Abstract
Single-cell mass spectrometry (MS) empowers metabolomic investigations by decreasing analytical dimensions to the size of individual cells and subcellular structures. We describe a protocol for investigating and quantifying metabolites in individual isolated neurons using single-cell capillary electrophoresis hyphenated to electrospray ionization time-of-flight MS. The protocol requires ~2 h for sample preparation, neuron isolation, and metabolite extraction, and 1 h for metabolic measurement. The approach was used to detect more than 300 distinct compounds in the mass range of typical metabolites in various individual neurons (25–500-µm in diameter) isolated from the sea slug (Aplysia californica) central and rat (Rattus norvegicus) peripheral nervous systems. A subset of identified compounds was sufficient to reveal metabolic differences among freshly isolated neurons of different types and changes in the metabolite profiles of cultured neurons. The protocol can be applied to the characterization of the metabolome in a variety of smaller cells and/or subcellular domains.
Categories: Neuroscience, Metabolomics, Spectroscopy, Analytical Chemistry, Biochemistry
Keywords: Sea slug, rat, mass spectrometry, single cell mass spectrometry, neuron, capillary electrophoresis
INTRODUCTION
The metabolome, which encompasses the suite of metabolites in a biological system, is an indicator of the physiological, as well as pathological, condition of a cell. Concentrations and localizations of intracellular metabolites are in a dynamic balance that can change rapidly (e.g., in minutes to seconds). Intracellular metabolites are structurally diverse and are found over a broad range of concentrations, e.g., from picomolar to millimolar. The intricate complexity of the metabolome is in part reflected by the number of documented metabolites. For example, the human metabolome database1, which currently contains more than 8,000 entries, is continually expanding. It is therefore understandable that characterization of this chemical milieu within the small volume of a single cell places challenges on analytical measurement systems and benefits from developing measurement protocols tailored for metabolites. Despite significant progresses in the field, further developments are needed in the technology and methodology for single-cell analysis (SCA) to facilitate interrogation of the metabolome with sufficient detection sensitivity and metabolite coverage.
Mass spectrometry (MS) is at the forefront of the bioanalytical technologies that facilitate investigations of various levels of cellular and molecular organization, including the metabolome. MS complements single-cell and subcellular interrogations via established and emerging techniques such as light microscopy2–4 (e.g., Bessel beam plane microscopy5 and light endoscopy via nanowires6). MS detection provides rich chemical information and usually does not require analyte labeling. The approach can be quantitative, and allows the user to observe diverse types of unknown and/or unpredicted compounds. Advances in small-volume sampling and MS measurement schemes have improved this powerful technique for the study of individual cells making detection of hundreds of metabolites possible.
The number of direct MS-based techniques capable of performing single-cell measurements is rapidly increasing. The current state of the field has been the focus of several recent reviews7–13. We previously described a number of approaches for characterizing peptides from a range of cells, from larger molluscan to smaller insect and mammalian neurons, using matrix-assisted laser desorption ionization (MALDI) MS14–16, including a detailed protocol adapted to pituitary cells17. Several other developments in MS technology were recently introduced and applied to SCA and have included investigation of individual metastatic breast cancer cells by nanostructure initiator mass spectrometry18,19, unicellular algae and bacteria by high-density microarray MS20, and yeast cells using nanostructure post arrays21. The distributions of vitamin E22, lipids, and lipid fragments at the soma-neurite junctions of neurons23, as well as lipid domain formation in mating unicellular organisms with ~100-nanometer resolution, were determined using secondary ionization MS imaging24. More recently, analyses of plant, animal, and human cells were extended to ambient conditions compatible with specimen viability in live video MS25, laser ablation electrospray ionization26,27 and femtosecond laser desorption ionization28. The chemical complexity inherent to single cells can potentially interfere with gas-phase ion generation, which in turn can hamper metabolite identification and/or quantitation. To expand the coverage of the metabolome and improve quantitation, additional steps, such as the separation of metabolites in small sample volumes, are needed.
The current protocol facilitates SCA by efficiently separating chemical species using capillary electrophoresis (CE) before electrospray ionization (ESI) MS detection. CE is amenable to multiple sampling approaches, offers relatively high peak capacity in separation, and operates with small sample volumes (e.g., a few nanoliters); thus, it has been adapted for individual cell measurements10,29–32. CE combined with the high information content of MS enables measurements of a range of metabolites in a single experiment. This technology also unites high detection sensitivity with a wide linear dynamic concentration range for quantitation (at least four orders of magnitude31).
Experimental workflow of single-cell CE-ESI MS
Here we demonstrate that CE-ESI-MS is capable of successfully profiling and quantifying the metabolic content of single cells isolated from model organisms in neuroscience and systems biology. Our SCA protocol was used to measure multiple metabolites including classical neurotransmitters in individual neurons collected from the sea slug (Aplysia californica) central nervous system (CNS) and rat (Rattus norvegicus) peripheral nervous system. We selected a set of biologically important cationic metabolites (e.g., carnitines and betaines) and characterized and differentiated neurons based on their underlying chemistries. The analytical workflow begins with sample preparation, followed by CE separation with mass spectrometric detection, and subsequent data analysis and quantitation.
The main goal of sample preparation is to identify, isolate, and stabilize a particular neuron of interest, and then extract its metabolites. A number of different approaches can be used for cell selection and isolation in combination with this protocol, including physical collection via manual manipulations, sorting of cells with select attributes by flow cytometry, and sampling tissue-bound or fixed individual cells and their organelles using laser capture microdissection10,29. During the initial stage of an experiment, the selected cells are isolated, optionally cultured and rinsed, and placed in a small volume of organic solution such as acidified aqueous methanol to facilitate extracting metabolites, quenching enzymatic activity, and reducing metabolic degradation during storage. Each step may influence the detectable portion of the intracellular metabolome, and in turn determine the ability to deduce the in vivo chemistry of the neuron. Our goal is to attain high analytical reproducibility in sample preparation, as well as use an adequate number of biological replicates (e.g., the same neuron type isolated from different members of the same species) and technical parallels (same extract measured multiple times) to ensure experimental success. The results presented here are shown for multiple biological replicates (at least five) measured in technical duplicates and are in agreement with genetic, metabolic, physiological data obtained in independent investigations. However, the exact number of biological and technical replicates that is required is strictly dependent on the sample type and the purpose of the study.
For this protocol, only a portion of the obtained single-cell extract is measured; the sampled volume corresponds to 0.1% of the volume of the extract, and is adjustable so that a greater fraction of the cell content is measured for smaller cells. Alternatively, the metabolite coverage can be improved by drying and then reconstituting the cell extract in a smaller volume, thus increasing the concentration of metabolites that are present at low abundance in the cell. To enable the detection of many metabolites, often present in broad concentration ranges, this protocol imposes stringent criteria on the performance and operation of the CE-ESI MS platform. Stable CE-ESI operation as well as the use of the internal standards and data analysis schemes discussed here (Box 1), improve the detection limit to about 100 amol for standards of neurotransmitters (e.g., acetylcholine and dopamine), electrophoretic migration reproducibility to 1% relative standard deviation (RSD), and mass accuracy to below 5 ppm31,33,34. By collecting higher-quality data, these measurements enable the identification and quantitation of more metabolites.
BOX 1 | CONSIDERATIONS TO ENSURE ROBUST SYSTEM OPERATION.
Successful experiments are facilitated by careful attention to the methodological details, precise calibrations, and frequent maintenance of the CE-ESI-MS system. Sample injection reproducibility is improved by handling the capillary with caution and consistency. Bending the inlet end should be avoided as the bare fused silica is fragile and can readily chip off or break. During sample loading and positioning of the capillary inlet into the background electrolyte, the polyimide coating of the capillary should not contact the solutions as droplets may be trapped at the silica-coating interface and transferred between vials, potentially causing cross-contamination or sample dilution. CE separation reproducibility and reliability is enhanced by frequent cleaning, as well as occasional conditioning of the capillary using a sodium hydroxide solution. If migration time reproducibility falls below ~15% RSD, the separation capillary is conditioned by flushing it with 100 mM of sodium hydroxide for ~5–10 min, followed by thorough rinsing with Sigma water and the background electrolyte for 10 min. The CE-ESI interface should also be thoroughly rinsed with the ES solvent, and its metal emitter frequently rinsed to remove salts and compound deposits. When the platform is not in use, a gentle flow of Sigma water through the capillary connections helps to avoid capillary and emitter clogging and prolong system lifetime. Additional temperature control of the separation capillary further enhances separation reproducibility.
