2 as compared with the T1 population, at least at the mRNA level (Fig. S5).
Figure 2. Characterization of tumor-associated myeloid cells in clear cell renal cell carcinoma. (A–C) Fresh primary clear cell renal cell carcinoma (ccRCC) and paired peripheral blood samples were collected and processed as described under Materials and Methods. (A) Dot plots display the staining of peripheral blood mononuclear cells (PBMCs, left histogram) and processed tumors (right histogram) for CD11b and CD163 after gating on CD45+CD3−CD19− cells. P1, T1 and T2 designate individual myeloid cell populations. The plots show a representative example of one patient. (B) Expression of different macrophage-associated molecules after gating on P1, T1 and T2 populations, displayed as the geometric mean of fluorescence intensity. Each symbol represents an individual patient; means and significant differences (*p < 0.05) are displayed. (C) qRT-PCR analysis of FACS-sorted P1 and T2 cell fractions. The genes displayed on the left side of the vertical line are related to M2 TAMs, on the right side to M1 TAMs. ∆Ct levels were calculated upon normalization to the endogenous control PPIA. Results are presented as fold change in expression level of T2 relative to P1; the geometric mean of each group is depicted. Fold differences in expression within the shaded area are considered as not significant. Symbols at the 0.0001 line on the y-axis represent samples for which the fold change could not be calculated, since the expression was only detected in the P1 fraction. Each symbol in (B) and (C) represents an individual patient.
The expression of M2-associated transcripts correlates with tumor progression
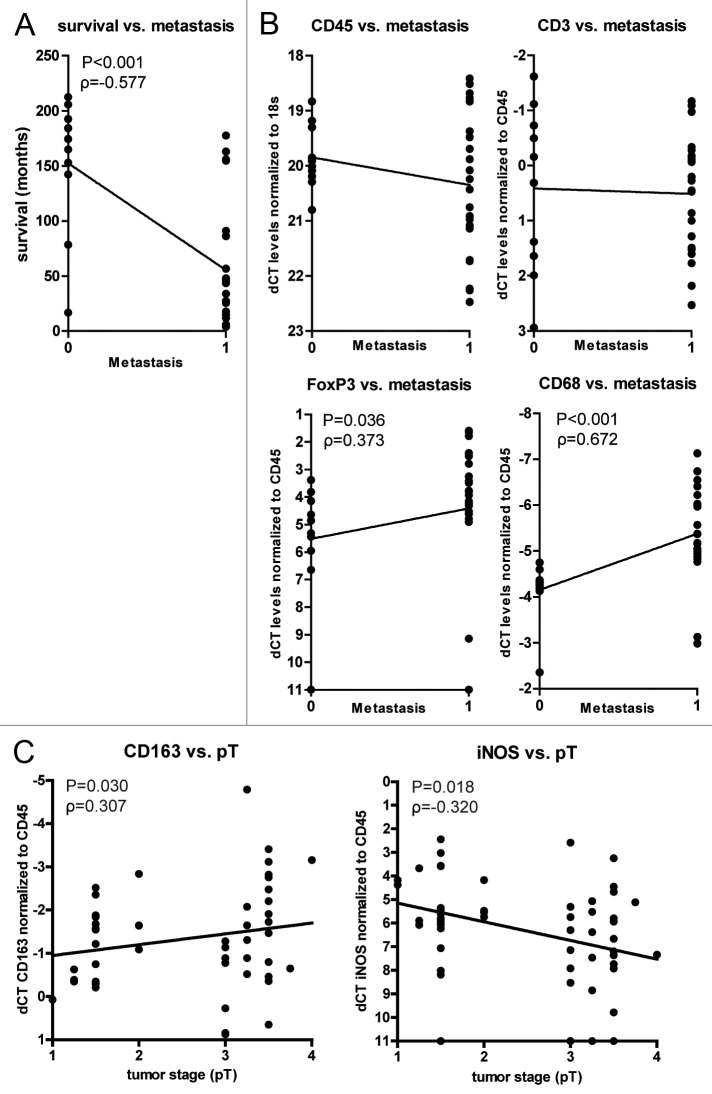
RCC patients who develop metastasis have a poor prognosis, with a five-year survival rate of less than 15%.23 We observed a similar correlation between reduced survival and the incidence of metastasis (Fig. 3A). High levels of FOXP3 or CD68 transcripts also correlated with the incidence of metastasis, whereas the expression of CD45 and CD3 did not (Fig. 3B). A correlation between tumor infiltration by macrophages and metastasis has been documented in various types of cancer.24 In this context, a role has been proposed for the paracrine and autocrine signaling axis involving colony-stimulating factor 1 (CSF1), epidermal growth factor (EGF)25 as well as the actin-binding protein mammalian enabled (MENA).26 However, in our cohort of primary ccRCC patients, we did not observed a significant correlation between the expression levels of CSF1, EGF, their receptors and MENA with survival (Fig. S1C).
Figure 3. Positive correlation of FOXP3 and CD68 gene expression with the incidence of metastasis and of M2-associated genes with tumor stage. (A) Survival in months after diagnosis versus incidence of metastasis. (B) The ∆Ct values of CD45, CD3, FOXP3 and CD68 correlated with the incidence of metastasis (0 = patient did not develop metastasis, 1 = patient developed metastasis). (C) The ∆Ct values of CD163 and iNOS correlated with tumor stage (pT). Each dot represents an individual patient. ∆Ct levels were calculated as in the legend of Figure 1. Only when correlations are significant, the results of the Spearman Rho correlation test are displayed. Tumor stage was defined as: 1 = pT1, 1.25 = pT1a, 1.5 = pT1b, 2 = pT2, 3 = pT3, 3.25 = pT3a, 3.5 = pT3b, 3.75 = pT3c and 4 = pT4.
