Abstract
G protein-coupled receptors (GPCRs) are critical for cardiovascular physiology. Cardiac cells express >100 nonchemosensory GPCRs, indicating that important physiological and potential therapeutic targets remain to be discovered. Moreover, there is a growing appreciation that members of the large, distinct taste and odorant GPCR families have specific functions in tissues beyond the oronasal cavity, including in the brain, gastrointestinal tract and respiratory system. To date, these chemosensory GPCRs have not been systematically studied in the heart. We performed RT-qPCR taste receptor screens in rodent and human heart tissues that revealed discrete subsets of type 2 taste receptors (TAS2/Tas2) as well as Tas1r1 and Tas1r3 (comprising the umami receptor) are expressed. These taste GPCRs are present in cultured cardiac myocytes and fibroblasts, and by in situ hybridization can be visualized across the myocardium in isolated cardiac cells. Tas1r1 gene-targeted mice (Tas1r1Cre/Rosa26tdRFP) strikingly recapitulated these data. In vivo taste receptor expression levels were developmentally regulated in the postnatal period. Intriguingly, several Tas2rs were upregulated in cultured rat myocytes and in mouse heart in vivo following starvation. The discovery of taste GPCRs in the heart opens an exciting new field of cardiac research. We predict that these taste receptors may function as nutrient sensors in the heart.
Introduction
G protein-coupled receptors (GPCRs) are seven transmembrane-spanning proteins that mediate cellular and physiological responses by converting extracellular stimuli into intracellular signals. GPCRs represent the largest receptor superfamily in the genome, recognizing and binding an array of sensory input and ligands, including photons, ions, odors/tastes, bioamines, lipids, peptides and proteins [1]. Many of the ligands for these receptors, including norepinephrine/epinephrine, endothelin and angiotensin have profound homeostatic and regulatory effects on the cardiovascular system. Not surprisingly, mutations and modifications of GPCRs, G proteins and their regulatory partners are linked to dysfunction and disease, with an estimated 40% of all drugs on the market eliciting their activity through GPCRs [2]. Although cardiovascular therapeutics are well established clinically, they target a very small fraction of cardiac-expressed GPCRs. Moreover, conservative estimates are that the heart expresses upwards of 100 different nonchemosensory GPCRs, yet over 30% of these have no known endogenous ligand, indicating that much biology and many potential targets remain to be discovered [3], [4].
A case-in-point is the chemosensory (odorant and taste) receptors, which account for over half of the GPCR repertoire. Previously considered exclusive mediators of olfaction and taste, these large GPCR families have been generally neglected as drug targets, with the exception of the fragrance and food industries [1]. However, it is becoming evident that chemosensory receptors are expressed in diverse tissues, where they perform additional functions and could represent important therapeutic targets. For instance, in the mouth, the taste receptor type 1 (TAS1 in humans; Tas1 in rodents) family sense the nutrient content of food and mediate sweet (TAS1R2-TAS1R3) and umami (TAS1R1-TAS1R3) taste. In other organ systems, such as in the brain [5] and gastrointestinal tract [6], [7], [8], Tas1 receptors have also been implicated in nutrient sensing and regulation of hormone release. In addition, intriguing recent work suggests that Tas1 GPCRs act as direct sensors to communicate amino acid availability to the mammalian target of rapamycin complex 1 (mTORC1) and regulate autophagy [9].
The taste receptor type 2 (TAS2/Tas2) GPCRs encode a family of ∼30 highly divergent receptors that mediate bitter taste [1]. Tas2rs expressed in taste papillae in the tongue detect and respond to a large number of structurally diverse aversive and toxic compounds, resulting in canonical avoidance and rejection responses [10]. Beyond the mouth, the Tas2rs are enigmatic, having been implicated in several distinct functions in the airways and throughout the gastrointestinal tract [11]. Specifically, Tas2rs have been identified in ‘solitary’ chemosensory cells throughout the respiratory epithelium, as well as in the airway smooth muscle, where they might respond to bitter/toxic compounds to mediate protective airway reflexes and bronchodilation, respectively [12], [13], [14]. Tas2rs have also been identified in specialized subpopulations of gastrointestinal cells, where they modulate hormone release and impact on gastric emptying [15], [16].
The expression of taste GPCRs is yet to be systematically studied in the heart. Here, we report the expression of TAS1/Tas1 and TAS2/Tas2 GPCRs in human and rodent heart, their cellular localization, developmental regulation and upregulation in conditions of starvation. We speculate that one function of cardiac cells expressing taste GPCRs might be as sentinels for cardiac nutrient sensing.
Materials and Methods
Ethics Statement
Human hearts were obtained immediately following explantation from five patients with terminal heart failure undergoing heart transplantation at The Prince Charles Hospital, Brisbane. Written informed consent was obtained from each patient (ethics approval number EC28114, Human Research Ethics Committee, Metro North Hospital and Health Service, The Prince Charles Hospital).
All animal procedures were carried out with specific approval from The University of Queensland Animal Welfare Unit (ethics approval numbers SBMS/095/11/NHMRC/NHF and ANAT DVB/658/07NHMRC) and follow the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Experimental Animals and Tissue Collection
Animals were maintained on a 12 hour light/dark cycle with ad libitum access to standard chow and water. Sprague-Dawley Rats were killed by intraperitoneal injection of an overdose of sodium pentobarbital. Tas1r1Cre/Rosa26tdRFP mice report the history of activity of the Tas1r1 promoter. Briefly, Tas1r1Cre mouse line was generated by replacing the entire Tas1r1 coding region by an expression cassette containing the coding sequence of barley lectin, an internal ribosomal entry site, and Cre recombinase. Heterozygote gene-targeted Tas1r1Cre mice were bred with Rosa26tdRFP mice [17], to generate Tas1r1Cre/Rosa26tdRFP mice. Tas1r1Cre/Rosa26tdRFP mice carry one recombinant Tas1r1 and one recombinant ROSA26 allele. The Cre recombinase is expressed under the control of the Tas1r1 promoter; therefore a Cre-mediated recombination leads to excision of a transcriptional stop signal flanked by Lox-P sites and thus activates expression of tandem dimer red fluorescent protein (tdRFP) exclusively in Tas1r1 expressing cells. Tas2r131Cre/Rosa26tauGFP mice report the history of activity of the Tas2r131 promoter. Briefly, Tas2r131Cre mouse line was generated by replacing the coding region of Tas2r131 by an expression cassette, containing the coding sequence of barley lectin, an internal ribosomal entry site, and Cre recombinase. Heterozygote gene-targeted Tas2r131Cre mice were bred with Rosa26tauGFP [18], mice to generate Tas2r131Cre/Rosa26tauGFP mouse line. These mice carry one recombinant Tas2r131 and one recombinant ROSA26 allele. Cre recombinase expression is controlled by Tas2r131 promoter activity. Activation of Tas2r131 promoter leads to the expression of Cre recombinase and therefore induces Cre-mediated recombination. This recombination event leads to the excision of a transcriptional stop signal flanked by Lox-P sites and thus activates expression of a green fluorescent protein (tauGFP) exclusively in Tas2r131 cells. So, the gene-targeted mouse lines (Tas1r1Cre/Rosa26tdRFP, Tas2r131Cre/Rosa26tauGFP) express both the Cre recombinase and the taste receptors in a heterozygote manner, as well as the fluorescent proteins tdRFP and tauGFP, respectively.
