Abstract
Automated image analysis of histopathology specimens could potentially provide support for early detection and improved characterization of breast cancer. Automated segmentation of the cells comprising imaged tissue microarrays (TMA) is a prerequisite for any subsequent quantitative analysis. Unfortunately, crowding and overlapping of cells present significant challenges for most traditional segmentation algorithms. In this paper, we propose a novel algorithm which can reliably separate touching cells in hematoxylin stained breast TMA specimens which have been acquired using a standard RGB camera. The algorithm is composed of two steps. It begins with a fast, reliable object center localization approach which utilizes single-path voting followed by mean-shift clustering. Next, the contour of each cell is obtained using a level set algorithm based on an interactive model. We compared the experimental results with those reported in the most current literature. Finally, performance was evaluated by comparing the pixel-wise accuracy provided by human experts with that produced by the new automated segmentation algorithm. The method was systematically tested on 234 image patches exhibiting dense overlap and containing more than 2200 cells. It was also tested on whole slide images including blood smears and tissue microarrays containing thousands of cells. Since the voting step of the seed detection algorithm is well suited for parallelization, a parallel version of the algorithm was implemented using graphic processing units (GPU) which resulted in significant speed-up over the C/C++ implementation.
Keywords: Mean Shift, Level Set, Segmentation, Seed Detection, Parallel Computing
I. Introduction
Breast cancer is one of the most frequently diagnosed cancers in women. Approximately 209, 060 new cases of invasive breast cancer were reported in women in the US during 2010. There is more than a 98% five-year relative survival rate when localized breast cancer is detected before it spreads to other parts of the body [1]. Tissue microarrays (TMAs) are a relatively new technology for arranging small histological sections (histospots) in a matrix configuration on a recipient paraffin block [2], [3]. TMAs provide an efficient method for preserving tissue while facilitating high-throughput of multiple tissue samples in parallel [4], [5], [6], [7], [8]. Digital microscopy is a complementary technology which is now generally accepted as a reliable tool for visualizing [8], archiving and sharing [9], [10], [11] pathology specimens including TMAs.
Segmenting individual cells in digitized histopathology specimens is usually the first step that is required in automatic image analysis. Recently, an unsupervised clustering approach which utilizes both color and texture features to segment prostate cancer specimens was proposed in [12]. A computationally efficient approach that exploits color and differential invariants to assign class posterior probabilities to delineate the epithelial nuclei, stroma and background regions in breast microarray was reported in [13]. Segmentation of color and multispectral images was later proposed by combining spatial clustering and vector level set active contours to assess prostate cancer samples in [14].
Each of these segmentation methods produced good results on regions exhibiting little or no cell crowding, however, they often failed to separate touching cells accurately. The watershed family of algorithms has become one of the most commonly used segmentation methods to address the challenge of touching cells. However the primary limitation of the watershed approaches is that they often result in over segmentation. Some algorithms such as marker-controlled watershed correction [15], [16], rule-based strategies [17], [18], [19] were developed to address this problem by merging over-segmented regions, but it is difficult to derive a generalized rule to merge over-segmented patches across image ensembles. Evolving generalized Voronoi diagrams [15], [20], [21] which utilize image intensity and geometric information was recently investigated to segment 2D and 3D images containing overlapping cells. Li et. al. [22] presented a gradient flow tracking algorithm for segmenting cell nuclei in 3D microscopic images. Although those methods were designed to segment images where the nuclei were closely juxtaposed or touching slightly, they are not suitable for specimens containing large numbers of cells with extensive overlapping areas. A double threshold-based watershed and statistical analysis for clustered cell segmentation was proposed in [17]. This algorithm measures the quality of the resulting segmentation using statistical analysis and provides feedback to correct errors. However this algorithm relies on the assumption that all cells are similar or belong to a limited number of classes or cell types. Wen et. al. [23] later reported a study on decomposing clumps of nuclei using high level geometric constraints derived from maximum curvatures. This approach was very effective in separating touching objects. Unfortunately, its use is somewhat limited because within some touching cells, the common connecting regions do not exhibit local maximal curvatures.
Kothari et. al. [24] proposed a semi-automatic method for touching cell segmentation, which applied concavity detection at the edge of clusters to find the points of overlap between two nuclei. An ellipse-fitting technique was applied to segment the concavities between two nuclei with overlapping regions. However, the ellipse used in these studies is unable to accommodate the shape of some cells, especially irregularly shaped cancerous cells. Diaz et al [25] reported their study to split overlapping cells using template matching. In the paper, maximal correlation points between the overlapping and template shapes were determined by affine registration. The template size varied from 70% to 120% to find the “best match”. Our team [26] proposed an approach to address touching cell segmentation using concave vertex graphs, and several other graph based methods were proposed in [27], [28], [29] to segment touching stem cells in fluorescence microscopy images. Unfortunately, such graph-based methods generally require the image to exhibit a high contrast at the edges of the structure of interest which is often not the case, especially in cancerous regions of the tissue. Elter et. al. [30] had proposed a method called maximum-intensity-linking for segmenting touching cells. This approach is based on the idea of representing an image as a directed graph structure which is significantly faster than classic watershed algorithms, however, it still results in over-segmentation and requires complicated post-processing steps when used in our experiments. Al-Kofahi et. al. [31] reported an automatic segmentation of cell nuclei in histopathology images. Their seed detection results are used as initialization markers for nuclear segmentation. Their approach achieved very good results on heterogeneous regions.
Level set based deformable methods have been widely used for cell segmentation [32], [33], [34] and different terms [35], [36], [37], [38] were inserted into original Mumford-Shah function to try to address the overlapping object segmentation problem. Parametric texture adaptive snakes were proposed in [21] for cell segmentation and tracking. However, for the level set based algorithm or parametric snake to work properly, a good initialization is required to locate each touching object.