Conversely, we found a significant correlation between low iNOS expression levels and increased tumor stage and a similar trend for high levels of CD163 transcripts (Fig. 3C). These data suggest a progressive accumulation of, or a conversion to, CD163high M2 macrophages at the expense of iNOShigh M1 macrophages within ccRCC, which correlates with reduced patient survival.
ccRCC cells contribute to the M1 → M2 conversion of myeloid cells
To investigate whether freshly isolated ccRCC tumor cells can induce tumor-promoting changes in myeloid cells, we co-incubated P1 cells (sorted from the peripheral blood) with autologous CD45- cells (obtained from tumor samples) for 48 h at a 1:3 ratio, followed by the analysis of P1 cells. Such a co-culture induced the upregulation of CD163 and MR in blood-derived myeloid cells, as observed at the protein level (Fig. S6A). At the mRNA level, a slight increase in the expression of M2-related genes coding for CD163, c-MYC and IL-10 was observed, coupled to a strong elevation in FN-1 (Fig. S6B). Of note, the M1-associated factor iNOS was upregulated in two samples, while IL-12 was not expressed in two samples and lost in one upon co-culture with tumor cells. Because we only had sufficient material from a limited number of patients, we could not investigate this phenomenon in a larger series of samples.
In summary, the CD45+CD3−CD19−CD68+CD11b+CD163high T2 population, which is found in ccRCCs but not in paired peripheral blood samples, shows features of tumor-promoting TAMs with an M2-like signature. Our findings indicating that the intratumoral levels of CD163 transcripts correlate with reduced survival and increased tumor grade lend further support to this notion. Furthermore, we demonstrated that malignant cells favor the M1 → M2 skewing of TAMs, which is in line with the progressive accumulation of T2 TAMs within ccRCC lesions.
The ccRCC microenvironment impacts on the phenotype and function of T cells
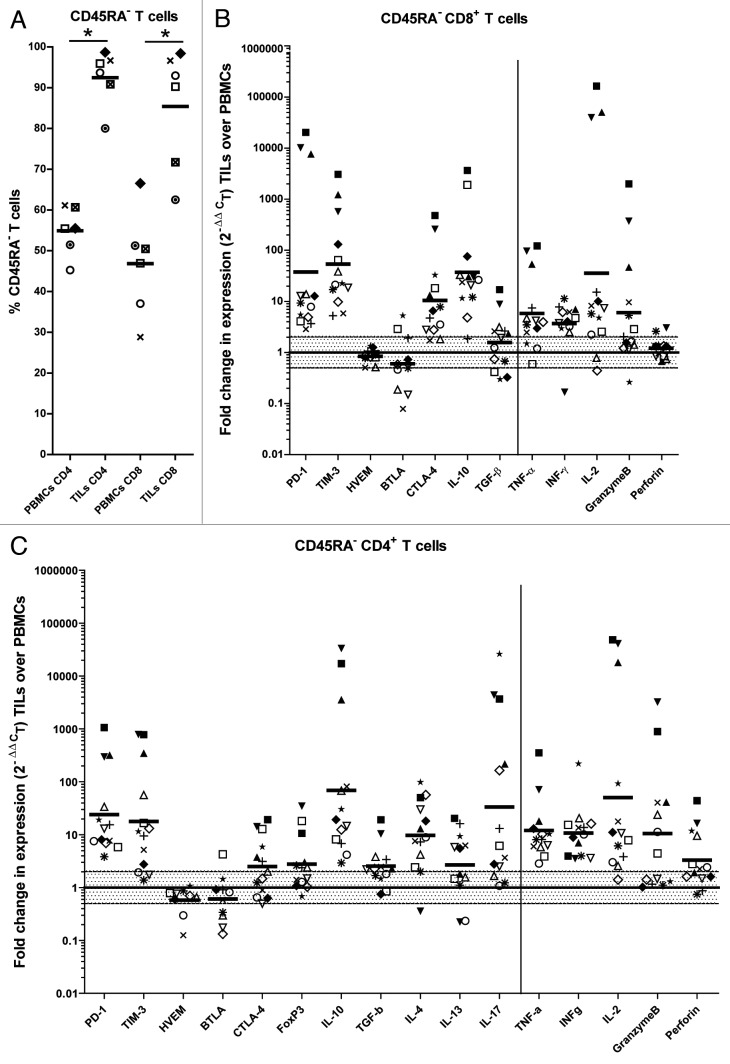
We noticed a correlation between high tumor stage and elevated FOXP3 or IL-10 expression levels (Fig. S7), suggesting a progressive accumulation of Tregs and IL-10 in the tumor. To investigate the impact of the immunoregulatory tumor microenvironment on the phenotype and function of T cells we sorted CD45RA−CD4+ and CD45RA−CD8+ T cells from fresh ccRCCs (Table S3) and from paired peripheral blood samples. We sorted CD45RA− (antigen-experienced) T cells because the proportion of antigen-experienced T cells in tumors was much higher than in paired blood samples (Fig. 4A) and these T cells have a lower threshold for in vitro cytokine production upon stimulation than naïve T cells.27 The abundance of transcripts coding for effector cytokines including TNFα, interferon γ (IFNγ), IL-2 and for the cytotoxic molecule granzyme B was higher in tumor-derived CD4+ and CD8+ T cells than in blood-derived CD4+ and CD8+ T cells (Fig. 4B and C). Alongside, however, tumor-derived T cells contained elevated levels of transcripts coding for immunoregulatory molecules including programmed death 1 (PD-1), T-cell immunoglobulin mucin 3 (TIM-3) and IL-10. Furthermore, tumor-derived CD4+ T cells expressed higher levels of FOXP3, IL-17 and the TH2 cytokines IL-4 and IL-13. Only a slight increase in TGFβ was observed in TILs, which may be explained by the fact that TGFβ is mainly regulated at a post-translational level (Fig. 4B and C). By flow cytometry, we confirmed that TIL-derived T cells were in an activated state, as demonstrated by the strong expression of the activation marker CD69, and confirmed the increased expression of PD-1, TIM-3, CTLA-4 and FOXP3 at the protein level (Fig. 5A and B; Fig. S8). Altogether, these data suggest that ccRCCs are sites characterized by an active immune response, which manifests features reminiscent of immune regulation or chronic inflammation.28,29
Figure 4. Tumor-derived T cells show an antigen-experienced and regulated phenotype. (A) The percentage of CD45RA− (antigen-experienced) T cells in the peripheral blood and tumors of clear cell renal cell carcinoma (ccRCC) patients was determined by flow cytometry; the means of each group and significant differences (*p < 0.05) are displayed. (B and C) Gene expression analysis was performed by qRT-PCR on FACS-sorted CD45RA−CD8+ (B) and CD45RA−CD4+ (C) cells from the peripheral blood and tumors of ccRCC patients. Ct values were normalized to the endogenous control PPIA. The genes displayed on the left side of the vertical lines are related to immunoregulation, on the right side to effector functions. Results are presented as fold change in expression level of tumor-derived relative to peripheral blood-derived T cells; the geometric mean of each group is depicted; fold differences in expression within the shaded area are considered as not significant. Each symbol represents an individual patient.
