Letter to the Editor
ABL is the main target of several tyrosine kinase inhibitors (TKIs) used in the treatment of chronic myeloid leukemia (CML) patients. ABL tyrosine kinase domain (TKD) point mutations are the main mechanism of acquired TKI resistance(1–3). Although approximately 90 ABL TKD point mutations at 57 residues have been described, substitutions at seven amino acid residues account for a large proportion of point mutations detected. The T315I and the ATP-binding loop (p-loop) mutations have been associated with poor outcomes on imatinib mesylate (IM) therapy in several studies(2,3). Furthermore, the “gatekeeper” mutation, T315I, is resistant to currently available ABL tyrosine kinase inhibitors.
Direct nucleotide sequencing is the primary method of ABL TKD detection used in clinical practice. This technique has a sensitivity of 20%–25% (that is, to be detected the mutant clone must comprise at least 20% of the sample tested). Sensitivity can be improved slightly by pyrosequencing or denaturing high performance liquid chromatography methods. Because of this limited senstivity, ABL TKD mutations are typically detected at relapse, rather than early in the course of the emergence of a mutated ABL clone (2).
We have devised a mutation-specific PCR assay using a novel nanofluidic platform to identify and quantify T315I ABL in 34 CML patient samples from 28 patients(4). The Fluidigm BioMark™ Real-Time PCR System and a nanofluidic chip, the BioMark™ digital array (Fluidigm, South San Francisco, CA) were used in conjunction with mutation-specific PCR to detect the T315I mutation. The digital array uses nanoscale channels, valves, and pumps to partition samples into 12 panels, each panel containing either 765 or 1,104 chambers (depending on the platform design). Improved rare mutation detection occurs as a consequence of sample partitioning prior to PCR. For example, if a mixture containing 1 molecule of T315I ABL within 100,000 molecules of unmutated ABL is partitioned into 1,000 independent chambers, the chamber containing the single mutant molecule now only contains ~100 molecules of unmutated ABL. This 1,000-fold increase in relative concentration theoretically allows for a 1,000-fold improvement in the detection sensitivity of PCR reactions, thereby facilitating rare copy detection in limited patient samples.
Extensive experimental details are available at http://www.fhcrc.org/science/labs/radich/. In general, the experimental protocol first requires quantification of total amplifiable mRNA, specifically total ABL mRNA, and subsequently quantification of T315I mutated ABL using mutation-specific PCR. For mutation-specific primers the ultimate 3’ base pair was specific for each mutation, and specificity was increased by placing another mismatch 4 base pairs from the 3’ end. ABL, rather than BCR-ABL, was analyzed due to the product size constraints when using quantitative PCR. A pre-amplification step (Applied Biosystems, Foster City, CA) was performed for all samples where remaining cDNA was extremely limited. Comparisons utilizing model systems demonstrated that no bias was introduced through the use of pre-amplification prior to mutation detection.
Total ABL was quantified across three panels of 765 reactions per panel (total 2,295 reactions), using the average number of positive chambers, the dilution factor, and Poisson distribution to calculate the original number of ABL molecules. Subsequently, for each T315I assay, equal numbers of ABL molecules were loaded into three panels (samples 5–34) or one panel (samples 1–4). The number of T315I mutant molecules in each panel was calculated using Digital PCR Analysis software (Fluidigm, South San Francisco, CA) as described above. The sensitivity and specificity of the reaction was determined using serially diluted plasmids and cell lines containing the T315I mutation in a background of unmutated ABL. The assay detected as few as three T315I mutated molecules in a total background of 100,000 unmutated ABL molecules (Figure 1).
Figure 1. Mutation detection sensitivity.
T315I mutated ABL plasmids were diluted serially into a background of 100,000 unmutated ABL plasmids. The number of mutations was calculated using a Poisson distribution. The plot shows the actual number of T315I molecules loaded (Y axis) versus the number of T315I molecules detected (X axis). Single molecules of T315I can be detected in a background of unmutated ABL. From 3 to 30,000 T315I cDNA molecules were detected in a highly reproducible manner in replicate experiments.