A preliminary version of our work was presented in the 2010 High Performance Computing Workshop associated with Medical Image Computing and Computer Aided Intervention [39]. Compared to the shorter conference version which focused on parallel computing, each step of the proposed algorithm is explained in detail in the journal paper. Epithelial region extraction and connect component analysis have been newly introduced in the journal version for more efficient parallelization of cellular level segmentation to support whole slide images. Different tissue types prepared with a range of different stain were used to test the effectiveness of the algorithm through exhaustive experiments. The studies were completely redesigned to compare the proposed algorithm with five state-of-arts segmentation methods. An extensive evaluation of seed detection and the selection of a feasible range of parameters were investigated and added in the journal paper. The algorithm has now been tested on a set of comprehensive, large scale datasets. The contributions are:
A computationally efficient mean shift based single-pass voting algorithm which provides accurate seed detection and robust touching object localization, used as an initialization for the repulsive level set model;
A segmentation framework which can successfully separate cells residing in a densely touching regions. The algorithm has been tested on a large clinical dataset using a range of different tissues and stain preparations.
The algorithm is designed for easy parallelization as a result of data independence. The GPU parallel version of the seed detection part in the algorithm can process a 1392 × 1040 image which contains hundreds of touching cells in less than 0.2 second.
II. Touching Cell Segmentation
We propose a novel algorithm for separating touching cells, which is not limited to a specific type of staining preparation. Breast TMAs and blood smears have been acquired using standard RGB imaging with a 40× magnification objective. Standard hematoxylin staining was applied to breast TMA specimens, and the commonly used Giemsa stain was used to prepare peripheral blood smears. Please note that Giensa stain is a mixture of methylene blue, eosin and azure B. It is the most dependable stain for differentiating nuclear and/or cytoplasmic morphology of platelets, RBCs, WBCs, etc.
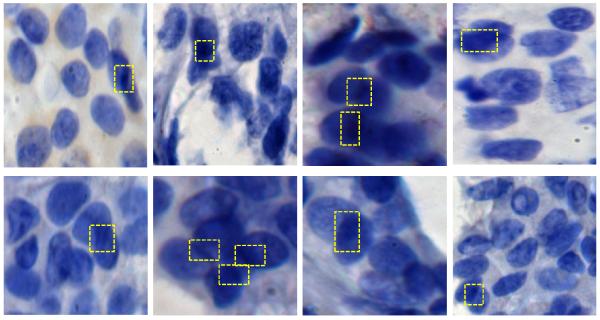
The algorithm was tested on whole slide digitized TMA specimens. In Figure 1 we show the entire procedure describing the method used to automatically zoom in and crop a single disc from the TMA array using our previous algorithm [40]. Figure 2 shows some typical overlapping patches in hemotoxylin stained breast TMA disc. Many patches are shown to contain overlapping regions which are darker than the intensity of non-overlapping regions. The overlapping regions in this figure are marked with yellow rectangles.
Fig. 1.
The procedure on how to automatically zoom in and crop a single disc from the TMA array. The TMA whole slide images were taken under 40× objective using Trestle MedMicro.
Fig. 2.
Representative examples of hemotoxylin stained breast TMA RGB images acquired using a 40× objective using a Nikon Microscope. Some cells overlap with each other and the intensity of overlapping regions are darker than the intensity of non-overlapping regions.The overlapping regions are marked with yellow rectangles.
The touching cell segmentation algorithm that we developed is composed of two steps. The first step is to automatically locate the geometric center of each cell using a novel single-pass voting with mean-shift based seed detection. The result of this step is used as an initial position for the second step which is touching cell segmentation, which extracts each contour of touching cells using a level set function with a repulsion force to penalize any object overlap. The flow chart of whole touching cell segmentation is shown in Figure 3. It contains the epithelium region segmentation [41], seed detection, connect component analysis and the level set based contour extraction. The connect component analysis step is intentionally added into the algorithm. By separating the whole disc into connected components, the algorithm can be run in parallel on multiple cores and the final segmentation result is an ensemble of all the connected components. Graphic processing unit (GPU) was applied to speed up the entire segmentation procedure.
Fig. 3.
The flow chart for the automated segmentation of overlapping cells in hemotoxylin stained breast TMA discs. From left to right, it contains input image; extracted epithelial region of input image; seed detection results with green dots representing the detected seeds; cell contours of the biggest connected component within the epithelial region; final segmentation results of an ensemble of all the connected components.
A. Seed Detection
Since the number and location of cells are not known, a-priori, it is difficult to directly segment cells from microscopic images especially when they touch one another. The geometric centers of cells is considered as a basic perceptual cue that is used by human experts to support the accurate separation of touching cells. Al-Kofahi et. al. [31] reported a distance-constrained LoG filtering method to identify the center of nuclei. Parvin et. al. [42] proposed an iterative voting method which was used to detect the centers of touching cells.
The method in [31] produced excellent results in detecting the nuclear seed points using a distance-map constrained multiscale Laplacian-of-Gaussian filtering. The algorithm could lead to under-segmentation for some homogenous regions with multiple nuclei. Therefore the authors provide a post-processing module which requires human interaction to “fix” some segmentation errors. The method in [42] provides excellent results in detecting the centers in touching cells exhibiting homogenous intensity, however, when the intensity of overlapping regions is brighter (or darker) than the non-overlapping regions within individual cells, a set of false seeds will be created in the overlapping regions. This is not surprising because the voting schema in [42] is biased towards the boundary of the object. The edges of overlapping regions contribute to the creation of false seeds within the overlapping regions using methods by Parvin et. al. [42].We will explain in detail how we are able to address this problem, and we will also show the significant improvement of the new algorithm which applies a shifted Gaussian kernel and mean shift onto single-pass voting to generate more accurate and quicker seed detection, for both synthetic and real testing datasets.