Figure 5. Tumor-derived T cells express higher levels of regulatory molecules than T cells derived from paired blood samples. (A and B) FACS analysis on tumor- and peripheral blood-derived cells after gating on CD8+CD45RA- T cells (A) or CD4+CD45RA− T cells (B). The graphs display the percentage of cells positive for a particular marker. A representative staining is shown in Figure S4. Each symbol represents an individual patient. The mean of each group and significant differences (*p < 0.05; **p < 0.005; ***p < 0.0005) are depicted.
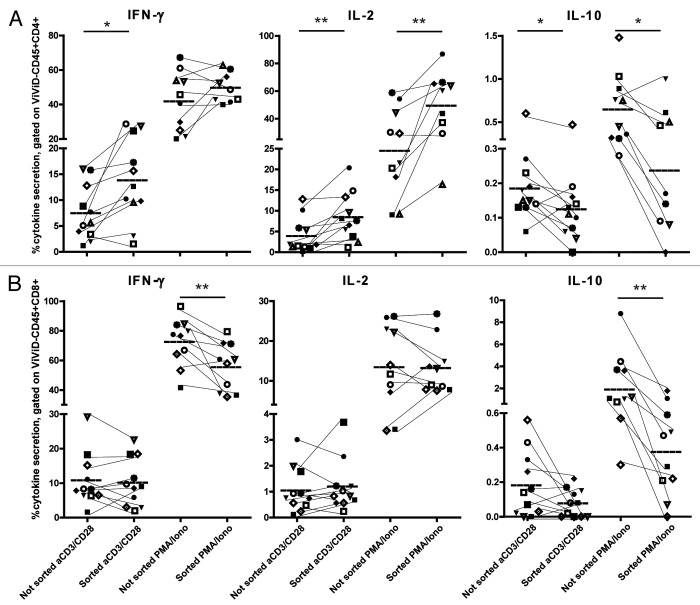
To confirm the alterations of the cytokine profile in tumor-derived T cells at the protein level, and to investigate the role of the tumor microenvironment on these functional deviations toward a more regulated phenotype, we polyclonally stimulated CD45RA- antigen-experienced T cells in the presence of the whole tumor singe-cell suspension and compared their cytokine production profile with that of CD45RA− T cells from the same source that were previously sorted. The production of the effector cytokines IFNγ and IL-2 by sorted CD4+ T cells was higher, whereas the production of the immunoregulatory cytokine IL-10 was lower, as compared with the same T cells in the presence of their natural tumor environment (Fig. 6A). The difference in effector cytokine production between sorted and non-sorted cells was less pronounced for CD8+ T cells, whereas the production of IL-10 was supported by the presence of the tumor (Fig. 6B). We detected a very low percentage of IL-17-producing cells, which was comparable in sorted and non-sorted cells (Fig. S9). We conclude that the tumor microenvironment induced the skewing toward a more regulated phenotype upon T-cell receptor (TCR)-dependent (anti-CD3/CD28-coated beads) as well as TCR-independent (phorbol 12-myristate 13-acetate plus ionomycin) stimulation.
Figure 6. The tumor microenvironment impacts on the cytokine profile of T cells. Intracellular staining for cytokines after 6h ex vivo stimulation with anti-CD3/CD28 beads or with phorbol 12-myristate 13-acetate (PMA) + ionomycin in the presence of brefeldin A and monensin. Cells were stimulated within the tumor single-cell suspension or after sorting of CD45+CD4+ T cells (A) or CD45+CD8+ T cells (B) and analyzed after gating on live CD45+CD4+ T cells (A) or live CD45+CD8+ T cells (B). Each symbol represents an individual patient. Results of unsorted and sorted T cells from the same patient are connected by a thin line. The mean of each group and significant differences (*p < 0.05; **p < 0.005) are depicted.