Stable ESI is a prerequisite for reproducible analyte quantitation. In a typical operation, the performance of the ESI is characterized continuously through multiple channels of observation. The temporal pattern of the total ion current and spray current (capillary current) are monitored using the mass spectrometer; the electrohydrodynamic behavior of the ES liquid meniscus is visually observed by a microscope. The operation mode of the ES is driven in the dripping-burst-pulsating-astable-cone-jet spraying regime realm, primarily by decreasing the ES emitter-to-sampling plate distance while keeping the spray voltage constant; cessation of hydrodynamic pulsation at the ES emitter tip and a sudden increase in ion signal intensity indicates onset of the cone-jet mode, usually at a distance below ~3 mm from the sampling-plate held at –1,700 V when the ES sheath solution is supplied at 750 nL/min through the ES emitter55. Note that changing spray potential and sheath flow rate and composition can further affect the spraying mode parameters. As CE-ESI-MS measurements span over a long time period, it is important to actively monitor and, if needed, regulate ES stability; among the governing factors that should be closely managed are the potential and geometry of the ES, the composition and flow rate of the solvent supply, as well as the integrity of connections.
The CE-ESI interface presented here can be operated in two distinct modes, each of which helps to assess system performance at different stages of the experiments. Initially, the ion source is maintained as an ESI-MS-only device—i.e., the CE voltage is held at ground potential—and the integrity of the solution supply and stability of the Taylor-cone are tested. Unstable spray current (e.g., high ion signal fluctuation and/or lack of signal) may indicate erroneous connections and/or improper anchoring of the liquid meniscus at the ES emitter tip (Figure S1, panel a), and/or variations in spraying modes. In the subsequent step, the ion source is operated as a CE-ESI device by applying the CE voltage across the separation capillary. Experimental conditions are routinely refined to mitigate excessive electrolysis and/or solvent heating in the CE region and/or electrical breakdowns (Figure S1, panel b) in the ESI-MS region of the setup. Representative examples of spray and total ion current measurements are demonstrated for erroneous and ideal conditions in Figure S1.
The overall performance of the CE-ESI-MS system is validated daily. A reproducibility in migration time with no more than ~15% RSD, a signal-to-noise ratio of at least 3 for the reference signals (test solution), and an error in absolute ion signal intensity within 20% RSD are preferred for 6 nL of the test solution analyzed. Careful optimization of variables, regular system maintenance, and data analysis can ensure errors of <1% RSD in migration time and 5% RSD in quantitation33. The CE-ESI-MS system, when optimized as described in this protocol, provides reproducible and stable CE separation and efficient ion generation over an extended period of time.
This protocol was validated using A. californica and rat individual neuronal cell bodies with diameters of ~25–500 µm. The metabolites detected were classical neurotransmitters (e.g., acetylcholine and histamine), energy carriers (e.g., adenosine), and osmolytes (e.g., betaines), among others33. The metabolic content allowed us to distinguish distinct neuron types based on chemistry33, and determine metabolic changes in single neurons placed in overnight culture34. Contemporary developments in cell isolation offer technological and methodological capabilities for the measurement of ever-smaller cells and organelles with increasing sample throughput. A number of sample preparation approaches can feasibly be combined with our CE-ESI MS platform. Examples include laser-capture microdissection (to harvest miniscule amounts of samples for extraction35,36), flow and chemical cytometries (to sort and analyze cells with exceptional throughput37–39), and single-cell microfluidic platforms (to automate sample preparation, cell lysis, and direct analysis40). It is anticipated that the presented protocol can be readily extended to SCA of smaller cells, subcellular structures, and cells of various species (see also Box 2).
BOX 2 | SYSTEM ADAPTABILITY.
The success of analyte identification and quantitation is enhanced by monitoring the quality of the analyte signal that is registered by the mass spectrometer. The present protocol profiles analytes over a wide concentration range (e.g., nM to mM concentrations) by addressing low signal-to-noise ratios and detector saturation at the low- and high concentration extremes, respectively. Measuring the appropriate signal intensities aids accurate mass determinations (m/z values) and obtaining informative tandem mass spectra, and also helps to expand the linear dynamic range of concentration for quantitation. As only a fraction of the cell extract is analyzed per this protocol, the sample can be measured more than once. Replicate measurements allow adjustment of the sample amounts that are loaded into the separation capillary by refining the duration of injection and/or vertical displacement between the inlet and outlet ends, thus producing different hydrodynamic injection forces. Analytes in aliquots of the sample may also be concentrated via air-drying or diluted in situ in the loading vial prior to measurement—the use of internal standards is especially recommended. In addition, various CE separation methodologies can be adapted to this protocol to enhance the separation efficiency of the platform (e.g., on-line sample stacking and micellar electrokinetic chromatography). Similar analytical considerations should be effective in measuring diverse types of small molecules and concentrations, while also providing an opportunity to expand detection to larger compound types such as peptides and proteins of single cells.
Advantages and limitations of single-cell CE-ESI MS
Single-cell CE-ESI MS provides a means to profile, identify, and quantify a representative part of the metabolome. Thus, selected cells that have real or suspected cell heterogeneity make key targets. Compared to many more traditional single-cell assays, advantages of the approach include an ability to detect both known and unknown compounds. CE separation is compatible with the volume regime of single cells, yields high separation power and an ability to work with metabolites of broad types. Furthermore, CE separation can tolerate biological samples with high salt concentration such as the individual neurons of A. californica and rat measured here.
This section discusses several challenges. The first involves creating a robust approach for selecting the cell of interest. Cell selection is defined by the specific question being addressed and will vary from model to model. As cell sampling is application- and cell-type-dependent, it is discussed further only for the specific cases covered here that use manual cell isolation. Note that the sample preparation and measurement process, analytical system setup, and routine operation all require judicious attention. Key parameters for optimization are discussed in Box 1 and include isolation of single cells and extraction of metabolites during sample preparation (Steps 1–11), collecting information-rich metabolic data with low limits of detection and high analytical reproducibility using the CE-ESI MS platform (Steps 12–29), and streamlining data analysis (Steps 30–5).
Another challenge relates to the applied aspects of ESI MS. A technical challenge is that ESI sources are somewhat intolerant of inorganic salts and complex buffers and exhibit low stability of Taylor-cone formation in the negative ion mode. Although this protocol is optimized for detecting metabolites in the positive ion mode, revising the ES source parameters can help to extend detection to the negative mode, thus potentially enhancing coverage of the single-cell metabolome (see REAGENT SETUP). A practical matter, as is the case with the majority of ‘omic approaches that yield high data content, is that some users may find manual data analysis to be particularly labor intensive. As a final point worth noting, the throughput of many single-cell measurements, including the CE-MS approaches highlighted here, is low enough that large numbers of cells are not normally probed and therefore, the cell selection step becomes especially critical.
Adapting our protocol to alternative platforms may help to mitigate some challenges. Interfering compounds (e.g., salts and buffers) may be diverted from the analytical path of ion generation via microreactors in microfluidic devices or capillary-based electrophoretic systems40,41. Alternatively, separated compounds may be ionized using ion sources with higher sample matrix tolerance or different operating principles; e.g., CE coupled to desorption electrospray ionization is compatible with 10–50 mM detergents and inorganic salts42, and similar advantages can be anticipated for other ambient ion sources11,43,44. Regarding data analysis, several software packages (e.g., metaXCMS45, MZmine,46 and MathDAMP47) can help to align electrophoretic data and automate metabolite identification, statistical, and multivariate analysis.
MATERIALS
REAGENTS
(Optional) Air dehumidifier
(Optional) Hygrometer and thermometer (Fisher Scientific, cat. no. 14-649-84)
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (Sigma, cat. no. H3375)
Acetic acid (Sigma, cat. no. A6283) ! CAUTION Acetic acid is highly corrosive, combustible, and toxic if inhaled. It readily absorbs upon contact and may be harmful. When handling, wear gloves, safety goggles, use a pipetting aid, and operate in a ventilated chemical hood.
Acetylcholine chloride (Sigma, cat. no. A6625) ! CAUTION Acetylcholine chloride causes irritation if contacted, inhaled, or swallowed. When handling, wear gloves, and operate in a ventilated chemical hood.
Calcium chloride dihydrate (Sigma, cat. no. C7902)
Dopamine hydrochloride (Sigma, cat. no. H8502) ! CAUTION Dopamine hydrochloride causes irritation if contacted or inhaled and is harmful if swallowed. When handling, wear gloves, and operate in a ventilated chemical hood.