Patient samples from CML patients (all phases) and normal patients were obtained from the University of California, Los Angeles (samples 1–4), Oregon Health and Sciences University (samples 5–22), and the Fred Hutchinson Cancer Research Center (samples 23–34). Prior approval for these studies was obtained from the Institutional Review Boards at each center. The negative control for each T315I assay consisted of 5,000 molecules per panel of unmutated ABL; and the positive control consisted of a spike-in of 2 molecules per panel (6 molecules total) of T315I mutated ABL in the background of 5,000 molecules of unmutated ABL. Receiver operating characteristic (ROC) curve analysis (Prism software, version 4, GraphPad Software, San Diego, CA) was used to determine the diagnostic performance of the assays using Area Under the Curve (AUC) analysis computed using a non-parametric method (5). The negative and positive controls run on each chip were used to calculate the false positive and true positive rates, respectively, and to generate the ROC curves. To ensure specificity the negative control was used to set the cycle threshold at which patient data would be analyzed. Data were only accepted if the negative control showed no amplification below the established cycle threshold. The area under the ROC curve at cycle 36 was 0.88 (95% CI, 0.77–1.0; p< 0.0001).
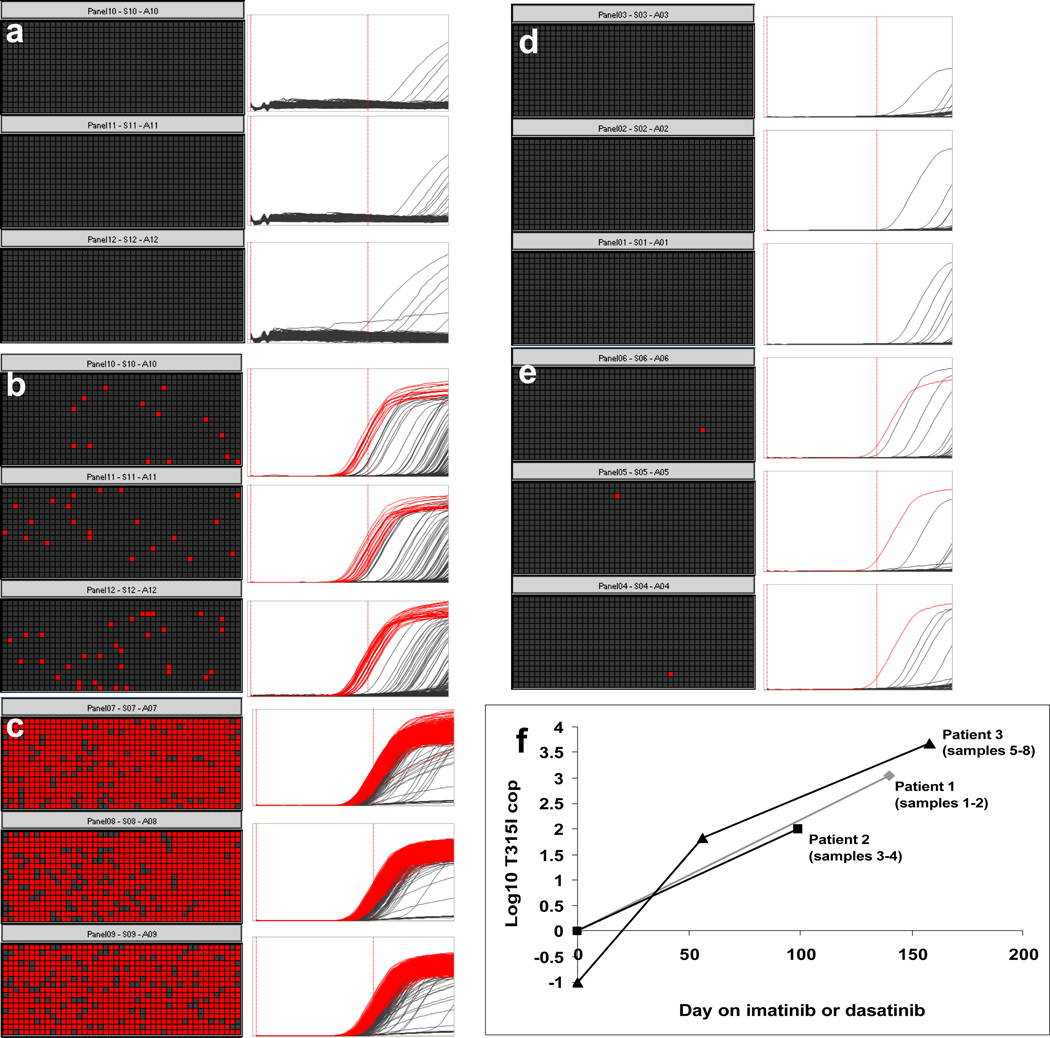
Analyses of 34 samples from 28 patients, taken both before and at the time of relapse, were performed in a blinded manner. As shown in Table 1 and Figure 2, the T315I mutation was detected in all 8 patients in whom it was ultimately detected at the time of relapse. The T315I mutation was not detected in 8 CP CML patients who maintained complete cytogenetic responses (CCRs) on IM, in 3 patients who failed IM without evidence of point mutations, in 2 patients with no clinical information available but no evidence of point mutations, or in 3 CML patients who developed other ABL TKD point mutations. Lastly, the T315I was not detected in one bone marrow (BM) sample and two peripheral blood (PB) samples from healthy volunteers without leukemia. We then compared the performance characteristics of our method directly to allele-specific oligonucleotide PCR (ASO-PCR), where detection is qualitative (yes vs. no), rather than quantitative(6). Samples 6–22 comprised archival residual material from the same patient samples examined by Willis et al.