M2 TAMs impact on the cytokine profile of CD4+ T cells
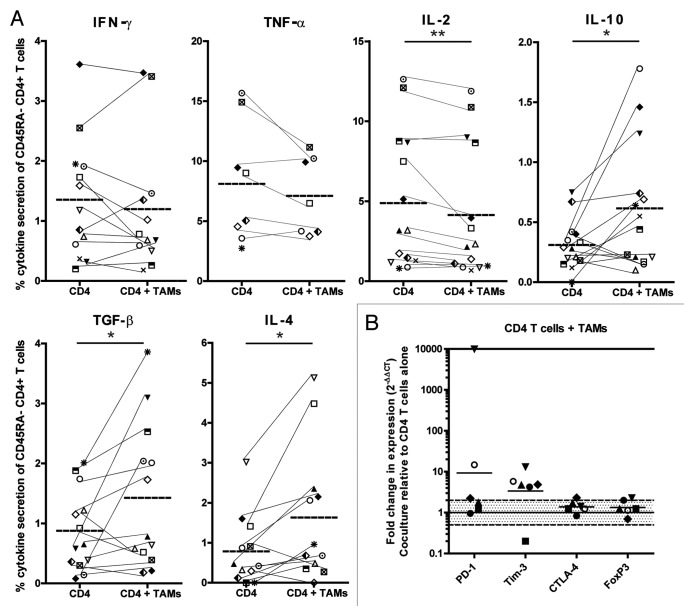
TAMs have been described to suppress tumor-specific immunity.30,31 To investigate whether the intratumoral T2 population contributes to the immune subversion of tumor-infiltrating antigen-experienced T cells, we stimulated sorted, blood-derived CD45RA−CD4+ cells with anti-CD3/CD28-coated beads in the presence or absence of autologous, sorted, tumor-derived T2 (CD45+CD2−CD19−CD11b+CD163high) cells (Fig. S10). Because of the limited availability of material, we could only perform TCR-dependent stimulation for CD4+ T cells. Peripheral blood-derived CD4+ T cells produced significantly less IL-2 and significantly more TGF-β, IL-10 and IL-4 upon the addition of autologous T2 TAMs (Fig. 7A). The production of the effector cytokines IFNγ und TNFα followed the same trend as IL-2; although the effect was not statistically significant. Furthermore, the co-incubation with T2 TAMs resulted in an upregulation of the transcripts coding for PD-1 and TIM-3 (Fig. 7B). No changes in T-cell function were observed when T cells were co-cultured with the sorted T1 fraction (CD45+CD2−CD19−CD11b+CD163low) (data not shown). There was a trend toward a reduced proliferation of T cells in the presence of the T2 fraction, but results were not consistent (data not shown). These results indicate that CD45+CD2−CD19−CD11b+CD163high cells contribute to functional and phenotypic immune subversion of tumor-infiltrating T cells in ccRCC patients.
Figure 7. Clear cell renal cell carcinoma-derived M2 tumor-associated macrophages skew T cells toward a more regulated phenotype. (A and B) Sorted peripheral blood-derived CD4+ T cells were cultured for 48 h with or without sorted autologous T2 cells (CD45+CD2−CD19−CD11b+CD163high) in a ratio of 1:1. (A) The production of cytokines was assessed via intracellular cytokine stating 6 h after ex vivo stimulation with anti-CD3/CD28 beads in the presence of brefeldin A and monensin upon gating on live CD45+CD11b−CD4+ cells. Results of the same patient are connected by a thin line. The mean of each group and significant differences (* p < 0.05; ** p < 0.005) are depicted. (B) The expression of regulatory molecules was assessed by qRT-PCR. ∆Ct levels were calculated by normalizing the Ct values of the target genes to the Ct values of CD4. Results are presented as fold change in expression level of CD4+ T cells co-cultured in the presence of autologous T2 (CD45+CD2−CD19−CD11b+CD163high) cells relative to CD4+ T cells cultured in their absence. Fold differences in expression within the shaded area are considered as not significant. The symbol at the 10,000 line on the y-axis represents a sample of which fold change could not be calculated, since expression was only detected after co-culture. Each symbol represents an individual patient.
Discussion
Growing evidence indicates that the immunoregulatory milieu that is associated with most tumors precludes protective antitumor immunity and compromises the efficacy of immunotherapy.32 To better understand local immunoregulatory processes that impact on ccRCC progression, we correlated intratumoral immunological profiles with survival in patients affected by primary ccRCC. In addition, we performed functional experiments to dissect the mutual influences of tumor cells, TAMs and T cells.
We retrospectively analyzed the transcripts coding for 31 immune response-related genes in 54 FFPE primary ccRCC samples, and correlated the expression of each of these genes with survival. In line with the finding that the subtype function and location of leukocytes, rather than the degree of infiltration are relevant to disease outcome,1 we did not observe a correlation between the extent of leukocyte (CD45+ mRNA level) infiltration and survival. Data on the impact of tumor infiltrating leukocytes (TILs) on ccRCC outcome are conflicting. Whereas one study has shown that an increased frequency of CD8+ T cells correlates with poor prognosis, except when these T cells are proliferating,33 another report shows a correlation between increased TH1 responses and improved prognosis.34 Furthermore, it has recently been shown that an increased abundance of intratumoral lymphocytes negatively impacts the overall survival of ccRCC patients.21 Among all lymphocyte-associated transcripts, only the expression levels of FOXP3, coding for a Treg-specific transcription factor35 correlated with decreased survival, which is in line with previous findings.36,37 Tregs are a subpopulation of T cells that play an important physiological role in suppressing effector responses to self-antigens, thereby preventing autoimmunity.38 Furthermore, Tregs suppress antitumor immunity.7,39 A correlation between increased numbers of intratumoral Tregs or a low ratio between effector T cells and Tregs and reduced survival has been shown in several cancer types, including ccRCC.39,40
We discovered that increased levels of transcripts coding for the macrophage marker CD68 correlate significantly with reduced survival in primary ccRCC patients. Although most often an increased frequency of TAMs has been correlated with unfavorable prognosis in patients affected by breast, prostate, bladder or kidney cancer, some studies involving patients bearing melanoma, gastric and colocrectal cancer report opposite findings (summarized in ref. 8). A possible explanation for these conflicting observations may be the use of the pan-macrophage marker CD68, which recognizes all the subsets of this highly heterogeneous and plastic cell population41 and notably does not differentiate between functionally different M1 and M2 macrophages.42,43 Pro-inflammatory M1 macrophages are induced by IFNγ and lipopolysaccharide (LPS), whereas alternatively activated, anti-inflammatory M2 macrophages differentiate in the presence of IL-4, IL-13 and IL-10.5 M2 macrophages dampen immune and inflammatory responses to prevent tissue damage in the case of chronic inflammation.44 Moreover, M2 macrophages promote tumor progression, metastasis and immunosuppression.43 For this reason we expanded our retrospective analysis with 12 