Defining I(x, y) as the original image, the image gradient ∇I(x, y) and the magnitude ||∇I(x, y)|| are subsequently calculated. Because typically the background of original images can be converted to black or darker than the intensity of objects of interest, the direction from outside of object to center of object is negative for this definition. For each pixel (x, y), the voting direction α(x, y) is defined as the negative gradient direction where θ is the angle of the gradient direction with respect to x axis. The voting area A(x, y; rmin, rmax,Δ) of each pixel is defined by a cone-shape with its vertex at (x, y). A cone-shape voting area was chosen for two reasons. First, the center of the cell is far away from its boundary, thus more voting points are located within the region closest to the center rather than within the region closest to the edge of the cell. Second, a cone-shape voting area greatly reduces the time requirement by reducing the number of calculations since there are fewer voting points in total. The rmin, rmax, and voting area of pixel (x, y) are illustrated in Figure 4. We define a 2D Gaussian kernel g(x, y, μx, μy, σ) with its mean (μx, μy) located at the center of the voting area and oriented in the voting direction α(x, y). The shifted Gaussian kernel is defined as
| (1) |
where and . We designed the kernel in this manner so that voting is amplified at the center of the targeted object.
Fig. 4.
Cone shape voting area with the Gaussian kernel overlapped at the center of the voting area.
We define V(x, y; rmin, rmax,Δ) as the voting image, which has the same dimensions as the original image I(x, y). Setting an initialization of V as zero for all pixels (x, y), for each pixel (x, y), we update the voting image in a single-pass approach as
| (2) |
Using this single-pass voting approach, the geometric centers of objects are determined by executing mean shift on the sum of the voting images. The detailed algorithm is listed in Algorithm 1.
Although our work was initially motivated by [42], the method we propose differs because of several significant aspects: 1) For each point (x, y) with high gradient, we define
Algorithm 1.
Single-pass voting with mean shift based seed detection
| 1. Initialize the parameters: d is the estimated average diameter of cells within the image, rmin = 0.5d, rmax = 1.5d, Δ = 30. |
| 2. Calculate the Gaussian blurred gradient image and the orientation of the gradient at each pixel (x, y). Record the set of (x, y) with large gradient magnitude as S. |
| 3. For each point (x, y) ∈ S, calculate the voting image V (x, y) in a single-path way. |
| 4. for R = 0.3, 0.4, …, 0.9 do |
| 5. Record all the points (x, y) in the voting image with voting number larger than max(V (x, y)) × R. |
| 6. end for |
| 7. Sum all the voting images and run mean shift to generate the final list of the seeds. The bandwidth of the mean shift is defined as 1/3 of the estimated average diameter of the cells. |
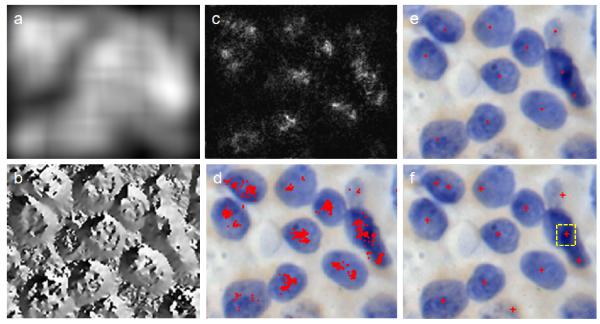
a shifted Gaussian kernel at the center of the voting area instead of (x, y). This is a critical step which enables the new algorithm to provide accurate results for overlapping cells in histopathology specimens. As the center of the object is usually far from the boundary, the shifted Gaussian kernel encourages the voting towards the center of the object and thereby avoids false seeds in overlapping regions (We revisit this issue in the experimental section for clarity). For the overlapping regions, the overlapped edges always exhibit higher gradient intensity values than other regions. The shifted Gaussian kernel causes high voting outside of overlapping regions instead of inside, thus the algorithm reduces false seeds. 2) Instead of using iterative voting as reported in [42], we calculate the centers of overlapping objects by running mean shift on the single-pass voting images. This step dramatically reduces the computational time of processing as many iteration steps are avoided. More importantly, using single-pass voting with mean shift to replace iterative voting facilitates subsequent parallelization. In Figure 5 we show the entire process of the seed detection applying single-pass voting with mean shift to generate the final seed. The false seed on the overlapping region is marked with a yellow square (using diameter = 50, Sigma = 3 and minimal voting = 550 in the most recent release of ImageJ plugin).
Fig. 5.
The whole process of the seed detection applying single-pass voting with mean shift to generate the final seeds. (a) The magnitude of the gradient image. (b) The angular image of the gradient direction with respect to x axis. (c) The summed voting images, and the white points show the number of candidate seed points. (d) The voting points superposed on its original image before mean shift. (e) The final detected seeds superposed on the original image after mean shift. (f) The detected seeds superposed on the original image using Pravin’s algorithm [42].
B. Parallelization of the Seed Detection on the Graphic Processing Unit (GPU)
During the seed detection step, each voting pixel (x, y) utilizes a cone-shaped voting area A(x, y; rmin, rmax, Δ). Each pixel in the final voting image, V, is then updated by equation (2).