For studies on the effects of starvation on taste GPCR expression, C57BL/6J mice were deprived of food for 48 hours with ad libitum access to drinking water, as described previously [19].
Heart and tongue tissues were immediately dissected and either rapidly frozen in liquid nitrogen for RNA extraction or fixed with 4% paraformaldehyde for cryosectioning.
Human Heart Tissue Samples
Following explantation from terminal heart failure patients undergoing heart transplantation, human hearts were immersed in preoxygenated (95% O2−5% CO2) modified Krebs’ solution containing (mmol/L) Na+125, K+5, Ca2+2.25, Mg2+0.5, Cl− 98.5, SO42− 0.5, HCO3 − 29, HPO4 2− 1, and EDTA 0.04 [20], and samples of right atrial free wall and left ventricle were rapidly dissected and frozen in liquid nitrogen.
Cardiomyocyte and Cardiac Fibroblast Isolation and Culture
Ventricular cardiomyocytes and fibroblasts were isolated, enzymatically digested and purified from one-day-old Sprague-Dawley rat pups, as described previously [21]. Briefly, neonates were killed by decapitation and the dissected ventricles were enzymatically digested with pancreatin and collagenase in a cell stirrer. Dissociated cardiac myocytes and fibroblasts were purified through a Percoll gradient to >99% homogeneity. Cardiomyocytes were plated on gelatin-coated tissue culture plates in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with sodium bicarbonate, L-glutamine, essential and non-essential amino acids, vitamins, penicillin-streptomycin, amphotericin B, bromodeoxyuridine (BrDu) and 10% (v/v) new born calf serum. Myocytes were subsequently cultured in DMEM media supplemented with sodium bicarbonate, vitamins, essential and non-essential amino acids, sodium pyruvate, insulin, apo-transferrin, penicillin-streptomycin, amphotericin B, BrDu and 50 mmol/L KCl. Cardiac fibroblasts were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum, sodium bicarbonate, penicillin-streptomycin and amphotericin B, and were grown to confluence in 6-well plates for RNA extraction. Nutrient deprivation experiments were performed for 24 hours on cultured myocytes in glucose-free DMEM media, supplemented as described above.
RNA Isolation and Reverse Transcription Quantitative PCR (RT-qPCR)
Total RNA was extracted from human and rodent heart tissue at various time points, as well as from isolated cardiomyocytes and fibroblasts using TRIzol reagent (Life Technologies, Melbourne, VIC, Australia) following homogenization with a Polytron homogenizer, as per the manufacturer’s recommendations. RNA was DNase-treated to remove genomic DNA contaminants and then cDNA was synthesized from 1–6 µg of total RNA using Superscript III (Life Technologies, Melbourne, VIC, Australia) according to the manufacturer’s protocols. DNase treatment is particularly important for RT-qPCR experiments involving single exon genes, such as the TAS2Rs. Reactions without reverse transcriptase were performed in parallel to control for contamination with chromosomal DNA (Figure S1A and Table S3).
RT-qPCR was used to determine mRNA expression levels of cardiac GPCRs, taste GPCRs and taste-associated genes. 18S ribosomal RNA and Gapdh were used as stably expressed endogenous control genes for samples as indicated. mRNA expression levels were measured for all human and rat genes using Taqman probe chemistry with a StepOne Plus Real-Time PCR System or SYBR green chemistry on an ABI7000 instrument (Applied Biosystems, Melbourne, VIC, Australia). Primers and probes for human and rat GPCRs and taste-associated genes, 18S and Gapdh endogenous primers were from Applied Biosystems, while mouse taste receptor primers were from qPrimerDepot (accessible online: http://mouseprimerdepot.nci.nih.gov) [22]. All primer sets and part numbers, and primer sequences are shown in Table S1. Where possible, primers span exon boundaries, although this is not possible for the single exon Tas2/TAS2 genes. PCR products were run on native PAGE gels to confirm correct amplicon size (Figure S1B). For experiments using SYBR green chemistry, melt curve analysis was performed at the completion of each assay to determine the specificity of the PCR reaction. Expression of the gene of interest was normalized to Gapdh and 18S expression using the 2−ΔΔCt method [23].
Tissue Preparation and in situ Hybridization
Tissues were fixed in 4% paraformaldehyde overnight at 4°C, rinsed in phosphate-buffered saline and incubated in 30% sucrose solution for 6 hours. Following embedding and freezing in Tissue-Tek OCT (Sakura Finetek, Melbourne, VIC, Australia), 10 µm cryosections were collected on Superfrost (+) slides, and stored at −70°C. In situ hybridization was performed on rat heart using digoxigenin (DIG) labeled-cRNA probes as described previously [24]. See Table S2 for T3 and T7 RNA polymerase-containing primer sequences used for in situ probe synthesis. 10 µm cryosections were fixed, treated with proteinase K, washed in PBS, acetylated and hybridized with gene-specific probes overnight at 65°C. Sections were probed with anti-DIG antibody (Roche Diagnostics, Sydney, NSW, Australia) overnight at 4°C. Color was developed at room temperature using NBT/BCIP (Promega, Sydney, NSW, Australia).
Data Analysis
Data expressed as mean±SEM. Statistical analyses were performed using one-way analysis of variance (ANOVA) with a Dunnett’s post-test or an unpaired student’s t-test, as indicated. P values below 0.05 were considered significant and are indicated as follows: *: P<0.05, **: P<0.01, ***P<0.001.