TAM-associated genes and found a correlation between the M1-associated factor iNOS and prolonged survival as well as between an increased expression of M2-associated genes (CD163, FN1 and IRF4) and reduced survival. iNOS has previously been correlated with increased survival in colorectal cancer patients.45 Furthermore, a correlation between CD163 and reduced survival has been recently observed in different types of cancer, including ccRCC.46,47 Subsequent analysis of prospectively collected fresh tumor samples showed the presence of two different myeloid fractions in primary ccRCC lesions: CD11b+CD68+CD163high (T2) and CD11b+CD68+CD163low (T1) cells, whereas we only found CD11b+CD68+CD163low cells in paired peripheral blood samples (P1 cells). The T2 population expressed high levels of MR, FN1 and IL-10 and low levels of iNOS, thus uncovering CD163 as a marker for TAMs with an M2-like signature in ccRCC.5,44 T2 cells—but neither T1 nor P1 cells—showed increased expression of molecules that are involved in immunosuppression, including PD-L1 and IL-10. We noticed that differences in the CD163 signal between the different fractions were not as pronounced at the mRNA level as they were at the protein level. One explanation for this observation may be that CD163 transcripts are less stable than the corresponding protein. The high level of MHC Class II molecules on the T2 fraction clearly discriminates them from CD11b+MHC-II−/low MDSC.6,48
TAMs can support tumor progression in multiple ways, e.g., by promoting the survival of tumor cells, invasion, metastasis and angiogenesis.8 Our analyses showed a correlation between CD68 transcripts and the incidence of metastases. We observed a correlation between increased CD163 levels and tumor stage, while the opposite held true for iNOS. This suggests a progressive accumulation of M2 TAMs in ccRCC lesions, which may be due to an increased influx of M2 TAMs10 or to the conversion of M1 to M2 macrophages in situ.9,48,49 Our data support the second scenario, since we observed the induction of M2-associated markers on peripheral blood-derived myeloid cells upon co-culture with autologous tumor cells. These findings are in line with another study showing that the supernatants of ccRCC cell lines induce the expression of M2 markers by human macrophages.49
The observations that the levels of the transcripts coding for CD163, FOXP3 and IL-10 positively correlate with tumor stage suggests that the immune regulation increased during tumor progression.7,50,51 Along this line, transcripts coding for the effector molecules TNFα and perforin positively correlated with survival, whereas the opposite was true for those encoding CTLA-4 and IL-10. Functional TIL analysis confirmed these observations: although TILs showed an antigen-experienced (CD45RO+) and activated (CD69+, IFNγ+, TNFα+) phenotype, they simultaneously expressed high levels of PD-1, TIM-3, CTLA-4 and IL-10, all of which are indicative of ongoing immunoregulation. The increased expression of PD-1 and TIM-3 was more pronounced in tumor-derived CD8+ T cells than on their CD4+ counterparts, whereas CTLA-4 and FOXP3 expression was more prominent in tumor-derived CD4+ T cells than in CD8+ T cells. This suggests that CD4+ and CD8+ T cells may be subject to different mechanisms of immunoregulation in the tumor. In particular, the significant increase of the PD-1+/TIM-3+ population is indicative of an exhausted T-cell phenotype52 and implies reduced effector functions of CD8+ T cells within ccRCCs. The tumor environment itself, or (more specifically) TAMs, contribute to the regulated phenotype of T cells because the effector functions of the latter was partially rescued by sorting CD4+ and CD8+ T cells out of the tumor. The reverse experiment, that is, the addition of sorted CD11b+CD163high cells (T2 fraction) from primary ccRCC tumor samples to autologous blood-derived CD4+ T cells, confirmed the negative impact of TAMs on T-cell effector function.
In conclusion, we found that an intense infiltration of primary ccRCC by Tregs and M2 TAMs correlates with reduced survival. Furthermore, we showed a progressive accumulation of (and/or the local conversion to) M2 macrophages, which is supported by tumor cells. M2 TAMs in turn induce the skewing of tumor-infiltrating T cells toward a more regulated phenotype at the expense of protective effector functions.
Materials and Methods
Paraffin embedded patient material
Formalin-fixed, paraffin-embedded material from 60 patients bearing ccRCC collected from 1993 to 2003 was identified in the archives of the Department of Pathology, University Hospital Zurich. All patients met the following criteria: (1) a new renal tumor diagnosis with histological confirmation of a clear cell renal tumor subtype according to the 2004 WHO classification; (2) no prior treatment for renal cancer. The local ethical committee approved this study (KEK ZH Ref. nr, STV38-2005). Six patients died of causes not related to cancer and were excluded from the analysis. A summary of the patients’ characteristics can be found in Table S2.
Fresh ccRCC tumor samples and blood
Twenty renal cancer patients were enrolled in the study. The patients underwent full or partial nephrectomy as part of their standard treatment at the Department of Urology, University Hospital Zurich. Tumor samples and peripheral blood were prospectively collected following informed consent in accordance with the Declaration of Helsinki. The local ethics committee and Swissmedic approved the study (EK-1017). All patients met the criteria described above. A summary of the patients’ characteristics can be found in Table S3. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll (Ficoll-PaqueTM PLUS; GE Healthcare) density centrifugation. The tumor material was cut into multiple pieces of 2–4 mm3 and further dissociated with 6 U/mL DNase I Type IV (Sigma) and 1 mg/mL collagenase IV (Sigma) in DMEM (Gibco) plus 50 U/mL penicillin and 50 U/mL streptomycin (Gibco) for 1–2 h at 37°C. The digested material was afterwards filtered and single-cell suspensions and the PBMCs were cryopreserved at −80°C until further analysis.
