Abstract
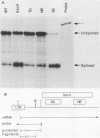
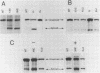
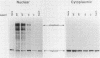
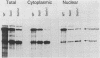
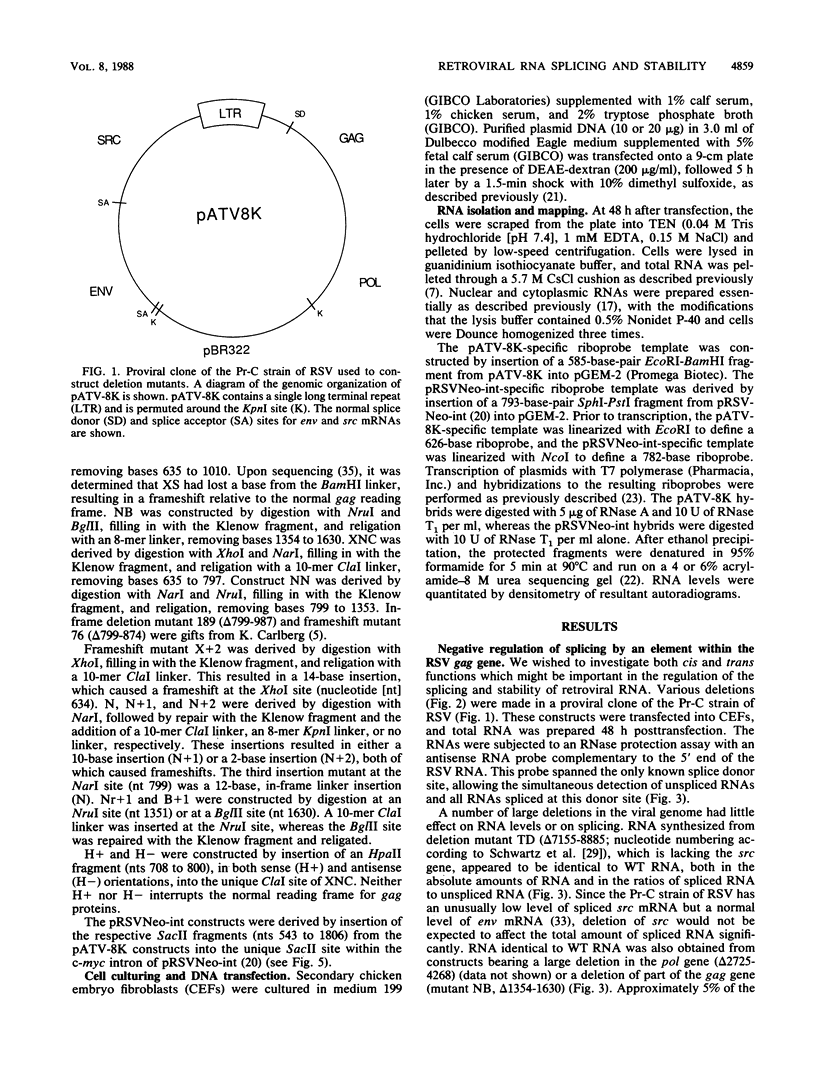
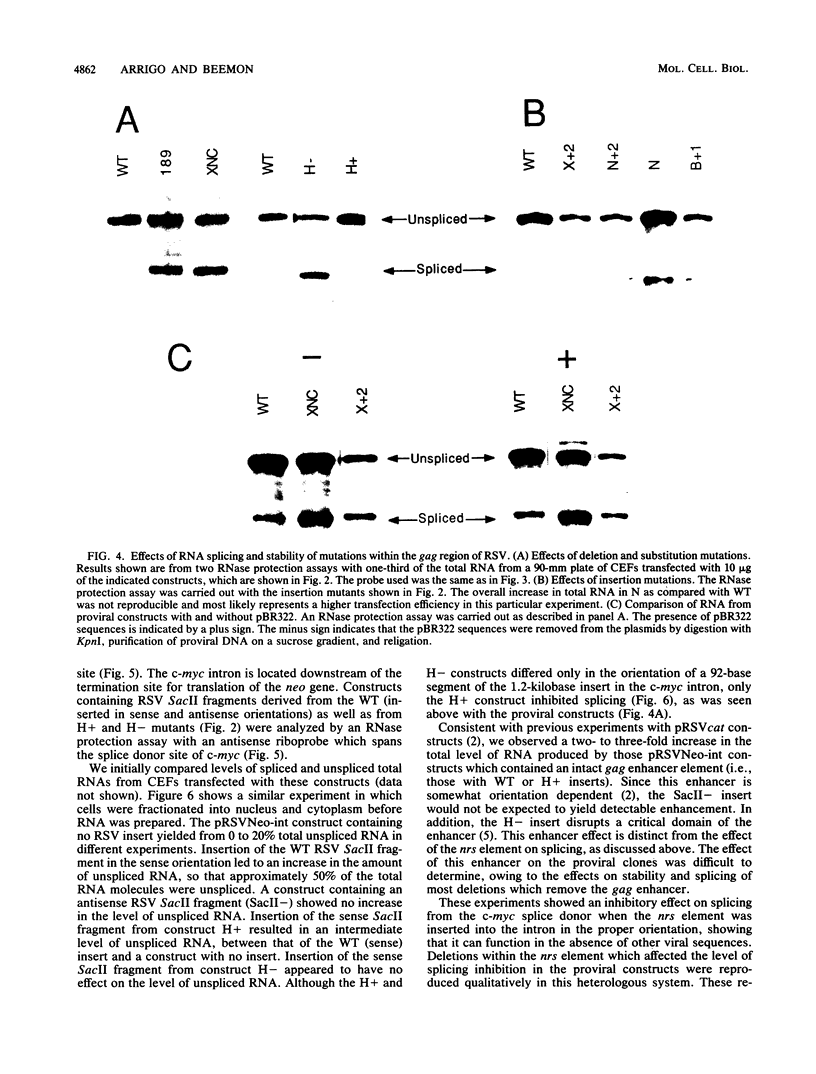
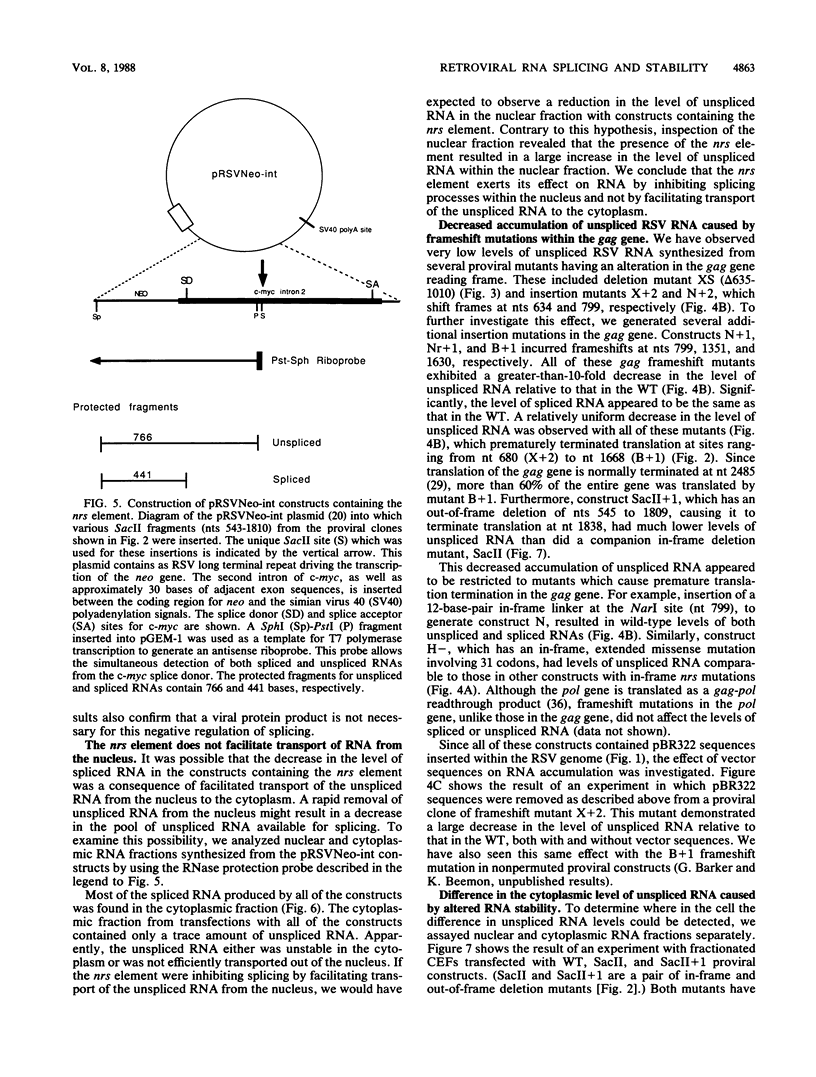
Only a fraction of retroviral primary transcripts are spliced to subgenomic mRNAs; the unspliced transcripts are transported to the cytoplasm for packaging into virions and for translation of the gag and pol genes. We identified cis-acting sequences within the gag gene of Rous sarcoma virus (RSV) which negatively regulate splicing in vivo. Mutations were generated downstream of the splice donor (base 397) in the intron of a proviral clone of RSV. Deletion of bases 708 to 800 or 874 to 987 resulted in a large increase in the level of spliced RSV RNA relative to unspliced RSV RNA. This negative regulator of splicing (nrs) also inhibited splicing of a heterologous splice donor and acceptor pair when inserted into the intron. The nrs element did not affect the level of spliced RNA by increasing the rate of transport of the unspliced RNA to the cytoplasm but interfered more directly with splicing. To investigate the possible role of gag proteins in splicing, we studied constructs carrying frameshift mutations in the gag gene. While these mutations, which caused premature termination of gag translation, did not affect the level of spliced RSV RNA, they resulted in a large decrease in the accumulation of unspliced RNA in the cytoplasm.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armentano D., Yu S. F., Kantoff P. W., von Ruden T., Anderson W. F., Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987 May;61(5):1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo S., Yun M., Beemon K. cis-acting regulatory elements within gag genes of avian retroviruses. Mol Cell Biol. 1987 Jan;7(1):388–397. doi: 10.1128/mcb.7.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2056–2060. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome S., Gilbert W. Rous sarcoma virus encodes a transcriptional activator. Cell. 1985 Mar;40(3):537–546. doi: 10.1016/0092-8674(85)90202-8. [DOI] [PubMed] [Google Scholar]