Formic acid (Thermo Scientific, cat. no. TS-28905) ! CAUTION Formic acid is highly corrosive, combustible, and toxic. It readily absorbs upon contact or inhalation. When handling, wear gloves, safety goggles, use a pipetting aid, and operate in a ventilated chemical hood.
Gentamicin sulfate (Sigma, cat. no. G1264) ! CAUTION Gentamicin sulfate may cause an allergic reaction on the skin or in the respiratory system. When handling, wear gloves, and operate in a ventilated chemical hood.
Glycerol (Sigma, cat. no. G7893) ! CAUTION Glycerol causes irritation to the eye and skin. When handling, wear gloves.
Histamine dihydrochloride (Sigma, cat. no. H7250) ! CAUTION Histamine dihydrochloride is toxic and causes irritation if contacted, inhaled, or swallowed. When handling, wear gloves, and operate in a ventilated chemical hood.
Isopropanol (Sigma, cat. no. 34965) ! CAUTION Isopropanol is highly flammable and is a mild skin- and serious eye-irritant. When handling, wear gloves, goggles, and operate in a ventilated chemical hood.
Magnesium chloride hexahydrate (Sigma, cat. no. M2393)
Magnesium sulfate heptahydrate (Sigma, cat. no. M2773)
Methanol (Sigma, cat. no. 34860) ! CAUTION Methanol is highly toxic and combustible. When handling, wear gloves, safety goggles, use a pipetting aid, and operate in a ventilated chemical hood.
Penicillin G sodiate (Sigma, cat. no. P3032) ! CAUTION Penicillin may cause an allergic reaction on the skin or in the respiratory system. When handling, wear gloves, and operate in a ventilated chemical hood.
Potassium chloride (Sigma, cat. no. P5405)
Serotonin hydrochloride (Sigma, cat. no. H9523) ! CAUTION Serotonin hydrochloride is highly toxic and causes irritation if contacted, inhaled, or swallowed. When handling, wear gloves, and operate in a ventilated chemical hood.
Sodium chloride (Sigma, cat. no. S5886)
Sodium hydroxide (Sigma, cat. no. 221465) ! CAUTION Sodium hydroxide causes severe skin burns and eye damage. When handling, wear gloves, and protective clothing and goggles.
Streptomycin sulfate (Sigma, cat. no. S9137)
Water for metabolite extraction and CE-ESI MS analysis (Sigma, cat. no. 39253). Note that this type of water is of high purity and is referred to as Sigma water in this protocol.
Deionized water for neuron isolation and culturing (Millipore Corp., Milli-Q water with resistivity of ≥18.2 MΩ cm). This type of water is referred to as Milli-Q water in this protocol.
EQUIPMENT
Alligator clips for making electrical connections
Binocular headband magnifier with 2.75× magnification (e.g., Donegan Optical DA-7)
Copper wire without insulation, 10 cm in length, thin and flexible
Data acquisition and control card with software (National Instruments Corp., model USB-6008 and LabVIEW 8.2 or later)
Digital multimeter (Fluke Corp., e.g., 79 series III)
Electrical wires with insulation
Electrospray (ES) mounting and positioning system (Thorlabs, cat. no. PT3/M or New Objective Ltd., cat. no. Pico View)
Fine glass pipettes made from thin-walled, single-barrel, standard borosilicate glass tubing with 1,000 µm OD and 750 µm ID (World Precision Instruments, cat. no. TW100F-4) using a micropipette puller (e.g., Sutter Instrument Co., model P-80)
Fused silica capillary with 105 µm OD and 40 µm ID and ~120 cm in length for separation (Polymicro Technologies, cat. no. TSP040105). Note that this capillary is referred to as separation capillary in this protocol. ! CAUTION Fused silica capillaries are flexible and have sharp ends; use protective eyewear when handling
Fused silica capillary with 365 µm OD and 75 µm ID and ~120 cm length for delivery of ES solvent (Polymicro Technologies, cat. no. TSP075375). Note that this capillary is referred to as sheath capillary in this protocol. ! CAUTION Fused silica capillaries are flexible and have sharp ends; use protective eyewear when handling
High-resolution, high-mass accuracy mass spectrometer equipped with software packages (Bruker Daltonics, micrOTOF TOF-MS and/or maXis Qq-TOF-MS/MS series and Compass software)
High-voltage power supply (HVPS) generating regulated 30 kV potential (Spellman High Voltage Electronics Corp., e.g., Bertan model 30R)
Inverted microscope (Carl Zeiss, model Axiovert 25) equipped with 2.5× and 10× objectives, AxioCam MRc camera (Carl Zeiss), and digital image processing software for widefield microscopy (Carl Zeiss, AxioVision; freeware edition of AxioVision LE is downloadable from www.zeiss.de)
PCR tubes (MidSci)
Petri dish filled with solidified Sylgard 184 silicone elastomer (Dow Corning)
Petri dishes (plastic) with 36-mm diameter for A. californica cell culturing
Powder-free nitrile gloves, goggles and low-lint absorbent wipes (Kimwipes, Kimberly-Clark Inc., cat. no. 34155)
Resistor (10 kΩ)
Scissors with pointed tips, 4- to 6-inch in length
Scribe for fused silica cleaving (New Objective Ltd., cat. no. SCRIBE)
Sharp tungsten needles (World Precision Instruments Inc.) ! CAUTION Needles have sharp ends; handle with care and use protective equipment
Small animal guillotine (e.g., NEMI Scientific Inc., model 701)
Stainless steel needle (Small Parts Inc., stainless steel tubing with 210 µm OD and 165 µm ID, or Hamilton Company, cat. nos. 21030A or 21031A blunt-tip needles)
Soldering station with solder
Standards solution containing ESI tuning mix (Agilent, cat. no. G1969-85000 or Bruker, cat. no. 18220)
Stereomicroscope with 7.9:1 adjustable zoom (Leica Microsystems Inc., cat. no. MZ 7.5) equipped with a digital camera
Stereo-zoom microscope on boom stand capable of 1–6.3× adjustable magnification with long-working range (Olympus Corp., cat. no. SZ6045) for visual monitoring of ES stability
Stopwatch (Sigma, cat. no. Z253294)
Surgical instruments, including fine forceps and microscissors (e.g., Fine Science Tools, Dumont #5 fine forceps, cat. no. 11253-20 and Cohan-Vannas spring scissors with 4 mm cutting edge, cat. no. 15018-10)
Syringe pump (Harvard Apparatus, model 70–2000 or 55–2222) with 500 µL and 5 mL gas-tight syringes (Hamilton, part nos. 81220 and 81520, respectively) equipped with 22-gauge needles (Hamilton Company, cat. no. 90134)
Tubes, protein LoBind, Safe-Lock, 0.5 mL, PCR clean (Eppendorf)
Tubes, plastic, ~2.5 mL (e.g., Eppendorf, cat. no. 05–402-7 and Wheaton, cat. no. 225415)
Tubing sleeves with 635 µm OD and IDs of 125 µm, 330 µm, and 395 µm (IDEX Health & Science, cat. nos. F-180, F-184, and F-185, respectively)
T-union (IDEX Health & Science, cat. no. P-875)
REAGENT SETUP
Animals
Adult A. californica (175–250 g) (National Resource for Aplysia, Rosenstiel School of Marine & Atmospheric Science), are maintained in continuously circulated and aerated artificial seawater cooled to 14 °C. Sprague Dawley outbred rats (Rattus norvegicus), 2.5 to 3 months old (Harlan Laboratories, Inc.), are housed on a 12 hour light cycle and fed ad libitum. Animal euthanasia is performed in accordance with the appropriate institutional animal care guidelines, and in full compliance with federal guidelines for the humane care and treatment of animals. ! CAUTION Experiments involving animals must follow institutional and federal regulations. Rats are sacrificed by decapitation using a sharp guillotine. 120 mL of ice cold mGBSS (see below) is injected immediately into the subepidermal areas close to the dorsal root ganglia (DRG) of interest. Injected rat trunks are placed on ice where all surgical manipulations are made. DRGs are surgically isolated during the ~10 min dissection procedure and placed into ~2 mL of cell stabilization mixture containing (v/v) 33% glycerol and 67% mGBSS prepared with Milli-Q water. Studies involving animals should be planned in accordance with the ARRIVE guidelines48.
Artificial seawater (ASW)
460 mM NaCl, 10 mM KCl, 10 mM CaCl2, 22 mM MgCl2, 26 mM MgSO4, and 10 mM HEPES in Milli-Q water with the pH adjusted to 7.8 using 1 M NaOH in Milli-Q water. The solution can be stored at 14 °C for up to one month.