Flow cytometry for phenotypic analysis
Surface staining with fluorochrome-conjugated antibodies was performed in PBS together with live/dead staining (LIVE/DEAD® Fixable Violet Dead Cell Stain Kit, ViVid, Invitrogen) for 20 min at room temperature. Intracellular staining for CD68 and FOXP3 was performed according to manufacturer’s instructions (eBioscience). Staining was measured with a CyAn ADP 9 flow cytometer (Beckman Coulter) and data were analyzed with FlowJo software (TreeStar). All antibodies used in this study (listed in Supplementary Materials and Methods) were purchased from BioLegend, BD Biosciences, Beckman Coulter or eBioscience.
FACS sorting of T cells and TAMs from TILs and PBMCs
TILs and PBMCs were stained in sorting buffer (SB: PBS supplemented with 2% pooled human serum (PAA Laboratories) and 6 U/mL DNase I Type IV (Sigma)) and stained with a mixture of fluorochrome-conjugated antibodies plus live-dead staining (LIVE/DEAD® Fixable Violet Dead Cell Stain Kit). After staining for 20 min at 4°C cells were washed and resuspended in SB. Gates were either placed on CD45+CD8+ T cells or on CD45+CD4+ T cells, or—in the case of the simultaneous sorting of T cells and myeloid cells—as shown in the gating strategy (Fig. S10), and the different populations were sorted using a FACSAria III cell sorter (BD Biosciences).
Polyclonal stimulation and intracellular cytokine staining (ICS)
After sorting, cells were allowed to rest overnight in a 96-well round-bottom plate in TC-RPMI (RPMI) (Gibco), supplemented with 2 g/L NaHCO3 (Sigma), 2 mM L-glutamine (Sigma), 50 U/mL penicillin and 50 U/mL streptomycin (Gibco), 1x MEM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 100 μM β-mercaptoethanol (Sigma) and 10% pooled human serum plus 6 U/mL DNase I Type IV (Sigma). T cells (at least 10,000 per well) were either cultured alone or with the tumor single-cell suspension or with TAMs (ratio 1:1). After the overnight resting period, T cells were stimulated in 96-well plates in the presence or absence of the tumor digest or TAMs with anti-CD3/CD28 Dynabeads (Invitrogen, 111.41D) (Ratio T cells:beads = 1:3) or with 50 ng/mL PMA (Sigma) plus 500 ng/mL ionomycin (Sigma). Stimulation was performed for 6 h at 37°C in TC-RPMI supplemented with 10 µg/mL brefeldin A (Sigma) and 6 U/mL DNase I Type IV (Sigma). Cells were surface-stained with a fluorochrome-conjugated antibody cocktail including live-dead staining (LIVE/DEAD® Fixable Violet Dead Cell Stain Kit) in PBS for 20 min at room temperature in the dark. Subsequently, cells were fixed in 4% formalin (Kantonsapotheke Zürich) and permeabilized with permeabilization buffer (PB: PBS supplemented with 2% FCS, 20 mM EDTA, 0.05% NaN3 and 0.1% saponin). For intracellular staining, cells were incubated with a mix of fluorescent-conjugated antibodies in PB for 20 min at room temperature in the dark. Cells were washed once with PB and resuspended in PBS containing 1% formalin before analysis by flow cytometry, as described above. The antibodies employed in this study are listed in Supplementary Materials and Methods. When the stimulation of T cells was performed in presence or absence of the tumor microenvironment, cytokine production was measured upon gating on ViViD−CD45+CD14−CD16−CD19−CD8+ or ViViD−CD45+CD14−CD16-CD19−CD4+ T cells, and—in presence or absence of TAMs—upon gating on ViViD−CD45+CD11b−CD4+ T cells.
RNA isolation
After FACS sorting or after co-culture experiments, a fraction of the cells (at least 500 cells) was resuspended in TRI Reagent (Ambion) and frozen at −80°C until RNA isolation. RNA was isolated using the MagMAXTM-96 for Microarrays Total RNA isolation kit (AM1839 Ambion) by the No-Spin procedure, according to the manufacturer’s instructions. RNA isolation from FFPE material was performed using Trizol, as described in Supplementary Materials and Methods.
Quantitative reverse transcription real-time PCR (qRT-PCR)
The concentration and purity of RNA was evaluated using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Five-hundred ng RNA were reverse transcribed using the high-capacity cDNA Reverse Transcription Kit (Applied Biosystems). Obtained cDNAs were stored at −20°C until qRT-PCR analysis. Because of limited material, cDNA of sorted cells and paraffin punches was pre-amplified. Pre-amplification was performed for 14 cycles according to manufacturer’s instructions (TaqMan® PreAmp Master Mix Kit, Applied Biosystems). We tested the correlation of expression analyzed on original cDNA with that analyzed on pre-amplified cDNA for selected highly (CD45, CD68, CD163, TNFα, MHC-II) and lowly (iNOS, FOXP3, IL-10, IDO) expressed genes, and found an excellent correlation for all, confirming uniform pre-amplification for all samples (CD68 and CD163 are shown in Fig. S3). qRT-PCR was done on a Rotor-Gene Q real-time PCR cycler (Qiagen) using commercially available pre-developed TaqMan reagents with optimized primer and probe concentrations (TaqMan® gene expression assays, Applied Biosystems) (Table S1).