Results
Taste GPCRs are Expressed in the Rodent and Human Heart
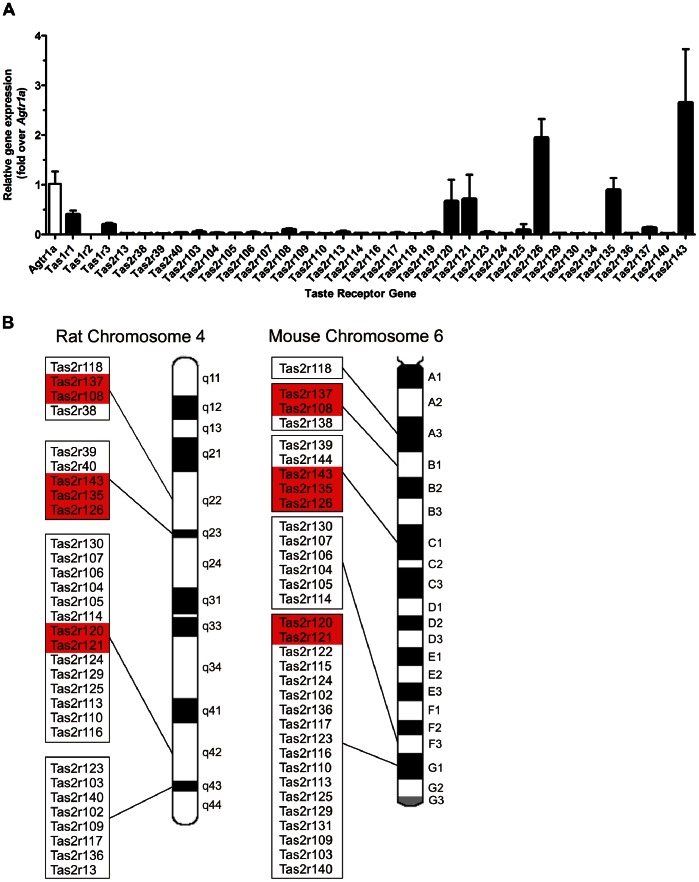
An RT-qPCR taste receptor screen in rat heart showed seven Tas2 receptors (Tas2r108, Tas2r120, Tas2r121, Tas2r126, Tas2r135, Tas2r137, Tas2r143), as well as two members of the Tas1 receptor family, Tas1r1 and Tas1r3, were expressed in neonatal whole hearts, notably at levels comparable to another important cardiovascular GPCR, the angiotensin II type 1a receptor (Agtr1a)(Figure 1A). The seven cardiac-expressed Tas2rs do not display obvious sequence homology, however they cluster into three sub-groups of proximal receptors located within the larger group of Tas2rs on chromosome 4 in rat (or 6 in mouse), indicating possible common upstream regulatory elements (Figure 1B). Moreover, although the majority of rodent Tas2rs do not have corresponding human orthologs [25], each of the cardiac-expressed Tas2 receptors described herein does, suggesting that the cardiac significance/function of these GPCRs may be evolutionarily conserved (Figure S2 and Table 1).
Figure 1. Taste GPCRs are expressed in the rodent heart.
A RT-qPCR screen of taste GPCRs in neonatal rat whole heart (mean±SEM, n = 4, normalized for Gapdh, presented as fold change over the angiotensin II type 1a receptor (Agtr1a)). B Rodent Tas2rs are predominantly clustered on chromosome 4 (rat) and chromosome 6 (mouse). The taste GPCRs highlighted in red are detected in the heart, suggesting that these receptors may be under transcriptional regulation of a common regulatory element.
Table 1. Homology of taste GPCRs in human and rodents.
| Human | Mouse | Rat | ||||
| Gene symbol | Human vs. Mouse identity (%) | Gene symbol | Mouse vs. Rat identity (%) | Gene symbol | ||
| Protein | DNA | Protein | DNA | |||
| TAS1R1 | 74 | 79.6 | Tas1r1 | 90.2 | 91.3 | Tas1r1 |
| TAS1R2 | 69.9 | 77.6 | Tas1r2 | Tas1r2 | ||
| TAS1R3 | 73.6 | 75 | Tas1r3 | 92.9 | 93.5 | Tas1r3 |
| TAS2R1 | 51.9 | 66.2 | Tas2r119 | 85.3 | 89.7 | Tas2r119 |
| TAS2R3 | 64.2 | 77.8 | Tas2r137 | 89.1 | 92.4 | Tas2r137 |
| TAS2R4 | 67 | 76.7 | Tas2r108 | 88.2 | 91.1 | Tas2r108 |
| TAS2R5* | ||||||
| TAS2R7 | 68.3 | 79.6 | Tas2r130 | 93.3 | 92.3 | Tas2r130 |
| TAS2R8* | ||||||
| TAS2R9* | ||||||
| TAS2R10 | 57 | 71.2 | Tas2r114 | 84.5 | 91.2 | Tas2r114 |
| TAS2R13 | 59 | 72.3 | Tas2r121 | 82.6 | 88 | Tas2r121 |
| TAS2R14 | 48.9 | 66.6 | Tas2r140 | 78.8 | 86.9 | Tas2r140 |
| TAS2R16 | 53.8 | 70.5 | Tas2r118 | 92 | 93 | Tas2r118 |
| TAS2R19* | ||||||
| TAS2R20 | 51.2 | 66.4 | Tas2r120 | 82.7 | 88.8 | Tas2r120 |
| TAS2R30* | ||||||
| TAS2R31* | ||||||
| TAS2R38 | 65.6 | 76.5 | Tas2r138 | 87.9 | 91.3 | Tas2r38 |
| TAS2R39 | 57.5 | 72.4 | Tas2r139 | 85 | 89.6 | Tas2r39 |
| TAS2R40 | 66.4 | 78.5 | Tas2r144 | 88.1 | 91.5 | Tas2r40 |
| TAS2R41 | 68.7 | 76.3 | Tas2r126 | 90.9 | 92.6 | Tas2r126 |
| TAS2R42 | 50.5 | 66.4 | Tas2r131 | 80 | 87.6 | LOC100363053 |
| TAS2R43* | ||||||
| TAS2R45* | ||||||
| TAS2R46* | ||||||
| TAS2R50* | ||||||
| TAS2R60 | 58.8 | 72 | Tas2r135 | 93.1 | 94.1 | Tas2r135 |
| TAS2R62P | 70.4 | Tas2r143 | 87.4 | 91.7 | Tas2r143 | |
| Tas2r102 | 79.3 | 89 | Tas2r13 | |||
| Tas2r103 | 76.6 | 86.4 | Tas2r103 | |||
| Tas2r104 | 86.1 | 91.9 | Tas2r104 | |||
| Tas2r105 | 83.9 | 88.7 | Tas2r105 | |||
| Tas2r106 | 84.4 | 91 | Tas2r106 | |||
| Tas2r107 | 85.3 | 89.6 | Tas2r107 | |||
| Tas2r109 | 75 | 85.2 | Tas2r109 | |||
| Tas2r110 | 77.3 | 85.6 | Tas2r110 | |||
| Tas2r113 | 79 | 88.3 | Tas2r113 | |||
| Tas2r116 | 73.4 | 85 | Tas2r116 | |||
| Tas2r117 | 77 | 87.8 | Tas2r117 | |||
| Tas2r123 | 80.4 | 87 | Tas2r123 | |||
| Tas2r124 | 81.6 | 88.3 | Tas2r124 | |||
| Tas2r125 | 76.1 | 85.6 | Tas2r125 | |||
| Tas2r129 | 78.1 | 87 | Tas2r129 | |||
| Tas2r134 | 82 | 88.7 | Tas2r134 | |||
| Tas2r136 | 75.5 | 85.4 | Tas2r136 | |||
Human taste receptors are shown on the left, with the corresponding mouse and rat taste receptor and percentage identity at the protein and DNA level, taken from the NCBI database (http://www.ncbi.nlm.nih.gov/homologene). Where human taste receptor homologs do exist, they are shown aligned to their rodent counterparts, with protein and DNA identity to mouse and rat sequences. Human specific taste GPCRs are denoted with an asterisk (*).