After calculating the execution time profile for each step in the proposed seed detection algorithm, we found that the most computationally expensive part is the calculation of the voting image (90% of the whole procedure). Because our voting image is calculated using single-pass with mean shift rather than iterative voting, it is much more computationally efficient. Furthermore, because the voting algorithm is a pixel-based method which has an advantage of easy parallelization as a result of data independence, this part can be accelerated by introducing parallelization on a graphics processing unit (GPU). In our algorithm, we utilized eight blocks for a GPU. Within each block, we create 128 threads. Each voting pixel was assigned to one thread to calculate its corresponding voting image. In total 1024 threads were created simultaneously. In this way the GPU accelerated the voting image calculation dramatically and therefore increased the entire seed detection procedure dramatically.
C. Cell Segmentation
Because of the accuracy of the detected seeds (the geometric centers of cells), the touching cell segmentation process was performed using level set based on an interactive model. The interactive model includes two types of mechanisms: (1) a repulsion term to prevent the contours of adjacent cells from overlapping and separating the touching cell boundaries; (2) the competition term to determine the membership of each pixel which is assigned to the cell producing the smallest difference. Considering an image I that has N cells, let Ci(i = 1, …, N) denote the contours that evolve towards the boundaries. Please note that each cell is represented by its own level set energy function. Instead of examining each contour independently, the interactive between neighboring contours was integrated into the level set energy function. The energy function E for cell segmentation combines the repulsion and competition terms and can be expressed as:
| (3) |
where Ai denotes region of cell {Ai|i = 1, 2, …, N} and Ωb is the background which represents the region outside all the cells out(C1) ∩ out(C2) ∩ … ∩ out(CN). The operator in() and out() represent the regions inside and outside of cells, respectively. The ci and cb are the mean intensities of the cell region and background region respectively. The λ0, λb, and u are the fixed weighting parameters. Function g is chosen to be a sigmoid function
| (4) |
where α is used to control the slope of the output curve and β controls the window size. By penalizing the union of the overlapped region {Ai|i = 1, …, N} enclosed by contours Ci(i = 1, …, N), the last item in Energy function E is the repulsion term which is used to represent the repulsion force between each adjacent touching object and the ω is the regulation parameter.
Segmentation is achieved by minimizing the energy function E using the evolution of the level set. In order to express the energy function using level set, we introduced the regularized Heaviside function H [43]
| (5) |
where ε is the regulation parameter of the Heaviside function and Delta function is defined as
| (6) |
The energy function can be minimized by iteratively employing the gradient descent method. The evolution equation for each energy function Ψi(t, x, y) is then obtained by deducing the associated Euler-Lagrange equation as
| (7) |
After evolving the level set contours, the means ci and cb of the cell and background regions are iteratively updated. This method was proposed and proved to be quite effective and accurate for RNAi fluorescent cellular image segmentation in [38]. Throughout the experiments, the parameters that we selected were: λ0 = 1, λb = 0.3, μ = 0.5, γ = 0.2, ω = 0.6, ε = 1, α = 1, β = 7 empirically.
III. Experimental Results
Hematoxylin stained breast TMA specimen images were captured at a high magnification objective (40 ×) using a Nikon Microscope. In total there were 234 image patches containing more than 2200 image cells.
A. Seed Detection
To illustrate the new seed detection method that we developed, an example of a synthetic image with five overlapping objects is shown in Figure 6. Figure 6a is the original synthetic image, with two and three overlapping cells, respectively. Figure 6b is the seed detection results using the iterative voting method in [42], which created false seeds in two overlapping areas. Figure 6c is the intermediate results of our method before applying mean shift clustering, and Figure 6d is the final detected seeds using our method. From this experiment, it can be seen that the iterative voting method [42] tends to put the seeds at the overlapping regions (shown in Figure 6b) when overlapping regions have brighter/darker intensity than its corresponding touching objects. Using our method as shown in Figure 6d, the detected seeds are approximately located in the centers of the objects and no seeds were misdetected in the overlapping regions. In the real dataset (hematoxylin stained pathology specimens), there are cases where the overlapping areas are darker than the intensity of the non-touching cells as shown in Figure 2.
Fig. 6.
Seed detection results for a representative synthetic image. The red crosses denote detected seeds. (a) the original synthetic image. (b) the seed detection results using the iterative voting method in [42]. (c) the intermediate results of our method. (d) the final detected seeds using our single-pass with mean shift based seed detection method.
In Figure 7, hundreds of cells were accurately detected on the epithelial region. For better illustration, we cropped several patches from the whole dataset and show their comparative seed detection results in Figure 8 using [42] and our method. It is obvious that the method in [42] creates false seeds in the overlapping regions; while the algorithm we developed avoids these errors and provides reliable seed detection results. In Figure 8, the first and the fourth rows are the original hematoxylin stained breast TMA images; the second and fifth rows are the seed detection results using [42]. The regions containing false detected seeds are marked in yellow rectangles; the third and sixth rows are the seed detection result using our method. All of the detected seeds are marked with red crosses. In Figure 9 we show the seed detection results on the whole slide scanned blood smear slide where thousands of cells are successfully detected. The blood smear image was acquired using 40× objective on a Trestle MedMicro.
Fig. 7.
The TMA whole disc seed detection results using our proposed method. The green dots represent the detected seeds on a whole TMA breast core acquired using a 10× objective. Please note that these are the seed detection results overlayed on the segmented epithelial region mask as was shown in the first step in Figure 3. Those seeds falling outside the epithelial region masks were intentionally excluded and are not the result of a missed detection.
Fig. 8.
Seed detection results of hematoxylin stained pathology specimens. The first and the fourth rows are the region of interest (ROI) of the original images; the second and fifth rows are the seed detection results using [42]; the third and sixth rows are the seed detection results using our method. The seeds are marked with red color. The false detected seeds are marked with yellow rectangles.