(6). A pre-amplification step prior to total ABL quantification was necessary as remaining sample quantities were very limited. Within this group, the 5 patients who ultimately developed a T315I mutation were correctly identified, including three patients in whom the T315I mutation was detected while they maintained complete cytogenetic responses. Overall, the assays were concordant in 14 of 17 cases. One sample (sample 6) was found positive for the T315I mutation by ASO-PCR, but negative by the Fluidigm assay. This archival sample was the diagnostic bone marrow sample from a patient in whom the T315I mutation was ultimately detected by direct nucleotide sequencing at the time of relapse on day 158. The Fluidigm assay detected the T315I mutation in two patients (samples 11 and 13) in whom it was not detected by direct nucleotide sequencing or by ASO-PCR. Explanations for these discordant results include very limited sample quantities, differences in sensitivity, and limited patient follow-up after the original studies were completed. Serial monitoring for the T315I mutation was possible in 8 samples from 3 patients (Table 1, Figure 2). The T315I mutation was detected 99, 102, and 144 days before it was detected by direct nucleotide sequencing in these 3 patients. As shown in Figure 2(f) each of 3 patients demonstrated a similar 2 to 3-log10 increase in mutated T315I ABL from the time of detection until overt clinical relapse.
Table 1. Patient characteristics, outcomes, and mutation detection.
The T315I mutation was detected 99, 102, and 144 days before it was detected by direct nucleotide sequencing (samples 1–8). The T315I mutation was not detected in 8 CP CML patients maintaining CCRs on IM before allogeneic transplantation, in 3 patients who failed IM without evidence of point mutations, in 2 patients with no clinical information available but no evidence of point mutations, or in 3 CML patients with other ABL TKD point mutations. The T315I was not detected in one bone marrow sample and two peripheral blood samples from individuals without leukemia. The T315I was also detected, concordantly with ASO-PCR, in 3 patients maintaining CCRs on IM. Mutation status is reported by ASO-PCR (samples 5–22) or by direct nucleotide sequencing (remaining samples). CCR indicates complete cytogenetic response; MCR indicates major cytogenetic response; and CHR indicates complete hematologic response. BM and PBL indicates bone marrow and peripheral blood, respectively.