To begin to empirically test this possibility, we obtained heart tissue from failing human hearts and performed an RT-qPCR screen for the taste GPCRs (Fig. 2A). Transcripts for more than half of the TAS2R family were detected in these human left ventricle samples, at comparatively high levels relative to the angiotensin II type 1 receptor (AGTR1). Remarkably, TAS2R14 is expressed at comparable levels to the β1-adrenergic receptor (ADRB1), an abundantly expressed GPCR that is a critical mediator of chronotropy and ionotropy in the heart. The same receptors and relative expression patterns were observed for right atrial tissue (Figure S3). It is also interesting to note that the human TAS2Rs detected in heart are clustered in close genomic proximity on chromosomes 7 and 12. As in rodents, this genomic organization seems to correlate with the expression of particular subsets of TAS2Rs in the heart (Figure 2B).
Figure 2. Taste GPCRs are expressed in the human heart.
A RT-qPCR screen of taste GPCRs in human left ventricle (mean±SEM, n = 5, normalized for 18S, presented as fold change over the angiotensin II type 1 receptor (AGTR1)). The abundantly expressed β1-adrenergic receptor (ADRB1) is shown as a comparator. B Human TASRs are localized on chromosomes 1, 5, 7 and 12, with the majority of TAS2Rs expressed in the heart (highlighted in red) present in a cluster on chromosome 12.
Localization of Taste GPCRs in the Rodent Heart
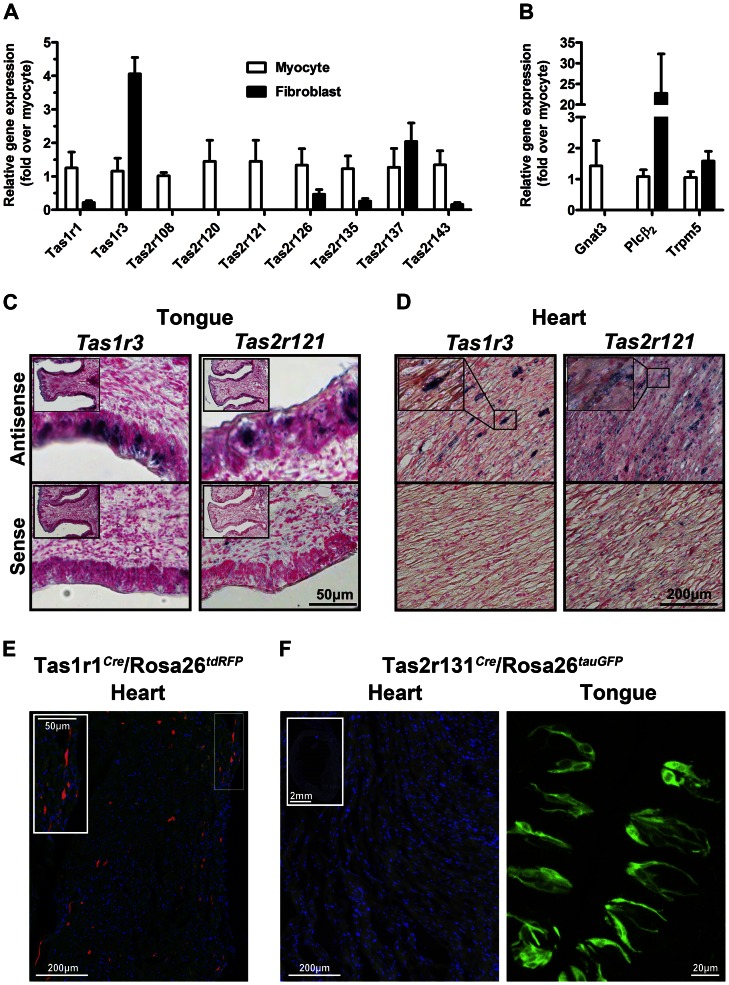
To further characterize the taste GPCRs that were identified in our initial RT-qPCR screen, we investigated their localization in cardiac cells. Several taste GPCRs were readily detected in primary cultures of both isolated, purified neonatal rat cardiac cells, and with the exception of Tas1r3, were enriched in cardiomyocytes over fibroblasts (Figure 3A). Several genes commonly implicated in the taste receptor signaling cascade, specifically Gnat3, Plcβ2 and Trpm5, were also expressed in cardiomyocytes (Figure 3B).
Figure 3. Taste GPCRs are localized in isolated cardiac cells.
A Taste receptors were detected in cultured neonatal cardiomyocytes and fibroblasts by RT-qPCR (mean±SEM, n = 4, normalized for Gapdh and expressed relative to myocytes). B The taste receptor signal transduction genes, including G protein (Gnat3), second messenger (PLCβ2) and channel (Trpm5) are also expressed in myocytes and fibroblasts (mean±SEM, n = 4, normalized for Gapdh and expressed relative to myocytes). C In situ hybridization using digoxigenin-labeled cRNA probes specific for Tas1r3 (left panels) and Tas2r121 (right panels) show expression in the taste buds of the circumvallate papillae (inset shows low magnification) and D in heart tissue (inset shows higher magnification of boxed region). Specific labeling (blue/black) is observed using antisense probes, but not sense probes. E Tas1r1 is expressed in mouse ventricular tissue from Tas1r1Cre/Rosa26tdRFP mice, where red fluorescent cells report activity of the Tas1r1 promoter. F In contrast, in heart sections from the Tas2r131Cre/Rosa26tauGFP reporter mouse line, there are no fluorescently-labeled myocytes (inset shows low magnification view of heart). Tas2r131–expressing cells are clearly labeled in the circumvallate papillae as control. Scale bars are as indicated.