Fig. 9.
The whole slide blood smear seed detection results using our proposed method. The stained blood smear image was taken under 40× objective using Trestle MedMicro. Even the image contains thousands of cells, we can still provide accurate detection results.
In order to gauge the accuracy of seed detection, an error function is defined as the pixelwise distance E between seeds manually located and seeds extracted by seed detection methods. Table I shows the quantitative results of our algorithm and [42] both compared with the ground truth annotation. The 80% column in Table I represents the sorted 80% accuracy among all the seed detection results. Meanwhile a number of missing and false seeds are calculated. When compared with the ground-truth annotation, the mean of the number of missing seeds and false seeds using our algorithm were 0.2 and 0.8 respectively. The mean of the number of missing seeds and false seeds using [42] were 1 and 1.2, respectively.
TABLE I.
The quantification of the pixelwise seed detection errors of our algorithm and ([42]), both compared with the ground truth annotation of the seeds.
| Mean | Variance | Median | Min | Max | 80% | |
|---|---|---|---|---|---|---|
| Pixelwise detection error E (Our method) | 6.63 | 4.53 | 6.81 | 3.05 | 10.31 | 5.08 |
| Pixelwise detection error E ([42]) | 7.46 | 7.29 | 7.75 | 3.32 | 12.55 | 5.23 |
Due to the minimal diameter rm in of cells and the bandwidth of mean shift played a significant role in our seed detection method, the error function defined above was calculated to test the parameter sensitivity of the algorithm. The range of minimal diameter is defined as minus and plus 10 of the estimated minimal diameter of the cell. The range of the bandwidth of mean shift is from 5 to 19. Figure 10 shows three examples of the average errors respect to the minimal diameter of cells (horizontal axis) and the bandwidth of mean shift (vertical axis). Each row represents one test image and its mean pixelwise detection errors with respect to two parameters: the minimal diameter of cells and the bandwidth of mean-shift. From the results in Figure 10, it is apparent that the errors are relatively insensitive to specific values. Within a feasible range, an average error smaller than 5 pixels is achieved.
Fig. 10.
Three examples of the average error respect to the minimal diameter of cells and the bandwidth of mean shift. The first column is the RGB images; the second column is their corresponding mean errors with respect to the minimal diameter of cells and the bandwidth of mean shift. Within a feasible range, an average error smaller than 5 pixels can be achieved.
B. Segmentation
In Figure 11, the performance of the proposed touching cell segmentation method is compared with the level-set based on an interactive model using the seeds detected by [42]. Each column represents two testing samples. The contours are presented using red lines. Yellow squares are over-segmented regions that arise due to false seeds. The first and the fifth rows are the original hematoxylin stained pathology specimens; the second and the sixth rows are the ground-truth annotation as provided by human experts; the third and seventh rows are the segmentation results generated by the interactive level set using the seed provided by [42]; the fourth and eighth rows are the segmentation results using single-pass with mean shift based seed detection and interactive level set.
Fig. 11.
Segmentation results illustrated using eight images of hematoxylin stained pathology specimens. Each column represents two testing samples. The contours are presented using red lines. Yellow squares are over-segmented regions due to false seeds. The first and the fifth rows are the original hematoxylin stained pathology specimens; the second and the sixth rows are the ground truth annotation by human experts; the third and seventh rows are the segmentation results by interactive level set using the seeds provided by [42]; the fourth and eighth rows are the segmentation results using single-pass with mean shift based seed detection and interactive level set.
In order to quantitatively measure the accuracy of this approach, precision (P ) and recall (R) [44] were calculated for the newly developed touching cell segmentation algorithm and [42], both were compared with the ground-truth annotations. The P is defined as the intersection between the segmentation results and the manually annotation results divided by the segmentation results. The R is defined as the intersection between the segmentation results and manually annotation results divided by the manually annotation results. The mean and standard deviation of P and R for our touching cell segmentation algorithm is 0.90 ± 0.02 and 0.78 ± 0.01, respectively, which indicates a good agreement between the manual and ground truth annotations. The mean and standard deviation of P and R for interactive level set using the seed provided by [42] is 0.84 ± 0.04 and 0.64 ± 0.02, respectively.
In Figure 12, we provide the segmentation results on a whole TMA disc, for illustration purposes we zoom in and crop several representative patches. In Figure 13, we show four representative patch image segmentation results of our method as compared to five other algorithms including marker-based watershed, mean shift, isoperimetric [45] and two methods presented in [42] and [31], respectively. Using our seeds, marker-based watershed tends to over-segment the whole cell in the image as it can’t handle the intensity variation within the cells. Mean shift, Isoperimetric [45] and method in [31] segmentation tends to under-segment the image by merging multiple cells into one object. Method in [42] tends to create false seeds in the overlapping region of cells, which leads to improperly segment cells in the overlapping regions. All the source codes or binaries of the other state-of-the-art method were implemented by their original authors and downloaded from their websites. In Table II, we show the detailed statistics of the quantitative segmentation results compared with the ground truth annotation. The 80% column in Table II represents the sorted 80% accuracy among all the 234 image patches, which contains around 2200 cells. From Table II, we achieved better segmentation result compared with other methods because of the reduction of false seeds.
Fig. 12.
The TMA whole disc segmentation results on epithelial regions. The first row is the hematoxylin stained whole disc breast TMA. The second row consists of regions which have been enlarged for detailed viewing.
Fig. 13.
The comparative segmentation results on four representative images patches. (a) The original image patches. (b) The marker-based watershed segmentation results using our detected seeds. (c) The mean shift results (d) The isoperimetric segmentation results [45]. (e) The segmentation results using method presented in [42]. (f) The segmentation results using the method described in [31]. (g) Segmentation results using the proposed method.