- Carlberg K., Ryden T. A., Beemon K. Localization and footprinting of an enhancer within the avian sarcoma virus gag gene. J Virol. 1988 May;62(5):1617–1624. doi: 10.1128/jvi.62.5.1617-1624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. S., Temin H. M. Substitution of 5' helper virus sequences into non-rel portion of reticuloendotheliosis virus strain T suppresses transformation of chicken spleen cells. Cell. 1982 Nov;31(1):111–120. doi: 10.1016/0092-8674(82)90410-x. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cizdziel P. E., de Mars M., Murphy E. C., Jr Exploitation of a thermosensitive splicing event to study pre-mRNA splicing in vivo. Mol Cell Biol. 1988 Apr;8(4):1558–1569. doi: 10.1128/mcb.8.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Katz R., Terry R., Skalka A. M., Leis J. Avian sarcoma and leukosis virus pol-endonuclease recognition of the tandem long terminal repeat junction: minimum site required for cleavage is also required for viral growth. J Virol. 1987 Jun;61(6):1999–2008. doi: 10.1128/jvi.61.6.1999-2008.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Kopchick J. J., Stacey D. W. Effect of intron size on splicing efficiency in retroviral transcripts. Nucleic Acids Res. 1982 Oct 11;10(19):6177–6190. doi: 10.1093/nar/10.19.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner A. L., Bernstein A. Retrovirus transduction: segregation of the viral transforming function and the herpes simplex virus tk gene in infectious Friend spleen focus-forming virus thymidine kinase vectors. Mol Cell Biol. 1983 Dec;3(12):2191–2202. doi: 10.1128/mcb.3.12.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. E., Beemon K. Inhibition of methylation at two internal N6-methyladenosine sites caused by GAC to GAU mutations. J Biol Chem. 1987 Mar 5;262(7):3422–3427. [PubMed] [Google Scholar]