Given the lack of selective and specific antibodies for the taste GPCRs, in situ hybridization was used to localize taste receptor expression in heart. In representative adult heart sections, Tas1 (Tas1r3) and Tas2 (Tas2r121) receptors showed distinct labeling in a small proportion of cardiac cells distributed throughout the myocardium (Figure 3C and D and inset). The probes labeled taste receptor-expressing cells in the circumvallate papillae of the tongue, whereas no staining was visible in sense controls. The expression of Tas1r1 in discrete cardiac cells within the myocardium was also confirmed in a Tas1r1 gene-targeted mouse (Tas1r1Cre/Rosa26tdRFP)(Figure 3E). Importantly, another gene-targeted mouse, Tas2r131Cre/Rosa26tauGFP exhibited strong labeling of taste receptor-expressing cells in the circumvallate papillae, but no cardiac expression, consistent with our RT-qPCR data (Figure 3F). Thus, specific taste receptor genes are expressed in cardiac cells.
Taste GPCRs are Developmentally Regulated in the Heart
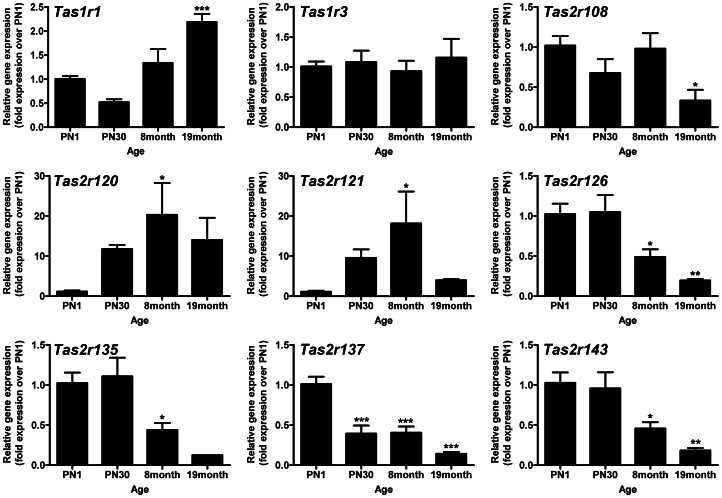
We characterized the ontogeny of taste receptor expression in the heart, at multiple time points from neonatal (postnatal day 1) to 19 months of age (Figure 4). Tas1r1 expression increased 2-fold in senescent rat hearts, whereas Tas1r3 expression levels remained constant throughout life. The Tas2rs displayed contrasting temporal patterns of expression. Tas2r120 and Tas2r121 expression increased nearly 20-fold in adult rats relative to neonate hearts. The remaining cardiac-expressed Tas2rs (108, 126, 135, 137 and 143) decreased in abundance with age (3–8 fold at 19 months).
Figure 4. Taste GPCRs are regulated in the heart during postnatal life in vivo.
Taste GPCR mRNA expression levels at different postnatal ages from postnatal day 1 (PN1) to 19 months. Gene expression levels are normalized to Gapdh and expressed relative to PN1 (mean±SEM, expressed relative to PN1, n = 4, *P<0.05; **P<0.01, ***P<0.001, one-way ANOVA with a Dunnett’s post-test).
Taste GPCRs are Regulated under Conditions of Starvation Both in vitro and in vivo
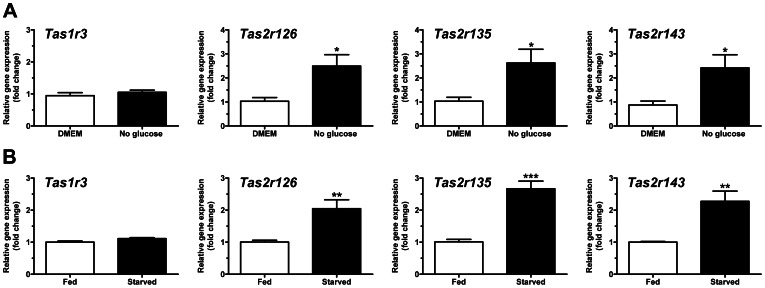
Given the purported role of Tas1 GPCRs in nutrient sensing, we investigated the effect of nutrient deprivation on expression of taste receptors in cultured rat cardiomyocytes. Contrary to our expectations, Tas1r1 and Tas1r3 expression levels were unchanged in myocytes following 24 hours glucose deprivation. Similarly, there was no effect of amino acid deprivation on taste GPCR expression (Figure S4). The regulation of Tas2 GPCRs has not been thoroughly investigated with respect to nutrient status. Intriguingly, the expression of Tas2r126, Tas2r135 and Tas2r143 increased 2–3 fold in the absence of glucose, whereas the remaining taste GPCRs expressed were unchanged (Figure 5A, Figure S5). These data were verified in vivo in a mouse model of starvation, where the same Tas2rs were upregulated after 48 hours food deprivation (Figure 5B). Taken together, these data suggest that the cardiac expression of Tas2 receptors could have physiological relevance as novel metabolic/nutrient sensors in the heart.
Figure 5. Taste GPCRs are regulated in the heart under conditions of starvation both in vitro and in vivo.
A Tas2r126, Tas2r135 and Tas2r143 mRNA expression levels in cultured neonatal rat ventricular myocytes deprived of glucose for 24 h (mean±SEM, n = 4, fold change over glucose-containing myocyte media conditions (DMEM)). B Tas2r126, Tas2r135 and Tas2r143 mRNA expression levels in hearts of mice starved for 48 h (mean±SEM, n = 4, fold change over fed (ad lib) controls, *P<0.05; **P<0.01, ***P<0.001, unpaired student’s t test).