TABLE II.
The segmentation accuracy compared with ground truth annotation, the watershed represents the marker based watershed algorithm.
| Precision(P) | Mean | Variance | Median | Min | Max | 80% | Recall(R) | Mean | Variance | Median | Min | Max | 80% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Our method | 0.90 | 0.02 | 0.94 | 0.22 | 1.00 | 1.00 | Our method | 0.78 | 0.01 | 0.80 | 0.20 | 0.96 | 0.85 |
| Iterative Voting [42] | 0.84 | 0.04 | 0.93 | 0.29 | 1.00 | 1.00 | [42] | 0.64 | 0.02 | 0.65 | 0.28 | 0.90 | 0.75 |
| Graph-cut & Coloring [31] | 0.82 | 0.04 | 0.90 | 0.18 | 1.00 | 0.99 | [31] | 0.81 | 0.02 | 0.87 | 0.20 | 0.99 | 0.93 |
| Isoperimetric [45] | 0.87 | 0.04 | 0.95 | 0.23 | 1.00 | 1.00 | [45] | 0.76 | 0.02 | 0.80 | 0.23 | 0.95 | 0.87 |
| Mean-shift | 0.73 | 0.07 | 0.85 | 0.17 | 1.00 | 0.99 | Mean-shift | 0.78 | 0.02 | 0.81 | 0.26 | 0.96 | 0.87 |
| Watershed | 0.73 | 0.03 | 0.99 | 0.07 | 1.00 | 1.00 | Watershed | 0.52 | 0.06 | 0.54 | 0.03 | 0.93 | 0.78 |
The code was parallelized on a Graphic Processing Unit (GPU). GPU is a massively parallel multi-core chip that can execute thousands of concurrent threads. The GPU cores (also called stream processors) are grouped into several streaming multiprocessors. They are managed by the thread manager. The GPU used in these experiments was a NVIDIA Quadro FX5800 which has 240 cores and 30 streaming multiprocessors, each of which contains 8 GPU cores. It supports both single and double float point precision; offers 933 GFlops single precision; and has memory bandwidth with 102 GB/sec. Unlike CPU threads that are heavy weight, GPU threads are light weight with little creation overhead, instant switching, and instruction and memory latency hiding. The Compute Unified Device Architecture (CUDA) environment from NVIDIA Corporate was utilized throughout the parallel implementations.
The experimental results show significant speed up (22 times faster when compared with the sequential implementation on CPU and thousands of times faster than the original Matlab implementation). In the parallel version of the algorithm, we can complete the seed detection procedure for an image with dimensionality 1392 × 1040 in 197 millisecond. The final repulsive level set algorithm, based on a C/C++ implementation, can process images in less than 5 seconds.
IV. Discussion and Conclusion
In this paper we present a touching cell segmentation algorithm using challenging clinical datasets to test performance. Accurate seed detection was shown to be a prerequisite for accurate segmentation. We demonstrated that single-pass coupled with mean shift based voting algorithm can accurately detect the centers of regions containing densely touching and overlapping cells.
The method in [42] was shown to achieve very good results for homogenous touching objects, even though it was sometimes necessary to tune the parameters. However, when we compared the newly developed seed detection method with the one in [42], where there existed touching cells with non-homogeneous intensities, the new method produced much better results. The accurate seed detection is the prerequisite to achieve an accurate cellular segmentation. We conducted comparison experiments using our method and Parvin’s [42] approach using the software posted on their group’s website at http://www.vision.lbl.gov/. These studies showed that our seed detection algorithm was not sensitive to parameters initialization as shown in Figure 10. Within a feasible range, an average errors smaller than 5 pixels can be achieved.
The new segmentation algorithm is not sensitive to the parameters in equation (7) [38]. Our experiments shown that there is only a slight variation in the performance even if the value of the repulsive weight ω changes more than 50%. Meanwhile, the algorithm performance varies less than 5% when the values of the parameters λ0 and λb vary about 20%. In addition, the parameters μ and γ play minor roles in the performance of the algorithm. However, due to the fact that active contours are local models, they are not robust to the initial positions which are determined by the location of the seeds. That is the central reason why the accurate seed detection is necessary before the level set method is applied to the cell segmentation.
For the cell overlapping regions, the edges have dramatic intensity variation. For the regions within the cells, the intensity variation within the cells is much milder compared to the intensity variation of overlapping regions. The third term in the equation (7) enhances the edge effects and the fourth term smoothes the contours, which is not sensitive to both noise and mild intensity variation within the cells, so that the algorithm can separate touching cells to create smooth and complete contour of each cell. Watershed algorithm can not handle intensity variation within cells very well, which leads to incomplete and jagged contours of touching cells. Consequently, the interactive model outperforms the watershed algorithm even when the watershed is provided with accurate seeds for initialization.
Given these estimated seeds, a level set active contour based on the interactive model can effectively separate each of touching cells. The improved segmentation results are achieved by accurately estimating the seeds (number of cells) and utilizing a repulsion term in the level set energy function to separate the touching boundaries. The method is automated requiring very little prior knowledge. Therefore it can be extended to other touching object segmentation applications including a broad range of applications requiring accurate spatial localization of multiple biomarkers which have been tagged with immunostains or quantum dot conjugates. The GPU implementation of the seed detection algorithm can handle one 2D image (1392 × 1040) in less than 0.2 second. The experimental results show that GPU is an efficient parallel platform for the proposed novel algorithm.
Acknowledgement
This research was funded, in part, by grants from the NIH through contract 9R01CA156386-05A1 from the National Cancer Institute and contracts 5R01LM009239-03 and 3R01LM009239-03S2 from the National Library of Medicine. Additional funds were provided by IBM through a Shared University Research Award. This research is also partially supported by UMDNJ Foundation grant #66-09.