Culturing solution
ASW freshly supplemented with 100 units/mL penicillin G, 100 µg/mL streptomycin, and 100 µg/mL gentamicin in Milli-Q water with the pH adjusted to 7.8 using 1 M NaOH in Milli-Q water. Solution temperature is maintained at 20–25 °C during culturing. (Optional) Nutrients and growth factors may be added depending on cell type and study goals. ! CAUTION Culturing media may influence the cellular metabolome34 and need optimization in specific studies.
Electrolyte
1% formic acid in Sigma water. Although this solution works well for the CE separation of many metabolites, its composition may be adjusted depending on the purpose of experiment. The solution can be stored in a tightly sealed container at room temperature for 1 month.
ES emitter
metal capillary (~35 mm length) with ends laser-cut perpendicular to the axis and fine-polished using 12-µm diamond lapping paper to yield a smooth surface. The capillary is thoroughly cleaned by sequential sonication in 50% isopropanol using Sigma water, Sigma water, 50% methanol in Sigma water, then methanol, and air-dried.
Enzyme solution for Aplysia neuron isolation
Freshly prepared 1% protease type IX in ASW supplemented with 100 units/mL penicillin G, 100 µg/mL streptomycin, and 100 µg/mL gentamicin.
Mass calibration solution for the mass spectrometer
15 mM sodium formate in Sigma water. The solution can be stored at room temperature up to 1 year. (Optional) The majority of the available mass calibration solutions for MS can be used in the protocol (e.g., polyethylene glycol and polypropylene glycol polymers).
Metabolite extraction solution
50% methanol prepared using 0.5% acetic acid and Sigma water. The solution can be stored in a tightly sealed container at room temperature for 1 month. (Optional) Standards may be added to the solution: migration time markers for CE data alignment (e.g., HEPES, glutathione, and peptides) and internal calibrants for mass spectrometer calibration, signal normalization, and/or quantitation (e.g., deuterated metabolite standards). The solution composition may be adjusted to improve extraction efficiency for certain classes of compounds; for example, increasing methanol concentration should help to better extract nonpolar compounds and adjusting the pH to basic conditions may better extract acidic metabolites for negative ion-mode detection.
Modified Gey's balanced salt solution (mGBSS)
1.5 mM CaCl2, 4.9 mM KCl, 0.2 mM KH2PO4, 11 mM MgCl2, 0.3 mM MgSO4, 138 mM NaCl, 27.7 mM NaHCO3, 0.8 mM Na2HPO4, and 25 mM HEPES dissolved in Milli-Q water with the pH adjusted to 7.2 using NaOH in Milli-Q water. The solution can be stored at 4 °C for 3 months without detectably affecting the outcome of metabolomic measurements in this protocol.
Sheath flow solution
50% methanol prepared with Sigma water containing 0.1% (v/v) formic acid. The solution can be stored in a tightly sealed container at room temperature for 1 month. (Optional) The composition of the sheath flow solution can be changed to optimize ion generation yield and stability. For example, replacing formic acid by volatile basic additives such as ammonium hydroxide facilitates detecting metabolites in the negative ion mode. Consult with the MS literature for alternative approaches.
Stabilizing solution for Aplysia neurons
mixture (v/v) of 33% glycerol and 67% ASW prepared with Milli-Q water. The solution can be stored at 4 °C for 3 months.
Stabilizing solution for DRG neurons
mixture (v/v) of 33% glycerol and 67% mGBSS prepared using Milli-Q water. The solution can be stored at 4 °C for 3 months.
Test solution
50 nM acetylcholine, 50 nM dopamine, 50 nM histamine, and 50 nM serotonin in 50% methanol prepared with 0.5% acetic acid and Sigma water. The solution can be stored in a tightly sealed secondary container at −20 °C for about 6 months without detecting significant changes in composition.
EQUIPMENT SETUP
Single neuron isolation
Microscissors, sharp metal needles, Petri dishes containing Sylgard 184, a stereomicroscope, and an inverted microscope are needed for cell isolation. A. californica neurons are cultured in plastic Petri dishes filled with the ASW-antibiotics solution. The same set of items is used for the isolation of the rat neurons. The setup utilized for the isolation of mammalian pituitary cells is discussed elsewhere17.
System for sampling and CE separation
The system includes a custom-designed stage capable of elevating a platform 15 cm within 1 s. Components of the platform are shown in Figure 1. This platform houses a capillary holder capable of transitioning in three degrees of freedom (element 3, Figure 1) and two stainless steel vials, one for the sample (element 7, Figure 1) and the other for the CE background electrolyte (element 8, Figure 1). The sample-loading vial is milled out conically to hold about 500 nL of solution, and the electrolyte-containing vial is cylindrical with an internal volume of 2 mL. Milling (in advance) 3–5 replicate vials for the CE background electrolyte, and 30–50 separate vials for sample loading, increases the overall measurement throughput of this protocol. During system use, both vials are held in a metal block and are removable for frequent cleaning. The metal block is connected through a 10 kΩ resistor to an HVPS, and a battery-operated digital multimeter connected in parallel measures the voltage drop across the resistor to deduce electrophoretic current during separation. An electrically insulating enclosure (e.g., Plexiglas), equipped with a safety interlock-enabled door, houses all components and overrides the power feed to the CE HVPS for added safety. The CE power supply is controlled by a personal computer executing the attached custom-written LabVIEW-based virtual instrument (vi; see Supplementary Sequence Archive.zip and Supplementary Methods for operation) that is capable of generating a temporally defined voltage and/or current program (see Supplementary Methods, Figure S2). An insulating plastic tube with an ~2 cm ID encloses and guides the separation capillary from the capillary holder in the enclosure into the CE-ESI interface (element 9, Figure 1). Additional details on the elements and operation of the system are available elsewhere31,33,44. (Optional) The CE HVPS may be operated manually. ! CAUTION High voltage and high current pose electrical shock hazards. Ground conductive parts and shield electrical connections to prevent contact. Provide warning signs to inform of the hazard. Make sure that no salt-containing solution spillage remains (even in dry form) inside the enclosure as buildup of contaminants can result in electrical breakdown.
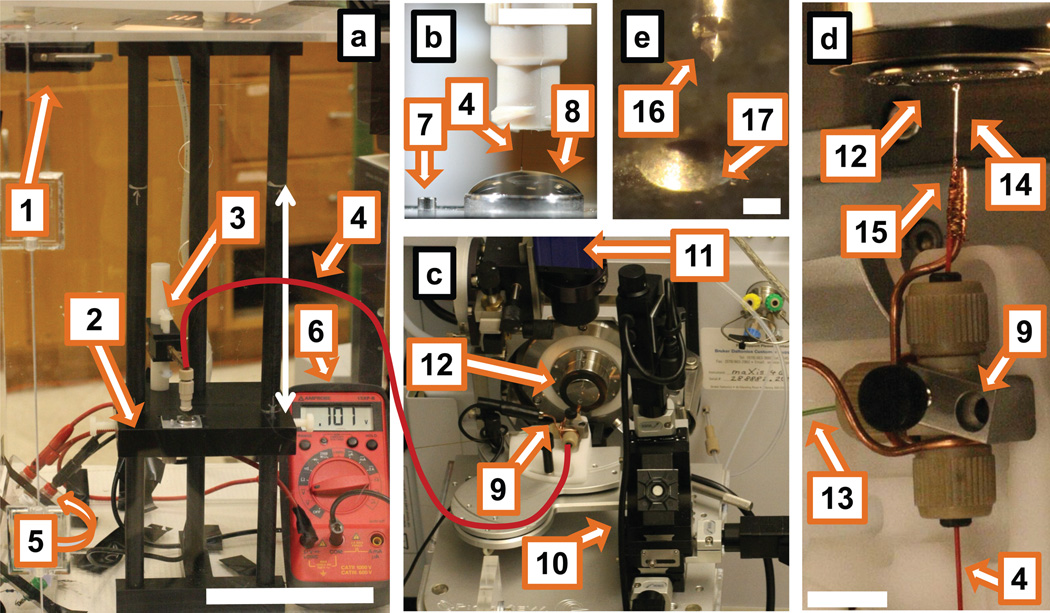
Figure 1.