Discussion
Using RT-qPCR, in situ hybridization, and gene-targeted reporter mice, we demonstrate, for the first time, the expression of individual Tas1 and Tas2 receptors in cardiomyocytes, fibroblasts and heart tissue. In rodents, these receptors are expressed throughout the myocardium in a small subset of cardiac cells and some receptors are upregulated at the transcript level in vitro and in vivo upon nutrient starvation. The cardiac-expressed Tas2rs are located in genomic clusters and share developmental and physiological expression patterns, suggesting common regulatory control. Each of these receptors described has a human homolog, implying conservation across eutheria. Indeed, in human heart tissue samples obtained from heart transplant patients, taste GPCR transcripts are readily detected, most strikingly the TAS2Rs. The delineation of Tas1/Tas2 GPCRs in heart opens up new avenues of cardiovascular research.
Recent large-scale efforts to profile and characterize GPCR expression in different tissues, demonstrate that the heart expresses more than 100 GPCRs [3], [4], [26], [27], although these studies have pragmatically excluded the chemosensory receptors. Collectively, these studies raise the possibility that we are overlooking important physiological and pathophysiological GPCR targets – specifically by focusing only on well characterized GPCRs with available, potent and selective ligands. Recent work by Insel and colleagues has epitomized this idea in demonstrating that a previously unheralded, albeit ubiquitously expressed GPCR (protease activated receptor 1, PAR1), may play an important role in cardiac fibrosis [28]. Our study is the first to systematically describe taste GPCRs in heart and supports the idea that previous studies may have been too narrow in their scope in profiling GPCR expression across tissues.
The type 1 (TAS1/Tas1) family taste receptors have a nutrient sensing role in the tongue, as well as other tissues, including the gastrointestinal tract and the brain [5], [29], [30]. Our specific identification in heart of Tas1r1/Tas1r3 is consistent with a nutrient-sensing role and may indicate a role in metabolic regulation. Indeed, Tas1r1/Tas1r3 have very recently been implicated as direct upstream amino acid sensors for the mTORC1-mediated autophagy pathway [9]. While not entirely surprising, given their well-described roles as broadly-tuned amino acid sensors, this newly attributed function to Tas1rs outside the mouth (and our discovery of them in heart) reinforces the notion that these GPCRs are involved in far more than taste.
Another major finding in our study is the prevalence of the type 2 “bitter” (TAS2/Tas2) GPCRs in both human and rodent hearts. We also detected the genes commonly associated with taste GPCR signaling, endowing the heart with taste-receptor like signaling potential, although there is no a priori reason to assume that these are the cardiac effectors of Tas2 activation. The endogenous complement of proteins in the heart would likely dictate the signal transduction and hence cardiac function of these GPCRs, although the downstream components remain to be delineated. While Tas2rs are predominantly located within multigene clusters in mouse, rat and human genomes, there is little known about their tissue-specific and transcriptional regulation [31]. Nevertheless, the proximal chromosomal organization and clustering of the subset of cardiac-expressed taste GPCRs, as well as their shared regulatory patterns, suggest possible transcriptional co-regulation.
What could be the potential function for a TAS2 receptor in the heart? The localization of taste GPCRs in discrete heart cells parallels that seen in the respiratory and gastrointestinal systems [13], [15], suggesting that these may function as specialized cells that sense the extracellular environment and influence surrounding tissue. In this regard, the upregulation of a subset of the Tas2 GPCRs in the heart following nutrient deprivation and starvation is interesting. These findings could reflect a potential function as nutrient sensors in heart. Interestingly, taste receptors are upregulated in several conditions where specific nutrients are depleted [5], [32], and a single nucleotide polymorphism in a human TAS2R has been linked to deficits in glucose homeostasis [33]. Similarly, gene variants in TAS2Rs are related to disease – a polymorphism in TAS2R38 contributes to an increased susceptibility to respiratory infection [34] and a mutation in TAS2R50 has been associated with cardiovascular disease in several population-based prospective studies [35], [36], but the mechanisms by which the variants of these TAS2 genes influence the pathophysiology of disease are mostly unknown. Although the present findings do not discriminate between increased TAS2 expression in a subpopulation of cells versus an increase in the number of cells expressing the receptor, the cardiac expression of taste GPCRs raises the possibility of a direct effect, beyond oral sensation [37].
An obvious consideration for the cardiac expression of Tas2rs is their ligand-mediated activation – from where do these ligands originate and how would they get to the heart? At present, the best candidates for activating TAS2Rs outside the gustatory system are inhaled irritants and toxins [15], [34]. It is possible that exogenous toxins could be taken into the circulatory system to target the cardiac-Tas2rs. An alternative possibility is that there are endogenous TAS2R agonists produced within the circulation and/or cardiovascular tissues. Indeed, there is a growing literature pertaining to the metabolite ligands for GPCRs in physiology and disease (recently reviewed in [38], [39]). Notably, metabolite ligands have been associated with a variety of receptor systems and may have potential therapeutic applications in a variety of metabolic diseases and in pain [40], [41], [42], [43], [44].
Clearly, the identification of TAS2Rs in the heart warrants further investigation in cardiac physiology and disease. We are currently working to identify potent and selective ligands for these Tas2rs in rodents, which will appreciably aid efforts towards their functional characterization in heart. In addition, given the interesting species similarities/differences in TAS2Rs, it will be important to also direct future studies to cells and tissues derived from humans, where the characterization of TAS2R ligand binding profiles [10] and pharmacological tools (e.g. small molecule TAS2R antagonists [45]) are slightly more advanced. Indeed, while chemosensory receptors represent the majority of GPCRs in our genome, we are yet to fully understand the extent of their biological importance in tissues beyond the nose and mouth. We agree with Munger and colleagues that the extraoral expression of TAS2Rs may be a potential confounder in the application and understanding of GPCR-based therapeutics [46], but reiterate that they could also be possible drug targets in their own right.
Supporting Information
A Representative RT-qPCR amplification plot for rat Tas2r143 in the presence (black traces) or absence (blue traces) of reverse transcriptase. As shown in Table S3, four independent samples of neonatal rat heart mRNA were assayed in triplicate. All +RT samples amplified with Ct values approximating 27.4, whereas 9 of 12 of the –RT replicates failed to amplify. The remaining 3 replicates amplified at an average CT value of 34.8, generally indistinguishable from background. B The correct amplicon sizes of cardiac-expressed TasRs were confirmed by running RT-qPCR samples (+ and – RT) on a 12% native PAGE gel. Shown are three representative rat Tas2rs (Tas2r126, Tas2r135 and Tas2r143) running at their expected molecular size (139 base pairs, 159 base pairs and 97 base pairs, respectively) relative to marker bands at 300, 150 and 50 base pairs, as indicated.