Contributor Information
Xin Qi, Dept. of Pathology and Laboratory Medicine, the Center for Biomedical Imaging & Informatics, and The Cancer Institute of New Jersey, UMDNJ-Robert Wood Johnson Medical School.
Fuyong Xing, Department of Electrical and Computer Engineering, Rutgers University, Piscataway, NJ, 08854.
David J. Foran, Dept. of Pathology and Laboratory Medicine, the Center for Biomedical Imaging & Informatics, and The Cancer Institute of New Jersey, UMDNJ-Robert Wood Johnson Medical School.
Lin Yang, Department of Radiology and the Center for Biomedical Imaging & Informatics, UMDNJ-Robert Wood Johnson Medical School, Piscataway, NJ, 08854.
References
- [1].Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics: 2010. CA Cancer Journal Clin. 2010 [Google Scholar]
- [2].Kononen J, Bubendorf L, Kallionimeni A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallionimeni OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature Medicine. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- [3].Rimm DL, Camp RL, Charette LA, Costa J, Olsen DA, Reiss M. Tissue microarray: A new technology for amplification of tissue resources. Cancer Journal. 2001;7:24–31. [PubMed] [Google Scholar]
- [4].Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab. Invest. 2000;80:1943–1949. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- [5].Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Human Molecular Genetics. 2001;10:657–662. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- [6].Parker RL, Huntsman DG, Lesack DW, Cupples JB, Grant DR, Akbari M, Gilks CB. Assessment of interlaboratory variation in the immunohistochemical determination of estrogen receptor status using a breast cancer tissue microarray. American Journal of Clinical Pathology. 2002;117:723–728. doi: 10.1309/PEF8-GL6F-YWMC-AG56. [DOI] [PubMed] [Google Scholar]
- [7].El-Rehim DMA, Ball G, Pinder SE, Rakha E, Paish C, Robertson JFR, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cdna expression analyses. Int. J. Cancer. 2005;116:340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- [8].Iorio MV, Ferracin M, Liu C, Veronese A, Spizzo R, Sabbioni EMS, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. Microrna gene expression deregulation in human breast cancer. Cancer Research. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- [9].Afework A, Beynon MD, Bustamante F, Cho S, Demarzo A, Ferreira R, Miller R, Silberman M, Saltz J, Sussman A, Tsang H. Digital dynamic telepathology - the virtual microscope. Proc AMIA Symp. 1998:912–916. [PMC free article] [PubMed] [Google Scholar]
- [10].Molnar B, Berczi L, Diczhazy C, Tagscherer A, Varga SV, Szende B, Tulassay Z. Digital slide and virtual microscopy based on routine and telepathology evaluation of routine gastrointestinal biopsy specimens. Journal of Clinical Pathology. 2003;56:433–438. doi: 10.1136/jcp.56.6.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lundin M, Lundin J, Isola J. A digital atlas of breast histopathology: an application of web based virtual microscopy. Journal of Clinical Pathology. 2004;57:1288–1291. doi: 10.1136/jcp.2004.018739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Datar M, Padfield D, Cline H. Color and texture based segmentation of molecular pathology images using hsoms. IEEE International Symposium on Biomedical Imaging. 2008:292–295. [Google Scholar]
- [13].Amaral T, McKenna S, Robertson K, Thompson A. Classification of breast-tissue microarry spots using colour and local invariants. IEEE International Symposium on Biomedical Imaging. 2008:999–1002. [Google Scholar]
- [14].Hafiane A, Bunyak F, Palaniappan K. Evaluation of level set-based histology image segmentation using geometric region criteria. IEEE International Symposium on Biomedical Imaging. 2009:1–4. [Google Scholar]
- [15].Zhou X, Liu KY, Bradley N, Perrimon N, Wong ST. Towards automated cellular image segmentation for rnai genome-wide screening. Medical Image Computing and Computer Assisted Intervention. 2005:885–892. doi: 10.1007/11566465_109. [DOI] [PubMed] [Google Scholar]
- [16].Rodríquez R, Alarcón TE, Pacheco O. A new strategy to obtain robust markers for blood vessels segmentation by using the watersheds method. Computers in Biology and Medicine. 2005;35(8):665–686. doi: 10.1016/j.compbiomed.2004.06.003. [DOI] [PubMed] [Google Scholar]
- [17].Wahlby C, Lindblad J, Vondrus M, Bengtsson E, Bjorkesten L. Algorithms for cytoplasm segmentation of fluorescence labelled cells. Analytical Cellular Pathology. 2002;24:101–111. doi: 10.1155/2002/821782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin G, Chawla MK, Olson K, Guzowski JF, Barnes CA, Roysam B. Hierarchical, model-based merging of multiple fragments for improved three-dimensional segmentation of nuclei. Cytometry. 2005;63:20–33. doi: 10.1002/cyto.a.20099. [DOI] [PubMed] [Google Scholar]
- [19].Yu W, Lee H, Hariharan S, Bu WY, Ahmed S. Quantitative neurite outgrowth measurement based on image segmentation with topological dependence. Cytometry. 2009;75:289–297. doi: 10.1002/cyto.a.20664. [DOI] [PubMed] [Google Scholar]
- [20].Yu W, Lee HK, Hariharan S, Bu W, Ahmed S. Evolving generalized voronoi diagrams for accurate cellular image segmentation. Cytometry. 2010;77:379–386. doi: 10.1002/cyto.a.20876. [DOI] [PubMed] [Google Scholar]
- [21].Jones TR, Carpenter A, Golland P. Voronoi-based segmentation of cell on image manifolds. Computer Vision for Biomedical Image Applications. 2005;3765:535–543. [Google Scholar]