- Katz R. A., Kotler M., Skalka A. M. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988 Aug;62(8):2686–2695. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Linial M. Creation of a processed pseudogene by retroviral infection. Cell. 1987 Apr 10;49(1):93–102. doi: 10.1016/0092-8674(87)90759-8. [DOI] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. K., Embretson J. E., Temin H. M. Transforming viruses spontaneously arise from nontransforming reticuloendotheliosis virus strain T-derived viruses as a result of increased accumulation of spliced viral RNA. J Virol. 1988 Apr;62(4):1219–1226. doi: 10.1128/jvi.62.4.1219-1226.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. Insertion of several different DNAs in reticuloendotheliosis virus strain T suppresses transformation by reducing the amount of subgenomic mRNA. J Virol. 1986 Apr;58(1):75–80. doi: 10.1128/jvi.58.1.75-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C., Spahr P. F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986 Nov;60(2):450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M., Lorenzen S. K., Berberich S. L. Noncoding region between the env and src genes of Rous sarcoma virus influences splicing efficiency at the src gene 3' splice site. J Virol. 1987 Jan;61(1):177–184. doi: 10.1128/jvi.61.1.177-184.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M. Synthesis and processing of avian sarcoma retrovirus RNA. Adv Virus Res. 1988;35:1–38. doi: 10.1016/s0065-3527(08)60707-1. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. mRNA precursor splicing in vivo: sequence requirements determined by deletion analysis of an intervening sequence. Proc Natl Acad Sci U S A. 1985 May;82(9):2885–2889. doi: 10.1073/pnas.82.9.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]