(TIF)
Schematic showing the genomic organization of the Tas2r143, 135 and 126 cluster of taste GPCRs in mouse, and the mammalian conservation. Generated using the UCSC Genome Browser database: http://genome.ucsc.edu/.
(TIF)
Taste GPCRs are expressed in the human right atria. RT-qPCR screen of taste GPCRs in human right atria (mean ± SEM, n = 5, normalized for 18S, presented as fold change over the angiotensin II type 1 receptor (AGTR1)). The abundantly expressed β1-adrenergic receptor (ADRB1) is shown as a comparator.
(TIF)
Amino acid deprivation (24 h) has no effect on Tas1 or Tas2 GPCR mRNA expression in cultured neonatal rat ventricular myocytes. Data expressed as mean±SEM, n = 4, fold change over amino acid-containing myocyte media conditions (DMEM).
(TIF)
Glucose deprivation (24 h) does not modulate the mRNA expression of a subset of taste GPCRs in cultured neonatal rat ventricular myocytes. Data expressed as mean±SEM, n = 4, fold change over glucose-containing myocyte media conditions (DMEM).
(TIF)
RT-qPCR primers and probes and their sequences/part numbers.
(DOCX)
In situ hybridization probe primer sequences, including T7 and T3 RNA polymerase binding sites.
(DOCX)
RT-qPCR experiments were performed in the presence and absence of reverse transcriptase following DNase treatment to confirm the specific amplification of cDNA, in contrast to genomic DNA.
(DOCX)
Acknowledgments
We gratefully acknowledge Dr Eric Olson (UT Southwestern Medical Center, Dallas, USA) for provision of heart samples from starved mice.
Funding Statement
This work was funded by the National Health and Medical Research Council of Australia (APP631443 to WGT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lagerstrom MC, Schioth HB (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7: 339–357. [DOI] [PubMed] [Google Scholar]
- 2. Overington JP, Al-Lazikani B, Hopkins AL (2006) How many drug targets are there? Nat Rev Drug Discov 5: 993–996. [DOI] [PubMed] [Google Scholar]
- 3. Insel PA, Snead A, Murray F, Zhang L, Yokouchi H, et al. (2012) GPCR expression in tissues and cells: Are the optimal receptors being used as drug targets? Br J Pharmacol 165: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moore-Morris T, Varrault A, Mangoni ME, Le Digarcher A, Negre V, et al. (2009) Identification of Potential Pharmacological Targets by Analysis of the Comprehensive G Protein-Coupled Receptor Repertoire in the Four Cardiac Chambers. Mol Pharmacol 75: 1108–1116. [DOI] [PubMed] [Google Scholar]
- 5. Ren X, Zhou L, Terwilliger R, Newton SS, de Araujo IE (2009) Sweet taste signaling functions as a hypothalamic glucose sensor. Front Integr Neurosci 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, et al. (2007) Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci U S A 104: 15069–15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mace OJ, Lister N, Morgan E, Shepherd E, Affleck J, et al. (2009) An energy supply network of nutrient absorption coordinated by calcium and T1R taste receptors in rat small intestine. J Physiol 587: 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Margolskee RF, Dyer J, Kokrashvili Z, Salmon KSH, Ilegems E, et al. (2007) T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci U S A 104: 15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wauson EM, Zaganjor E, Lee AY, Guerra ML, Ghosh AB, et al. (2012) The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol Cell 47: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, et al. (2010) The Molecular Receptive Ranges of Human TAS2R Bitter Taste Receptors. Chem Senses 35: 157–170. [DOI] [PubMed] [Google Scholar]
- 11. Finger TE, Kinnamon SC (2011) Taste isn't just for taste buds anymore. F1000 Biol Rep 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deshpande DA, Wang WC, McIlmoyle EL, Robinett KS, Schillinger RM, et al. (2010) Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med 16: 1299–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finger TE, Böttger B, Hansen A, Anderson KT, Alimohammadi H, et al. (2003) Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc Natl Acad Sci U S A 100: 8981–8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tizzano M, Cristofoletti M, Sbarbati A, Finger T (2011) Expression of taste receptors in Solitary Chemosensory Cells of rodent airways. BMC Pulm Med 11: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janssen S, Laermans J, Verhulst PJ, Thijs T, Tack J, et al. (2011) Bitter taste receptors and {alpha}-gustducin regulate the secretion of ghrelin with functional effects on food intake and gastric emptying. Proc Natl Acad Sci U S A 108: 2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeon TI, Seo YK, Osborne TF (2011) Gut Bitter Taste Receptor Signaling Induces ABCB1 through a Mechanism Involving CCK. Biochem J 438: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ (2007) Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol 37: 43–53. [DOI] [PubMed] [Google Scholar]
- 18. Wen S, Götze IN, Mai O, Schauer C, Leinders-Zufall T, et al. (2011) Genetic Identification of GnRH Receptor Neurons: A New Model for Studying Neural Circuits Underlying Reproductive Physiology in the Mouse Brain. Endocrinology 152: 1515–1526. [DOI] [PubMed] [Google Scholar]
- 19. Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaumann A, Bartel S, Molenaar P, Sanders L, Burrell K, et al. (1999) Activation of β2-Adrenergic Receptors Hastens Relaxation and Mediates Phosphorylation of Phospholamban, Troponin I, and C-Protein in Ventricular Myocardium From Patients With Terminal Heart Failure. Circulation 99: 65–72. [DOI] [PubMed] [Google Scholar]
- 21. Thomas WG, Brandenburger Y, Autelitano DJ, Pham T, Qian HW, et al. (2002) Adenoviral-directed expression of the type 1A angiotensin receptor promotes cardiomyocyte hypertrophy via transactivation of the epidermal growth factor receptor. Circ Res 90: 135–142. [DOI] [PubMed] [Google Scholar]
- 22. Cui WW, Taub DD, Gardner K (2007) qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res 35: D805–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 24. Simmons D, Rawn S, Davies A, Hughes M, Cross J (2008) Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus. BMC Genomics 9: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi P, Zhang J, Yang H, Zhang Y (2003) Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol 20: 805–814. [DOI] [PubMed] [Google Scholar]