Experimental setup of the single-cell CE-ESI-MS system. (a) Front view of the CE platform highlighting (1) the enclosure equipped with a safety-interlock enabled door, (2) platform for sample loading, which can be rapidly elevated by 15 cm (see arrow), (3) holder allowing manual positioning of the separation capillary in three degrees of freedom, (4) separation capillary (solid line in red), (5) resistor connected in series to a stable HVPS (Figure S2) and the CE platform, and (6) digital multimeter connected in parallel measuring voltage drop on the resistor. Scale = 10 cm. (b) Magnified view of the sample-loading platform consisting of (7) the sample-loading vial and (8) electrolyte-containing vial with the separation capillary positioned 2 mm below the electrolyte meniscus. Scale = 1 cm. (c) Distant view of the CE-ESI-MS ion source consisting of (9) the CE-ESI interface mounted on (10) the three-axis translation stage of a PicoView nanospray source, (11) CCD camera equipped with light-collimating and focusing lenses to record ES performance, and (12) a mass spectrometer equipped with a nanospray sampling plate. (d) Close-up view of the CE-ESI-MS ion source highlighting a T-union that houses fused silica capillaries for (4) CE separation and (13) ES sheath solution delivery as well as (14) a metal emitter grounded (earth) through a (15) thin copper wire. Scale = 1 cm. (e) Magnified view of (16) the stable Taylor-cone formed upon operating the ES in the cone-jet spraying mode (see reflected image) in front of (17) the orifice of the mass spectrometer sampling plate. Scale = 500 µm.
CE-ESI interface
The T-union incorporates the CE separation capillary, ES sheath supply capillary, and metal capillary in each end. The separation capillary outlet coaxially feeds through the metal capillary and protrudes ~20–200 µm beyond the emitter tip. A thin copper wire directly grounds (earth) the metal emitter. Instructions on assembling this interface are provided in Step 12, and further details are given elsewhere33,44. The CE-ESI interface is mounted on a three-axis translation stage, such as the PicoView nanospray source (Figure 1), and is fine-positioned in front of the sampling plate of the mass spectrometer. The performance of the CE-ESI interface is closely monitored, as detailed in Box 1.
MS and MS/MS
The single-stage time-of-flight (TOF) mass spectrometer (e.g., Bruker Daltonics, micrOTOF ESI-TOF-MS) for metabolic profiling, and/or the tandem TOF mass spectrometer for structural elucidation (Bruker Daltonics, maXis ESI-Qq-TOF-MS/MS series) are needed. The instruments are tuned for performance in sensitivity and mass-resolution (60,000 full width at half maximum below m/z 1,000) following vendor guidelines. In this protocol, the instruments are initially course-tuned below m/z 2,000 by electrospraying the ESI tuning mix, and then fine-tuned using the test solution below m/z 500; the resulting instrument operation conditions are saved as a tuning file, as detailed in the vendor’s manual. Afterwards, the commercial ion source is removed, and the sampling cone is replaced with a nanospray end plate (sampling plate; Bruker). The ion source interlock is disabled according to vendor instructions. In this protocol, both instruments are used and defaulted to the optimized tuning file. The sampling plate potential is set to –1,700 V relative to ground (earth) during the experiments, and 0 V otherwise, with a spectral acquisition rate of 2 Hz. The mass spectrometer is set to stand-by mode when not in use for more than a few hours. Note that this protocol can be implemented on most mass spectrometers equipped with an ESI atmospheric-pressure ion source. ! CAUTION The sampling plate presents a risk for electrical shock. Provide shielding to prevent contact, and post warning signs to inform of the hazard.
Online resources
Data analyses are facilitated via a number of online metabolic and peptidomic MS and MS/MS databases: Metlin (Scripps Center for Metabolomics, http://metlin.scripps.edu),49 the Human Metabolome Database (HMDB) (Genome Alberta and Genome Canada, http://www.hmdb.ca), LIPID Metabolites and Pathways Strategy resource (LIPID MAPS Consortium, http://www.lipidmaps.org), LipidBank (Japanese Conference on the Biochemistry of Lipids, http://www.lipidbank.jp), MassBank (http://www.massbank.jp), ChemSpider (http://www.chemspider.com/), Chemical Entities of Biological Interest (ChEBI) (European Bioinformatics Institute, http://www.ebi.ac.uk/chebi/), and NeuroPred for neuropeptides (UIUC Neuroproteomics Center on Cell-Cell Signaling, University of Illinois at Urbana–Champaign, http://neuroproteomics.scs.illinois.edu/neuropred.html)50.
PROCEDURE
Preparation for single neuron sampling ● TIMING ~2–28 hours
1| Prepare the ASW, mGBSS, and stabilization solutions as described in REAGENT SETUP.
2| Clean and sterilize surgical instruments. As documented in our earlier protocol17, ultrasonic treatment is recommended in SPECTRA-SONIC solution (Spectrum Surgical Instruments Co.), at pH 7, for 5–10 min. Autoclaving is performed according to the operating guidelines recommended by the manufacturer. Rinse the instruments with Milli-Q deionized water and allow them to air dry. This step minimizes the possibility of contaminating samples with bacteria and other metabolite sources.
3| Prepare gloves, goggles, Petri dishes, vials, paper, and the microscopes for neuron isolation as detailed in EQUIPMENT SETUP.
4| Isolate and culture the cells of interest. We have provided steps for isolation and culturing neurons from A. californica (option A) and for Rat DRG neuron isolation (option B).
A) A. californica neuron isolation and culturing ● TIMING 1–4 h (isolation), 8–24 h (culturing)
-
i.
Anesthetize A. californica by injecting a solution of 390 mM MgCl2 into the visceral cavity, equal by mass to approximately one-third of the animal’s body weight.
-
ii.
Open A. californica’s visceral cavity with sharp surgical scissors, dissect the ganglia of interest, and place them into 2.5 mL plastic tubes filled with the enzyme solution.
-
iii.
Incubate the A. californica ganglion in the enzyme solution at 34 °C for 20–120 min. This step helps to loosen or remove the connective tissues surrounding cells and the intercellular mechanical contacts. Depending on the age, size, and physiological condition of the animal, as well as the season and properties of the ganglion and cell of interest, treatment time should be optimized. Overtreated cells have a low rate of survival in culture, whereas undertreated neurons can be difficult to isolate or separate from glia.
-
iv.
Rinse the ganglion several times with fresh ASW for a total of 30–60 min to remove the enzyme, then incubate it for 10–15 min in the glycerol stabilization solution.
-
v.
Pin down the A. californica ganglion onto a Sylgard surface in a Petri dish. Working under the stereomicroscope, isolate the individual neurons of interest in ASW or the stabilization solution for Aplysia using sharp needles, forceps, or surgical scissors.
▲ CRITICAL STEP The MS measurement reproducibility is improved when the isolation is performed in the stabilization solution34. Although there are several established protocols and technologies to aid single-cell isolation from tissues and cell suspensions51–53, only one is described here.
? TROUBLESHOOTING
-
vi.
To culture neurons, aspirate the neurons one by one into a plastic micropipette, and transfer each neuron into a separate Petri dish filled with culturing solution. Culture the neurons overnight at room temperature. To extend the time frame of culturing (e.g., up to 3 d), reduce the temperature to 14 °C. Optimize the culturing conditions, including solution composition, nutrients, oxygen supply, temperature, and time of experiment, according to cell characteristics or the purpose of the study. After culturing, gradually replace the culturing solution with the stabilizing solution by sequentially removing the former and adding the latter in equal volume. ▲ CRITICAL STEP During this step, neurons should not contact the air to avoid cell lysis, or be exposed to the stabilizing solution for more than 3–10 min to preserve cell stability. (Optional) This step may be skipped if neuron culturing is not the goal of the study.
B) Rat DRG neuron isolation ● TIMING 35 min
-
i.
Euthanize rats by rapid decapitation. Cut the vertebral bones in the pedicle areas on both sides of the vertebrae, lift the dorsal side of the chain of cut vertebrae, and remove the spinal cord with forceps to expose the DRGs for isolation using fine forceps and scissors. Alternatively, multiple DRGs can be rapidly exposed by severing the dorsal and ventral roots via rapid hydro-extrusion to the spinal cord with cold physiological saline as described elsewhere54.
-
ii.
Place the isolated DRGs in the stabilization solution and incubate for at least 10 minutes.
-
iii.
Transfer the DRG onto a regular microscopy glass slide, and while monitoring the procedure under an inverted microscope (2.5× objective), use sharp metal needles to break the sheath of connective tissues surrounding the neurons.
-
iv.
Use the needles to pull small groups of DRG neurons out of the ganglia, and isolate the neuron of interest using the same needles while visually following the isolation procedure under a stereomicroscope (with 10–20× magnification).