After an initial hold for 2 min at 50°C and 10 min at 95°C, probes were cycled 45 times at 95°C for 15 sec and at 60°C for 60 sec. All PCR reactions were performed in triplicates. Threshold cycle (Ct) values were determined with the Rotor-Gene Q Series software 1.7. ΔCt values were calculated by normalizing the target mRNA levels to (1) the endogenous control 18S rRNA (Hs03928990_g1), (2) the endogenous control PPIA mRNA (Hs99999904_m1) or (3) to the levels of CD4 mRNA, (Ct target gene-Ct control gene) as stated in the figure legends. Immune response-related transcripts other than CD45 were normalized to the expression levels of CD45. Because there was no correlation between CD45 transcripts and survival (Fig. 1A), this normalization will not obscure potential correlations between the expression of other immune response-related genes and survival. In some cases ΔCt levels were expressed as relative to the appropriate control. Therefore, ΔΔCt values were determined (ΔCt target gene-ΔCt control) and fold change calculated with the equation 2−ΔΔCt. Ct values > 38 were interpreted such that the gene was not expressed. Changes in the expression levels lower than 2-fold (both as upregulation and as downregulation) were considered as not significant. This range is illustrated as a shaded area in the figures.
Statistical analyses
The relationship between the expression of target genes and patient survival was analyzed using a univariate Cox proportional hazard regression model with 95% confidence intervals. The prognostic effect of variables that showed a correlation with survival in univariate analysis was tested for dependence on age at operation and tumor stage by multivariate Cox regression analysis. Furthermore, data was dichotomized based on mean gene expression value of all analyzed samples, log-rank test performed and Kaplan-Meier curves generated in which probability of overall survival was plotted over time. Correlations between different parameters were assessed using the Spearman Rho correlation test (IBM SPSS statistics software). Wilcoxon matched-pairs signed rank test was used to analyze statistical significance between groups (GraphPad Prism Software). The criterion for significance was set at p ≤ 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Giovanni Sais and Prof.Dr. Maurizio Provenzano (Department of Urology, University Hospital Zurich) for help in setting up the qRT-PCR screen, Dr. Claudia Dumrese (Center for Microscopy and Image Analysis, University Zurich) for assistance in FACS sorting and Prof.Dr.Burkhardt Seifert (Division of Biostatistics, University Zurich) for advice in statistical analysis. This work was supported in part by the Cancer Research Institute/Cancer Vaccine Collaborative, the Hanne Liebermann Foundation, Dr. Leopold and Carmen Ellinger Foundation, Science Foundation Oncology SFO, Swiss National Science Foundation (3238BO, 10314531), the Hartmann Müller Foundation Zurich and Alumni Grant University Zurich.
Glossary
Abbreviations:
- ccRCC
clear cell renal cell carcinoma
- ICS
intracellular cytokine stain
- qRT-PCR
quantitative reverse transcription real-time PCR
- TAM
tumor-associated macrophage
- TIL
tumor-infiltrating leukocyte
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/23562
References
- 1.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 3.Beck C, Schreiber H, Rowley D. Role of TGF-beta in immune-evasion of cancer. Microsc Res Tech. 2001;52:387–95. doi: 10.1002/1097-0029(20010215)52:4<387::AID-JEMT1023>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 4.Kawamura K, Bahar R, Natsume W, Sakiyama S, Tagawa M. Secretion of interleukin-10 from murine colon carcinoma cells suppresses systemic antitumor immunity and impairs protective immunity induced against the tumors. Cancer Gene Ther. 2002;9:109–15. doi: 10.1038/sj.cgt.7700418. [DOI] [PubMed] [Google Scholar]
- 5.Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer. 2006;42:717–27. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Filipazzi P, Huber V, Rivoltini L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol Immunother. 2012;61:255–63. doi: 10.1007/s00262-011-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facciabene A, Motz GT, Coukos G. T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 2012;72:2162–71. doi: 10.1158/0008-5472.CAN-11-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 9.Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18:349–55. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Bailey C, Negus R, Morris A, Ziprin P, Goldin R, Allavena P, et al. Chemokine expression is associated with the accumulation of tumour associated macrophages (TAMs) and progression in human colorectal cancer. Clin Exp Metastasis. 2007;24:121–30. doi: 10.1007/s10585-007-9060-3. [DOI] [PubMed] [Google Scholar]
- 11.Krausgruber T, Saliba D, Ryzhakov G, Lanfrancotti A, Blazek K, Udalova IA. IRF5 is required for late-phase TNF secretion by human dendritic cells. Blood. 2010;115:4421–30. doi: 10.1182/blood-2010-01-263020. [DOI] [PubMed] [Google Scholar]
- 12.Keller R, Geiges M, Keist R. L-arginine-dependent reactive nitrogen intermediates as mediators of tumor cell killing by activated macrophages. Cancer Res. 1990;50:1421–5. [PubMed] [Google Scholar]
- 13.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–44. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 14.Lau SK, Chu PG, Weiss LM. CD163: a specific marker of macrophages in paraffin-embedded tissue samples. Am J Clin Pathol. 2004;122:794–801. doi: 10.1309/QHD6YFN81KQXUUH6. [DOI] [PubMed] [Google Scholar]
- 15.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–92. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 17.Kutikov A, Egleston BL, Wong YN, Uzzo RG. Evaluating overall survival and competing risks of death in patients with localized renal cell carcinoma using a comprehensive nomogram. J Clin Oncol. 2010;28:311–7. doi: 10.1200/JCO.2009.22.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers MM, Heng DY. Prognostic and predictive biomarkers in renal cell carcinoma. Target Oncol. 2010;5:85–94. doi: 10.1007/s11523-010-0143-8. [DOI] [PubMed] [Google Scholar]