| Patient Number |
Sample Number |
Disease | Therapeutic response | Mutation status by sequencing or ASO-PCR (#5–22) |
T315I molecules (Fluidigm) |
|---|---|---|---|---|---|
| 1 | 1 | CML, lymphoid BC | Initial sample, IM failure | None | 1 |
| 1 | 2 | CML, lymphoid BC | Relapse on dasatinib | T315I | 1104 |
| 2 | 3 | CML, myeloid BC | Initial sample, IM failure | None | 1 |
| 2 | 4 | CML, myeloid BC | Relapse on dasatinib | T315I | 99 |
| 3 | 5 | CML, AP | Initial sample, PBL | Not done | 0 |
| 3 | 6 | CML, AP | Initial sample, BM | T315I | 0 |
| 3 | 7 | CML, AP | Day 56, CHR | T315I | 68 |
| 3 | 8 | CML, AP | Day 366, relapse on IM | T315I | 4713 |
| 4 | 9 | CML, CP | Day 90, CCR on IM | T315I | 5 |
| 5 | 10 | CML | No information | None | 0 |
| 6 | 11 | CML | Relapse on IM, subsequent failure on dasatinib | None | 9, repeat 25 |
| 7 | 12 | CML, CP | Loss of MCR on IM | M244V | 0 |
| 7 | 13 | CML, CP | Loss of MCR on IM (later sample) | M244V and others, no T315I | 1 |
| 8 | 14 | CML, AP | Loss of CCR on IM | Y253H, G250E | 0 |
| 9 | 15 | CML, AP | Loss of CCR on IM | H396R | 0 |
| 10 | 16 | CML | No information | T315I | 39 |
| 11 | 17 | CML, CP | No CCR on IM | None | 0 |
| 12 | 18 | CML | CCR on IM | T315I | 112 |
| 13 | 19 | CML | CCR on IM | T315I | 1 |
| 14 | 20 | CML | No CCR on IM | None | 0 |
| 15 | 21 | CML | MCR only on IM | None | 0 |
| 16 | 22 | CML | No response to IM | None | 0 |
| 17 | 23 | CML, CP | Loss of response to IM | T315I | 14 |
| 18 | 24 | CML, CP | CCR on IM | None | 0 |
| 19 | 25 | CML, CP | CCR on IM | None | 0 |
| 20 | 26 | CML, CP | CCR on IM | None | 0 |
| 21 | 27 | CML, CP | CCR on IM | None | 0 |
| 22 | 28 | CML, CP | CCR on IM | None | 0 |
| 23 | 29 | CML, CP | CCR on IM | None | 0 |
| 24 | 30 | CML, CP | CCR on IM | None | 0 |
| 25 | 31 | CML, CP | CCR on IM | None | 0 |
| 26 | 32 | Normal BM | No drug | None | 0 |
| 27 | 33 | Normal PBL | No drug | None | 0 |
| 28 | 34 | Normal PBL | No drug | None | 0 |
Figure 2. T315I detection using the Fluidigm BioMark™ nanofluidic digital array.
Left panel (a–c). Detection of the T315I mutation in patient samples. The top 3 panels (a) show the detection of 0 molecules of T315I ABL in an AP CML patient by absolute quantification of the T315I mutation. The middle 3 panels (b) show the detection of 68 molecules of T315I ABL on day 56 after the initiation of IM at which time the patient had obtained a complete hematologic remission. No mutations were detected by direct nucleotide sequencing or by pyrosequencing at this time. The patient, however, relapsed 3 months later with the T315I mutation evident by direct nucleotide sequencing. On day 366 as shown in the bottom 3 panels (c) 4,713 molecules of T315I ABL were detected in this patient. A cycle cut-off of 36 and a threshold of 0.2 were used for this analysis.
Right panel top (d–e). Positive and negative controls present on each digital array. The top 3 panels show 5,000 molecules per panel of unmutated ABL amplified using the T315I primers. The bottom 3 panels show a spike-in of 2 molecules per panel (6 molecules total) of T315I mutated ABL in a background of 5,000 molecules of unmutated ABL. Data were used only if each control was appropriately negative or positive.
The kinetics of T315I ABL development in 3 patients who failed therapy (f). Serial measurements of the T315I mutation in 3 patients who failed dasatinib (black squares and gray diamonds), or imatinib (black triangles). Patient 1 was in lymphoid blast crisis, patient 2 in myeloid blast crisis, and patient 3 in accelerated phase. The mutation was detected 99, 144, and 102 days, respectively, before relapse at which time it was detected by direct nucleotide sequencing. T315I molecules are depicted on a log10 scale. T315I molecules of zero are depicted as −1.