Preparation of cell extracts and controls ● TIMING 30 s
▲ CRITICAL STEP Note that this protocol eliminates purposeful mechanical disruption to the cell as the extraction solution was found to reproducibly extract metabolites with sufficient efficiency. This conclusion was made from observing that the analysis of cell extracts from cells that are visually intact and damaged during transfer produce similar results. However, verification of this observation is needed for each new type of cell due to differences in cellular physicochemical characteristics as well as analyte and surrounding matrix properties. As a result, additional chemical and/or physical treatments may be implemented to influence analyte extraction and preservation.
5| Aspirate the isolated cell into a plastic micropipette with minimal ASW, and deposit it on the inner wall of a PCR vial. Use a pipette to gently reduce the volume of ASW surrounding the cell. Note that a small amount of extracellular solution is inherently transferred along with the neuron during this step and will be removed in Step 7.
▲ CRITICAL STEP Handle the cell gently and rapidly to minimize cell damage and accidental excessive leakage or release of metabolites.
6| (Optional) Collect this extracellular solution into a separate PCR vial as a negative control for characterizing the chemical environment of the cell and potential analyte loss due to its leakage or release (label: cell environment).
7| Rinse the neuron within 3 s by depositing ~1 µL of water on it and quickly withdrawing the solution into a pipette tip.
▲ CRITICAL STEP This step is especially important when incubating cells in culture media whose composition is not precisely known (e.g., serum). The purpose of rinsing is to minimize the influence of extracellular compounds on measurement results, thereby facilitating data interpretation. Although isolated cells are usually rinsed in this protocol, this step may be skipped with justification; for example, a cell prepared in the stabilization solution may transfer an insignificant volume of extracellular media.
8| (Optional) Collect this rinse solution into a PCR vial for the measurement of compounds removed from the cell environment (label: rinse solution).
▲ CRITICAL STEP Analyzing the rinse solution helps to establish the effect rinsing may have on the cell metabolome (e.g., rinse-stimulated endogenous compound release or leakage); systematic chemical biases in cell preparation may also be feasibly identified this way34.
9| Pipette 500 nL of the extraction solution onto a ~25 µm rat DRG neuron and 5 µL of the extraction solution onto a ~50–500 µm A. californica neuron (label: cell extract). The volume of the extraction solution may be adjusted depending on the size and chemistry of the cell as well as the purpose of the study. Alternatively, deposit glycerol-stabilized cells using pulled glass pipettes into said volumes of extraction solution preloaded into Protein LoBind or PCR tubes. Note that small glass pieces that may break into the extraction solution from the glass micropipette tip during cell transfer do not typically interfere with sample measurement. ▲ CRITICAL STEP Ensure that the cell is immersed in solution during metabolite extraction.
10| (Optional) Add standards to the extract: migration time markers for CE data alignment and internal calibrants for mass spectrometer calibration, signal normalization, and/or quantitation. Alternatively, the same standards may be added to the metabolite extraction solution (see REAGENT SETUP) prior to depositing it onto the cell.
11| Centrifuge vials and analyze cell extracts.
▪ PAUSE POINT Cells can be stored in the extraction solution at –20 °C for up to a week.
▲ CRITICAL STEP Ensure that cells are immersed in solution during storage. Other control samples, including the metabolite cell extraction, rinse, and test solutions, should be kept under identical conditions. (Optional) Keep solutions at –80 °C to reduce metabolite degradation for about a month. Method development should optimize storage conditions depending on the compounds of interest, cell type, and extraction parameters, among other factors.
Assembling the CE-ESI-MS platform ● TIMING 2–4 h
▲ CRITICAL STEP Use appropriate parts (e.g., sleeves, fittings, ferrules, and nuts) during system assembly to ensure the integrity of connections.
! CAUTION Bare fused silica is highly fragile and poses a poking hazard. When performing this task, wear protective eyewear and gloves.
12| Assemble the CE-ESI interface:
Mount the ES emitter into the T-union (see Figure 1d) and feed the separation capillary through the parallel inlet of the union until it protrudes ~15 cm past the emitter tip.
Cleave the separation capillary ends with a scribe and then rapidly burn about 3 mm of coating from both ends.
Wipe the capillary ends with isopropanol until the exposed fused silica ends appear clean under a microscope.
Gently retrieve the capillary through the ES emitter and finger-tighten the connections, allowing the outlet end to protrude 20–200 µm beyond the emitter tip as measured using a microscope.
Connect the sheath capillary to the perpendicular inlet of the T-union.
▲ CRITICAL STEP The protrusion length and position of the separation capillary within the ES emitter will affect the stability of the Taylor-cone during CE separation and should be optimized.
13| Mount the CE-ESI interface onto the three-axis translation stage and position the ES emitter perpendicular to the sampling plate at an ~20 mm distance. Figure 1c demonstrates the use of the PicoView nanospray source fitted for the maXis mass spectrometer.
14| Secure the separation capillary inlet into the capillary holder of the sample-loading platform (element 3, Figure 1), and connect the sheath capillary to the syringe driven by the syringe pump.
15| Clean the internal and external CE-ESI interface components for ~30 min after first assembly, ~10–15 min at the beginning of an experiment series, and at least 5 min between consecutive measurements according to the procedure in Box 3.
Box 3 | CLEANING CE-ESI INTERFACE COMPONENTS.
The purpose of these steps is to fill up the connections and remove potential contaminants, including metabolites that may be carried over between experiments. When appropriate, the steps may be performed in parallel to obtain higher throughput. Note that each measurement requires a new, dry clean sample-loading vial and replenishing of the electrolyte. The same electrolyte-containing vial can be utilized ~15 times before cleaning via sonication. To clean the interface:
-
1)
Turn on the mass spectrometer with the sampling plate potential set to 0 V
-
2)
Position the emitter ~15 mm from the sampling plate
-
3)
Flush the background electrolyte through the separation capillary using the syringe equipped with a rubber band applying gentle pressure on the syringe plunger (another syringe pump or gravity-driven solution supply may alternatively be used for this purpose)
-
4)
Supply the sheath solution at a 750 nL/min rate through the sheath capillary using a separate syringe and the syringe pump
▲ CRITICAL STEP The sheath flow supply rate should be optimized for system performance (Box 1) and kept stable throughout the protocol.
-
5)
Rinse the ES emitter tip with Sigma water and then methanol
-
6)
Using tweezers, remove a cleaned sample-loading vial from the storage solution, dry it with an adsorbent wipe (e.g., Kimwipe), and transfer it into the corresponding vial holder (element 7, Figure 1) of the sample loading platform to allow it to further air dry
-
7)
Replenish the background electrolyte in the electrolyte-containing vial (element 8, Figure 1) in the sample loading platform.
16| Remove the rubber band-operated syringe from the separation capillary, mount the capillary inlet into capillary mount of the platform, and translate this capillary mount to transfer the separation capillary inlet end ~2 mm below the background electrolyte liquid meniscus. Improve the accuracy of manual fine-positioning with a magnifier and adequate lighting. Ensuring the proper capillary dipping depth is important to offset the decreasing meniscus levels resulting from the evaporation of electrolyte. Although this protocol proposes an hour of separation, longer experiments are feasible by increasing the dipping depth.
▪ PAUSE POINT (Optional) In order to place the setup in stand-by mode for a short time (e.g., 1–2 h), apply –1,700 V on the sampling plate, and operate the ES in the burst-pulsating-dripping spraying regime realm55 by adjusting the ES emitter-to-sampling plate distance to ~5–15 mm; monitor the spraying mode using the microscope. See Box 1 and the Supplementary Methods for additional comments. (Optional) The system may be turned off by Step 29.
CE-ESI-MS measurement ● TIMING 1 h per measurement
17| Operate the ES as an ESI-MS-only device (Box 1) in the cone-jet mode by carefully decreasing the emitter-to-sampling plate distance to ~3 mm, and monitor and optimize system performance: upon first assembly of the CE-ESI-MS platform, optimize the signal intensity and stability by positioning the ES emitter in front of the sampling plate orifice in all directions, as well as refining the ES emitter axis-to-sampling plate angle to ~90°. The absolute location of the emitter is not crucial and can be adjusted, even during experiments, by a few millimeters in each direction without noticeably altering the absolute ion intensity. Monitor the stability of the electrified liquid meniscus by optical means and the ion signal using the mass spectrometer for about 45 min upon initial system assembly, 10 min at the beginning of an experiment series, and at least 1 min between consecutive measurements.