- 19.Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med. 1996;335:865–75. doi: 10.1056/NEJM199609193351207. [DOI] [PubMed] [Google Scholar]
- 20.Gouttefangeas C, Stenzl A, Stevanović S, Rammensee HG. Immunotherapy of renal cell carcinoma. Cancer Immunol Immunother. 2007;56:117–28. doi: 10.1007/s00262-006-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morra L, Rechsteiner M, Casagrande S, Duc Luu V, Santimaria R, Diener PA, et al. Relevance of periostin splice variants in renal cell carcinoma. Am J Pathol. 2011;179:1513–21. doi: 10.1016/j.ajpath.2011.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pello OM, De Pizzol M, Mirolo M, Soucek L, Zammataro L, Amabile A, et al. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–21. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- 23.Mulders PF, Brouwers AH, Hulsbergen-van der Kaa CA, van Lin EN, Osanto S, de Mulder PH. [Guideline ‘Renal cell carcinoma’] Ned Tijdschr Geneeskd. 2008;152:376–80. [PubMed] [Google Scholar]
- 24.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gertler F, Condeelis J. Metastasis: tumor cells becoming MENAcing. Trends Cell Biol. 2011;21:81–90. doi: 10.1016/j.tcb.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 28.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–84. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Attig S, Hennenlotter J, Pawelec G, Klein G, Koch SD, Pircher H, et al. Simultaneous infiltration of polyfunctional effector and suppressor T cells into renal cell carcinomas. Cancer Res. 2009;69:8412–9. doi: 10.1158/0008-5472.CAN-09-0852. [DOI] [PubMed] [Google Scholar]
- 30.Kuang DM, Zhao Q, Peng C, Xu J, Zhang JP, Wu C, et al. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206:1327–37. doi: 10.1084/jem.20082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savage ND, de Boer T, Walburg KV, Joosten SA, van Meijgaarden K, Geluk A, et al. Human anti-inflammatory macrophages induce Foxp3+ GITR+ CD25+ regulatory T cells, which suppress via membrane-bound TGFbeta-1. J Immunol. 2008;181:2220–6. doi: 10.4049/jimmunol.181.3.2220. [DOI] [PubMed] [Google Scholar]
- 32.Vasievich EA, Huang L. The suppressive tumor microenvironment: a challenge in cancer immunotherapy. Mol Pharm. 2011;8:635–41. doi: 10.1021/mp1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M, et al. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61:5132–6. [PubMed] [Google Scholar]
- 34.Kondo T, Nakazawa H, Ito F, Hashimoto Y, Osaka Y, Futatsuyama K, et al. Favorable prognosis of renal cell carcinoma with increased expression of chemokines associated with a Th1-type immune response. Cancer Sci. 2006;97:780–6. doi: 10.1111/j.1349-7006.2006.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi S. The origin of FOXP3-expressing CD4+ regulatory T cells: thymus or periphery. J Clin Invest. 2003;112:1310–2. doi: 10.1172/JCI20274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths RW, Elkord E, Gilham DE, Ramani V, Clarke N, Stern PL, et al. Frequency of regulatory T cells in renal cell carcinoma patients and investigation of correlation with survival. Cancer Immunol Immunother. 2007;56:1743–53. doi: 10.1007/s00262-007-0318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adotevi O, Pere H, Ravel P, Haicheur N, Badoual C, Merillon N, et al. A decrease of regulatory T cells correlates with overall survival after sunitinib-based antiangiogenic therapy in metastatic renal cancer patients. J Immunother. 2010;33:991–8. doi: 10.1097/CJI.0b013e3181f4c208. [DOI] [PubMed] [Google Scholar]
- 38.Hori S, Takahashi T, Sakaguchi S. Control of autoimmunity by naturally arising regulatory CD4+ T cells. Adv Immunol. 2003;81:331–71. doi: 10.1016/S0065-2776(03)81008-8. [DOI] [PubMed] [Google Scholar]
- 39.Siddiqui SA, Frigola X, Bonne-Annee S, Mercader M, Kuntz SM, Krambeck AE, et al. Tumor-infiltrating Foxp3-CD4+CD25+ T cells predict poor survival in renal cell carcinoma. Clin Cancer Res. 2007;13:2075–81. doi: 10.1158/1078-0432.CCR-06-2139. [DOI] [PubMed] [Google Scholar]
- 40.Li JF, Chu YW, Wang GM, Zhu TY, Rong RM, Hou J, et al. The prognostic value of peritumoral regulatory T cells and its correlation with intratumoral cyclooxygenase-2 expression in clear cell renal cell carcinoma. BJU Int. 2009;103:399–405. doi: 10.1111/j.1464-410X.2008.08151.x. [DOI] [PubMed] [Google Scholar]
- 41.Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81:1607–13. [PubMed] [Google Scholar]
- 42.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–73. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 43.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 44.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Ropponen KM, Kellokoski JK, Lipponen PK, Eskelinen MJ, Alanne L, Alhava EM, et al. Expression of inducible nitric oxide synthase in colorectal cancer and its association with prognosis. Scand J Gastroenterol. 2000;35:1204–11. doi: 10.1080/003655200750056709. [DOI] [PubMed] [Google Scholar]
- 46.Clear AJ, Lee AM, Calaminici M, Ramsay AG, Morris KJ, Hallam S, et al. Increased angiogenic sprouting in poor prognosis FL is associated with elevated numbers of CD163+ macrophages within the immediate sprouting microenvironment. Blood. 2010;115:5053–6. doi: 10.1182/blood-2009-11-253260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216:15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- 48.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–68. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komohara Y, Hasita H, Ohnishi K, Fujiwara Y, Suzu S, Eto M, et al. Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci. 2011;102:1424–31. doi: 10.1111/j.1349-7006.2011.01945.x. [DOI] [PubMed] [Google Scholar]
- 50.Sato T, Terai M, Tamura Y, Alexeev V, Mastrangelo MJ, Selvan SR. Interleukin 10 in the tumor microenvironment: a target for anticancer immunotherapy. Immunol Res. 2011;51:170–82. doi: 10.1007/s12026-011-8262-6. [DOI] [PubMed] [Google Scholar]
- 51.Daurkin I, Eruslanov E, Stoffs T, Perrin GQ, Algood C, Gilbert SM, et al. Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 2011;71:6400–9. doi: 10.1158/0008-5472.CAN-11-1261. [DOI] [PubMed] [Google Scholar]
- 52.Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. 2010;207:2175–86. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.