ABL TKD point mutation detection is an important component of therapeutic management in patients, particularly in patients who do not achieve cytogenetic and molecular milestones. Typically, ABL TKD mutations are detected at the time of cytogenetic relapse or at the time of disease progression, although increases in BCR-ABL copy number may indicate the presence of a mutation (7). While the significance of early ABL TKD mutation detection remains unclear (6), the early identification of the T315I mutation would seem to have clinical relevance; particularly as patients with the T315I mutation cannot be salvaged by nilotinib or dasatinib therapy. Thus, there may be a clinical niche for sensitive methods of ABL mutation detection. Recently a very sensitive method using a polymerase colony (or “polony”) assay was used to quantify ABL TKD mutations in 3 patients with assay sensitivities of up to 1:10,000 (8). The nanofluidic-based mutation-specific PCR assays described here have similar sensitivities, and have several unique features. First, the ability to partition the sample into thousands of independent reaction chambers increases the target to background ratio, and thus sensitivity increases as increased partitioning capacity is utilized. Second, small sample requirements enable a single sample to be screened for a battery of mutations even if sample amounts are limited. Third, this approach, unlike most other sensitive detection strategies, allows for the assessment of the kinetics of ABL TKD mutation development; thus, making it possible to address whether the kinetics of mutation development, rather than the presence or absence of a mutation, correlates more closely with outcome. Fourth, the potential flexibility of this platform allows for thousands of different reactions to be performed on a single sample or multiple samples using only one digital array, and we have designed assays for the simultaneous detection of other ABL TKD mutations. Such sensitive detection of the T315I mutation and other mutations may allow investigators to study the biology of clonal selection and evolution in the context of TKI therapy, resistance, and progression in CML.
Acknowledgments
Supported by the following grants: NCI CA106796, Leukemia and Lymphoma Society Translational Research Grant and V Foundation for Cancer Research V Scholar Grant (VGO); NCI CA18029 (JPR), and the Canary Foundation (SA and FM).
REFERENCES
- 1.Nicolini FE, Corm S, Le QH, Sorel N, Hayette S, Bories D, et al. Mutation status and clinical outcome of 89 imatinib mesylate-resistant chronic myelogenous leukemia patients: a retrospective analysis from the French intergroup of CML (Fi(phi)-LMC GROUP) Leukemia. 2006;20:1061–1066. doi: 10.1038/sj.leu.2404236. [DOI] [PubMed] [Google Scholar]
- 2.Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102:276–283. doi: 10.1182/blood-2002-09-2896. [DOI] [PubMed] [Google Scholar]
- 3.Soverini S, Martinelli G, Rosti G, Bassi S, Amabile M, Poerio A, et al. ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: a study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J Clin Oncol. 2005;23:4100–4109. doi: 10.1200/JCO.2005.05.531. [DOI] [PubMed] [Google Scholar]
- 4.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 5.Baker SG, Kramer BS, McIntosh M, Patterson BH, Shyr Y, Skates S. Evaluating markers for the early detection of cancer: overview of study designs and methods. Clinical Trials. 2006;3:43–56. doi: 10.1191/1740774506cn130oa. [DOI] [PubMed] [Google Scholar]
- 6.Willis SG, Lange T, Demehri S, Otto S, Crossman L, Niederwieser D, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106:2128–2137. doi: 10.1182/blood-2005-03-1036. [DOI] [PubMed] [Google Scholar]
- 7.Branford S, Rudzki Z, Parkinson I, Grigg A, Taylor K, Seymour JF, et al. Real-time quantitative PCR analysis can be used as a primary screen to identify patients with CML treated with imatinib who have BCR-ABL kinase domain mutations. Blood. 2004;104:2926–2932. doi: 10.1182/blood-2004-03-1134. [DOI] [PubMed] [Google Scholar]
- 8.Nardi V, Raz T, Cao X, Wu CJ, Stone RM, Cortes J, et al. Quantitative monitoring by polymerase colony assay of known mutations resistant to ABL kinase inhibitors. Oncogene. 2008;27:775–782. doi: 10.1038/sj.onc.1210698. [DOI] [PubMed] [Google Scholar]