▲ CRITICAL STEP The purpose of this step is multifold: the integrity of the connections and sheath supply are ensured, and the ES performance is characterized. Other experimental variables, including the flow rate and composition of the sheath flow, may be tuned during this step. Optimized variables, however, should not be altered between experiments.
18| (Optional) Refine the tuning of the mass spectrometer by electrospraying the test solution at ~300–1,000 nL/min through the CE-ESI interface as an ES-only device (see Supplementary Methods); register the new tuning file to default.
? TROUBLESHOOTING
19| Start the CE separation by gradually increasing the potential across the separation capillary to 20 kV within 10 s using the attached vi (see the Supplementary Methods). Test system performance in the ensuing CE-ESI-MS operation mode (Box 1): perform separation for 45 min upon initial system assembly, 10 min at the beginning of an experiment series, and at least 2 min between consecutive measurements, while monitoring the ES. The purpose of this step is to ensure stable ion generation for the sample analysis. (Optional) Measure the current-voltage characteristics of the CE-ESI system (Ohm’s plot) to minimize solvent heating and electrolysis (see Supplementary Information in reference33).
? TROUBLESHOOTING
20| Gather the sample and centrifuge it for 1 min at 2,000×g. This step helps to reduce cellular debris in the supernatant that may clog the separation capillary. (Optional) Thaw frozen sample extract and sonicate if needed prior to centrifuging. In the present protocol, thawing is accomplished at room temperature without homogenization. Temperature-sensitive samples may be thawed in an ice-water mixture at ~4 °C.
21| Stop the CE separation by gradually decreasing the potential across the separation capillary from 20 kV to 0 V (earth ground) within 10 s using the attached vi (see the Supplementary Methods). Open the door of the CE enclosure to clear access to the sample-loading platform. Note that this will automatically turn off the power supply to the CE HVPS (EQUIPMENT SETUP).
22| Transfer 500 nL of the sample supernatant into the clean and dry sample-loading vial installed in the CE platform per Step 15 (Box 1). (Optional) Due practice allows reproducible transfer of smaller sample volumes into the sample-loading vial. During this step, the sample may be modified depending on the purpose of the study, including dilution or concentration (see Box 2).
23| Load 6 nL of neuron extract into the separation capillary, as follows. Using the capillary holder of the CE platform, move the separation capillary inlet into the sample-loading vial, allowing it to submerge below the sample meniscus. Raise the inlet end of the separation capillary 15 cm above the outlet end by quickly elevating the CE stage (within 1 s). Wait 60 s to syphon ~6 nL of the extract into the separation capillary. Lower the stage immediately afterward (within 1 s), leveling it with the outlet end of the capillary.
▲ CRITICAL STEP Use magnifiers and work in a well-lit environment to increase reproducibility, and avoid bending the bare fused silica capillary inlet. (Optional) The inlet end of the separation capillary may be positioned 1 cm above the outlet end during all times to prevent sample loss during capillary positioning31. As discussed in Box 2, different neuron extract volumes may be sampled by adjusting the elevation or injection time.
? TROUBLESHOOTING
24| Using the capillary mount of the platform, quickly transfer the separation capillary over the electrolyte and secure its inlet end at least 2 mm below the liquid meniscus. Quickly close the CE enclosure door to automatically power the CE HVPS (EQUIPMENT SETUP).
25| Start CE separation by repeating Step 19, and simultaneously begin mass spectrometric acquisition in single- (MS) or tandem-stage (MS-MS) operation. It is advisable to continuously monitor the stability of the ES during separation (consult Box 1). The troubleshooting advice given in Steps 18 and 19 apply.
? TROUBLESHOOTING
26| Stop the CE separation and mass spectrometric data acquisition upon detecting the compounds of interest. Nontargeted metabolic and peptidomic analyses (e.g., profiling) can be enhanced by prolonging the CE separation and data acquisition. In this protocol, many amino acids, osmolites, and neurotransmitters of interest are separated and detected within 45 min31,33,34. Monitor the selected-ion electropherogram of an endogenous compound as an indicator of the end point in an experiment (e.g., similar to a targeted experiment). In the experiments shown here, CE-ESI-MS data was collected until detection of glutathione (protonated m/z 308.0916) with an ~40 min migration time. (Optional) Exogenous compounds spiked into the cell extracts prior to analysis can also help to mark the end point of the experiment (see REAGENT SETUP).
27| Clean the CE-ESI-MS system components by following Steps 15. Rinse the used sample-loading vial with Sigma water then methanol, and store it in 50% methanol prepared with Sigma water. It is practical to collect ~30 sample-loading vials in the same container for batch-cleaning via sonication in 50% methanol prepared with Sigma water.
28| By repeating Steps 15–27, analyze the test solution to assess system performance and the mass calibration solution to generate data for mass calibrating the mass spectrometer. (Optional) Mass calibration may be performed with alternative compounds (REAGENT SETUP).
▲ CRITICAL STEP Note that performing this step before and after measuring a series of sample extracts is recommended to help to identify potential instrumental biases (e.g., during the same day). It is also recommended that the cell environment and rinse solution samples be analyzed to aid in the interpretation of the results.
? TROUBLESHOOTING
29| To turn the system off:
Lower the mass spectrometer sampling plate potential to 0 V
Set the mass spectrometer to stand-by mode
Turn off the CE HVPS
Set the ES emitter-to-sampling plate distance to 15–20 mm
Clean the system components per Step 15
Begin flushing Sigma water through the sheath capillary using a syringe pump (750 nL/min rate or lower) and the separation capillary utilizing the rubber band-driven syringe (Step 15).
▲ CRITICAL STEP Maintain a gentle flow of deionized water in the separation and sheath capillaries until the next experiment to keep capillary walls hydrated and mitigate clogging of the capillaries.
▪ PAUSE POINT The system can be left turned off for about 1 week without a detectable change in performance.
Data analysis ● TIMING 5 min–1 d
30| Recalibrate the mass spectra to 5 ppm accuracy or better using the Compass software. In the present protocol, a selected-ion electropherogram is generated for the sodium formate dimer ion (m/z 158.9 ± 0.1), and a mass spectrum is integrated over the salt peak (9–11 min migration time). This spectrum typically contains an abundance of sodium formate cluster ions and is used to externally calibrate the mass spectra in the m/z 50–500 range. Alternatively, a calibration spectrum registered the same day for the mass calibration solution or a calibrated sample can serve as an external mass calibration. This protocol implements an enhanced quadratic equation for mass calibration (Compass), yielding a typical mass accuracy of 1 ppm.
▲ CRITICAL STEP Ensuring that data is collected with high mass accuracy is critical for successful identification of metabolites. (Optional) Although only one approach is discussed here, mass calibration may be performed by alternative means, or it can be skipped if the mass accuracy was sufficient during data acquisition; e.g., the mass accuracy of acquisition may be tested for known endogenous compounds detected at previously determined migration times (see Table S1).
31| Find molecular features and create a list of the measured ion masses of interest. Here, the PlotEIC.m (Bruker) script is executed in Compass to compute m/z-selected ion electropherograms between m/z 50–500 with a search window of 500 mDa, and the data are smoothed using a 3-point Gaussian function. Electrophoretic signals are high-pass filtered with an arbitrary threshold of 5×104 counts. Adducts, clusters, isotopes, and compounds not related to the neuron metabolome (e.g., polymers and plasticizers from vials and other contaminants from solvents) are manually eliminated, and the accurate masses of the remaining signals are determined by generating mass spectra across the corresponding electrophoretic peaks (see Figure 2b). (Optional) Advanced software packages, such as ProfileAnalysis (Bruker) or metaXCMS (Metlin), present automated protocols to find molecular features. Although the presented protocol aims for large-scale chemical profiling, this step may be skipped when performing a targeted analysis.