- 26. Regard JB, Sato IT, Coughlin SR (2008) Anatomical profiling of G protein-coupled receptor expression. Cell 135: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang CM, Insel PA (2004) GPCR expression in the heart – “New” receptors in myocytes and fibroblasts. Trends Cardiovasc Med 14: 94–99. [DOI] [PubMed] [Google Scholar]
- 28.Snead AN, Insel PA (2012) Defining the cellular repertoire of GPCRs identifies a profibrotic role for the most highly expressed receptor, protease-activated receptor 1, in cardiac fibroblasts. FASEB J. [DOI] [PMC free article] [PubMed]
- 29. Behrens M, Meyerhof W (2011) Gustatory and extragustatory functions of mammalian taste receptors. Physiol Behav 105: 4–13. [DOI] [PubMed] [Google Scholar]
- 30. Wellendorph P, Johansen LD, Brauner-Osborne H (2009) Molecular pharmacology of promiscuous seven transmembrane receptors sensing organic nutrients. Mol Pharmacol 76: 453–465. [DOI] [PubMed] [Google Scholar]
- 31. Toyono T, Seta Y, Kataoka S, Toyoshima K (2007) CCAAT/Enhancer-binding protein beta regulates expression of human T1R3 taste receptor gene in the bile duct carcinoma cell line, HuCCT1. Biochimica Et Biophysica Acta-Gene Structure and Expression 1769: 641–648. [DOI] [PubMed] [Google Scholar]
- 32. Young RL, Sutherland K, Pezos N, Brierley SM, Horowitz M, et al. (2009) Expression of taste molecules in the upper gastrointestinal tract in humans with and without type 2 diabetes. Gut 58: 337–346. [DOI] [PubMed] [Google Scholar]
- 33. Dotson CD, Zhang L, Xu H, Shin YK, Vigues S, et al. (2008) Bitter Taste Receptors Influence Glucose Homeostasis. PLoS One 3: e3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee RJ, Xiong G, Kofonow JM, Chen B, Lysenko A, et al.. (2012) T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. [DOI] [PMC free article] [PubMed]
- 35. Akao H, Polisecki E, Kajinami K, Trompet S, Robertson M, et al. (2012) KIF6, LPA, TAS2R50, and VAMP8 genetic variation, low density lipoprotein cholesterol lowering response to pravastatin, and heart disease risk reduction in the elderly. Atherosclerosis 220: 456–462. [DOI] [PubMed] [Google Scholar]
- 36. Shiffman D, O’Meara ES, Bare LA, Rowland CM, Louie JZ, et al. (2008) Association of Gene Variants With Incident Myocardial Infarction in the Cardiovascular Health Study. Arteriosclerosis, Thrombosis, and Vascular Biology 28: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duffy VB (2004) Associations between oral sensation, dietary behaviors and risk of cardiovascular disease (CVD). Appetite 43: 5–9. [DOI] [PubMed] [Google Scholar]
- 38.Smith NJ (2012) Low affinity GPCRs for metabolic intermediates: challenges for pharmacologists. Frontiers in Endocrinology 3. [DOI] [PMC free article] [PubMed]
- 39.Tonack S, Tang C, Offermanns S (2012) Endogenous metabolites as ligands for G-coupled receptors modulating risk factors for metabolic and cardiovascular disease. Am J Physiol Heart Circ Physiol. [DOI] [PubMed]
- 40.Deng H, Hu H, Fang Y (2012) Multiple tyrosine metabolites are GPR35 agonists. Sci Rep 2. [DOI] [PMC free article] [PubMed]
- 41. Patwardhan AM, Akopian AN, Ruparel NB, Diogenes A, Weintraub ST, et al. (2010) Heat generates oxidized linoleic acid metabolites that activate TRPV1 and produce pain in rodents. J Clin Invest 120: 1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sotnikova TD, Caron MG, Gainetdinov RR (2009) Trace Amine-Associated Receptors as Emerging Therapeutic Targets. Molecular Pharmacology 76: 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waku T, Shiraki T, Oyama T, Maebara K, Nakamori R, et al. (2010) The nuclear receptor PPARgamma individually responds to serotonin- and fatty acid-metabolites. EMBO J 29: 3395–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wootten D, Savage EE, Valant C, May LT, Sloop KW, et al. (2012) Allosteric modulation of endogenous metabolites as an avenue for drug discovery. Mol Pharmacol 82: 281–290. [DOI] [PubMed] [Google Scholar]
- 45. Slack JP, Brockhoff A, Batram C, Menzel S, Sonnabend C, et al. (2010) Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr Biol 20: 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clark AA, Liggett SB, Munger SD (2012) Extraoral bitter taste receptors as mediators of off-target drug effects. FASEB J 26: 4827–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A Representative RT-qPCR amplification plot for rat Tas2r143 in the presence (black traces) or absence (blue traces) of reverse transcriptase. As shown in Table S3, four independent samples of neonatal rat heart mRNA were assayed in triplicate. All +RT samples amplified with Ct values approximating 27.4, whereas 9 of 12 of the –RT replicates failed to amplify. The remaining 3 replicates amplified at an average CT value of 34.8, generally indistinguishable from background. B The correct amplicon sizes of cardiac-expressed TasRs were confirmed by running RT-qPCR samples (+ and – RT) on a 12% native PAGE gel. Shown are three representative rat Tas2rs (Tas2r126, Tas2r135 and Tas2r143) running at their expected molecular size (139 base pairs, 159 base pairs and 97 base pairs, respectively) relative to marker bands at 300, 150 and 50 base pairs, as indicated.
(TIF)
Schematic showing the genomic organization of the Tas2r143, 135 and 126 cluster of taste GPCRs in mouse, and the mammalian conservation. Generated using the UCSC Genome Browser database: http://genome.ucsc.edu/.
(TIF)
Taste GPCRs are expressed in the human right atria. RT-qPCR screen of taste GPCRs in human right atria (mean ± SEM, n = 5, normalized for 18S, presented as fold change over the angiotensin II type 1 receptor (AGTR1)). The abundantly expressed β1-adrenergic receptor (ADRB1) is shown as a comparator.
(TIF)
Amino acid deprivation (24 h) has no effect on Tas1 or Tas2 GPCR mRNA expression in cultured neonatal rat ventricular myocytes. Data expressed as mean±SEM, n = 4, fold change over amino acid-containing myocyte media conditions (DMEM).
(TIF)
Glucose deprivation (24 h) does not modulate the mRNA expression of a subset of taste GPCRs in cultured neonatal rat ventricular myocytes. Data expressed as mean±SEM, n = 4, fold change over glucose-containing myocyte media conditions (DMEM).
(TIF)
RT-qPCR primers and probes and their sequences/part numbers.
(DOCX)
In situ hybridization probe primer sequences, including T7 and T3 RNA polymerase binding sites.
(DOCX)
RT-qPCR experiments were performed in the presence and absence of reverse transcriptase following DNase treatment to confirm the specific amplification of cDNA, in contrast to genomic DNA.
(DOCX)