- [22].Li G, Liu T, Tarokh A, Nie J, Guo L, Mara A, Holley S, Wong STC. 3d cell nuclei segmentation based on gradient flow tracking. Bio Med Central Cell Biology. 2007;8:1–10. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wen Q, Chang H, Parvin B. A delaunary triangulation approach for segmenting clumps on nuclei. IEEE International Symposium on Biomedical Imaging. 2009:9–12. [Google Scholar]
- [24].Kothari S, Chaudry Q, Wang WD. Automated cell counting and cluster segmentation using convavity detection and ellipse fitting techniques. IEEE International Symposium on Biomedical Imaging. 2009:795–798. doi: 10.1109/ISBI.2009.5193169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Diaz G, Gonzalez F, Romero E. Automatic clump splitting for cell quantification in microscopical images. Progress in Pattern Recognition, Image Analysis and Applications. 2007;1:763–772. [Google Scholar]
- [26].Yang L, Tuzel O, Meer P, Foran DJ. Automatic image analysis of histopathology specimens using concave vertex graph. International Conference on Medical Image Computing and Computer Assisted Intervention. 2008;5241:833–841. doi: 10.1007/978-3-540-85988-8_99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Faustino GM, Gattass M, Rehen S, Lucena CJP. Automatic embryonic stem cells detection and counting method in fluorescence microscopy images. IEEE International Symposium on Biomedical Imaging. 2009:799–802. [Google Scholar]
- [28].Chen C, Li H, Zhou X, Wong STC. Constraint factor graph cut-based active contour method for automated cellular image segmentation in rnai screening. Journal of Microscopy. 2008;230:177–191. doi: 10.1111/j.1365-2818.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nasr-Isfahani S, Mirasfian A, Masoudi-Nejad A. A new approach for touching cells segmentation. International Conference on BioMedical Engineering and Informatics. 2008;1:816–820. [Google Scholar]
- [30].Elter M, Daum V, Wittenberg T. Maximum-intensity-linking for segmentation of fluorescence-stained cells. Proceedings of Microscopic Image Analysis with Applications in Biology. 2006:46–50. [Google Scholar]
- [31].Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Transaction on Biomedical Engineering. 2010;57(4):841–852. doi: 10.1109/TBME.2009.2035102. [DOI] [PubMed] [Google Scholar]
- [32].Malladi R, Sethian JA, Vermuri BC. Shape modeling with front propagation: A level set approach. IEEE Trans. Pattern Anal. Mach. Intell. 1995;17:158–174. [Google Scholar]
- [33].Zhao HK, Chan TF, Merriman B, Osher S. A variational level set approach to multiphase motion. J. Comput. Phys. 1996;127:179–195. [Google Scholar]
- [34].Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: algorithms based on Hamilton-Jacobi formulations. J. Comput. Phys. 1998;79:12–49. [Google Scholar]
- [35].Vese LA, Chan TF. A multiphase level set framework for image segmentation using the mumford and shah model. International Journal of Computer Vision. 2002;50:271–293. [Google Scholar]
- [36].Zhang B, Zimmer C, Olivo-Marin JC. Tracking fluorescent cells with coupled geometric active contours. International Symposium on Biomedical Imaging. 2004;1:476–479. [Google Scholar]
- [37].Dufour A, Shinin V, Tajbakhsh S, Guillen-Aghion N, Olivo-Marin JC, Zimmer C. Segmenting and tracking fluoresent cells in dynamic 3D microscopy with coupled active surfaces. IEEE Transaction on Image Processing. 2005;14:1396, 1410. doi: 10.1109/tip.2005.852790. [DOI] [PubMed] [Google Scholar]
- [38].Yan P, Zhou X, Shah M, Wong STC. Automatic segmentation of high-throughput RNAi fluorescent cellular images. IEEE Transaction on Information Technology in Biomedicine. 2008;12:109–117. doi: 10.1109/TITB.2007.898006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Qi X, Xing F, Foran DJ, Yang L. Gpu enabled parallel touching cell segmentation using mean shift based seed detection and repulsive level set. High Performance Computing (HP) workshop associated with Proc. International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); 2010. [Google Scholar]
- [40].Chen W, Reiss M, Foran DJ. A prototype for unsupervised analysis of tissue microarrays for cancer research and diagnostics. IEEE Transactions on Information Technology in Biomedicine. 2004;8(2):89–96. doi: 10.1109/titb.2004.828891. [DOI] [PubMed] [Google Scholar]
- [41].Foran DJ, Yang L, Tuzel O, Chen W, Hu J, Kurc T, Ferreira R, Saltz J. A cagrid-enabled, learning based image segmentation method for histopathology specimens. International Symposium of Biomedical Imaging; 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Parvin B, Yang Q, Han J, Chang H, Rydberg B, Barcellos-Hoff MH. Iterative voting for inference of structural saliency and characterization of subcellular events. IEEE Transactions on Image Processing. 2007;16:615–623. doi: 10.1109/tip.2007.891154. [DOI] [PubMed] [Google Scholar]
- [43].Chan TF, Vese LA. Active contours without edges. IEEE Transaction on Image Processing. 2001;10:266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- [44].Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. 1st ed Addison Wesley; 2005. [Google Scholar]
- [45].Grady L, Schwartz EL. Isoperimetric graph partitioning for image segmetentation. IEEE Transaction on Pattern Analysis and Machine Intelligence. 2006;28(1):469–475. doi: 10.1109/TPAMI.2006.57. [DOI] [PubMed] [Google Scholar]