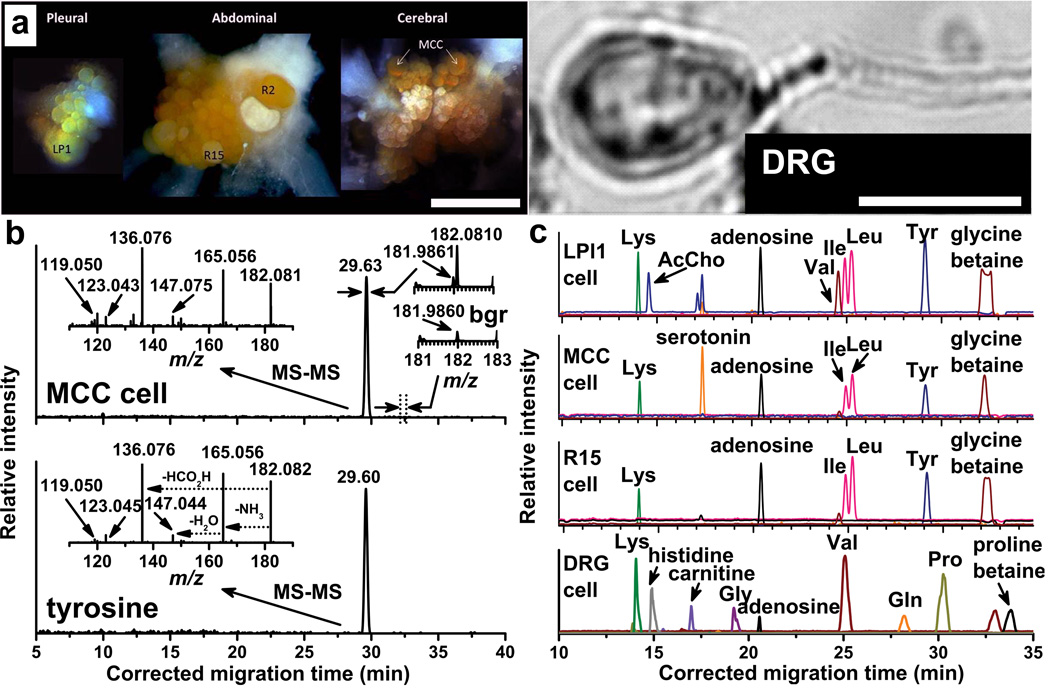
Figure 2.
Targeted CE-ESI-MS analysis of single neurons, 25–500 µm in diameter. (a) Optical images show (left panel) left pleural 1 (LPl1), R2, and R15 abdominal, and metacerebral (MCC) neurons, 150–350 µm in cell body diameter, located in the corresponding ganglia from the A. californica CNS, with other neurons and nerves located adjacently, and (right panel) a DRG neuron, 25 µm in cell body diameter, from the rat peripheral nervous system. Scales = 1 mm and 20 µm, respectively. (b) Multifaceted data analysis identifies a molecular feature for the ion with a nominal mass of m/z 182 ± 0.5 in the MCC neuron extract and determines the corresponding accurate mass of the compound (right insets, bgr = background spectrum). Comparison of the data against a mass spectrometric metabolite database yields tyrosine as a putative identification (based on accurate mass). Confirmation via tandem mass spectrometric and migration time measurements on the related standard (lower spectra) corroborates the finding. Migration times are aligned using internal standards and a quadratic equation. (c) In a targeted analysis, selected metabolites are profiled in single LPl1, MCC, and R15 neurons, and a 25-µm-diameter DRG neuron.
32| Calculate and tabulate the abundance and migration time of the measured signals of interest (m/z values). Here, the custom-written script (Figure S3, Supplementary Methods) is executed, generating mass (m/z)-selected ion electropherograms with a 5 mDa window for the accurate masses noted in Step 31, and also implementing 3-point Gaussian smoothing on the data. Peak areas are manually integrated and tabulated into mass–migration time brackets. Although laborious, this semi-automated approach provides complete oversight on data analysis, and is capable of profiling 144 endogenous compounds in over 60 different single-neuron extracts33. (Optional) ProfileAnalysis and metaXCMS can help to automate this step, thus significantly reducing the processing time. In a targeted study, limit analysis to the accurate mass of the compound of interest.
33| Evaluate the chemical signatures among the extracts via multivariate and statistical means to note signals (m/z values) of interest. In this protocol, the bracketed data of Step 32 are subjected to unsupervised principal component analysis (PCA) in Markerview (AB Sciex) (e.g., see Figure 3a). Various data preprocessing and scaling techniques are explored to appreciate the chemical signatures. Afterward, statistical analysis is performed on the data using Student’s t-test to confirm that the chemical signatures are of statistical significance (e.g., see Figure 3b). The accurate masses of the compounds of interest are eventually tabulated to proceed with chemical identification. Although only PCA and Student’s t-tests are implemented here, other chemometric and statistical analysis tools are available to help data processing (e.g., hierarchical cluster analysis, orthogonal projections to latent structures discriminant analysis (OPLS-DA), and analysis of variance (ANOVA)).
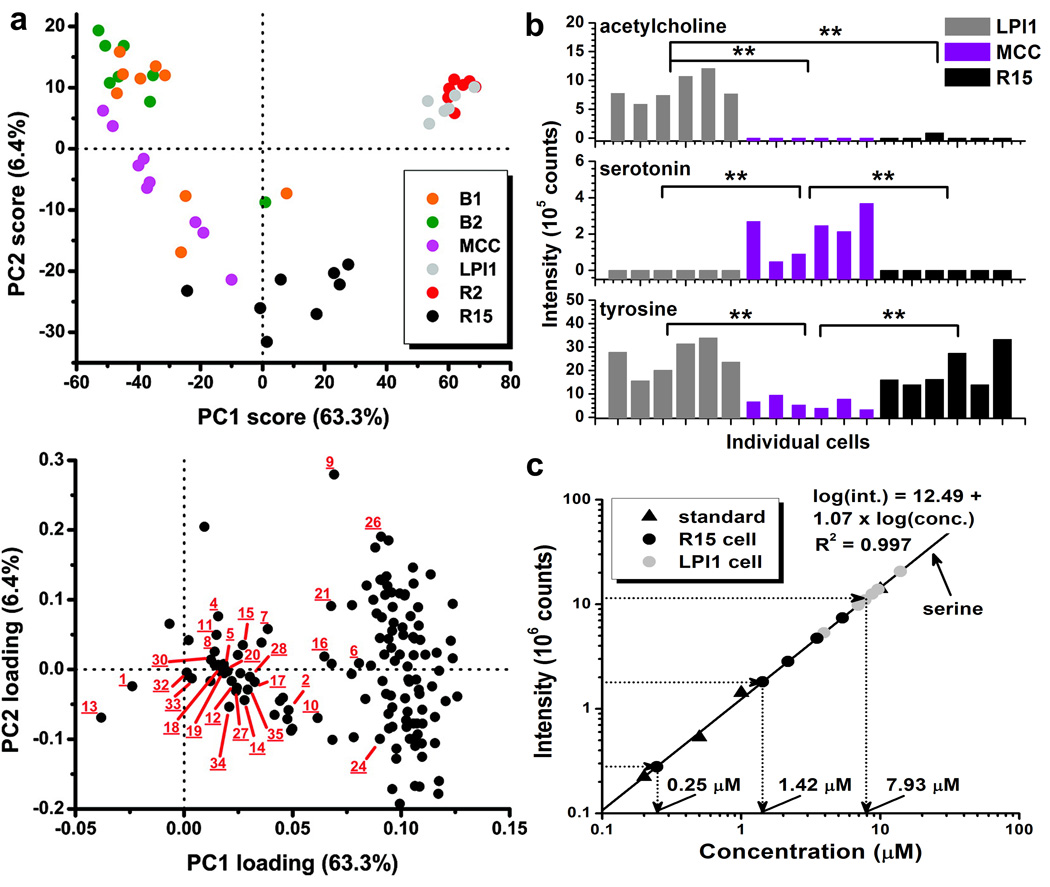
Figure 3.
Chemical profiling, quantitation, and comparison of metabolites among individual cells. (a) (Top panel) The unsupervised PCA score plot indicates metabolic differences between MCC, R15, B1 or B2, and R2 or LPl1 single neurons and uncovers similarities between B1 and B2 as well as R2 and LPl1 cells. Underlined numbers correspond to metabolites identified per reference30 and include histamine (1), acetylcholine (9), serotonin (13), glycine betaine (32), and proline betaine (33). (Bottom panel) The corresponding loading plot reveals metabolites with different abundance patterns among the extracts. (b) Student’s t-test identifies statistically significant differences in the acetylcholine, serotonin, and tyrosine levels measured in LPl1, MCC, and R15 single neurons; two asterisks (**) mark p < 0.005. (c) External concentration calibration with serine standard helps to quantify this endogenous metabolite in selected LPl1 and R15 neurons. Differences in serine concentration are apparent among the cell types as well as individual neurons of the same type. (Adapted with permission from reference33. Copyright 2011 American Chemical Society.)
34| Identify the compounds marked for evaluation (accurate m/z values from Step 33). This protocol implements multifaceted chemical identifications as discussed in depth elsewhere33. Briefly, accurate masses are searched against the Metlin metabolite tandem mass spectrometric database with 5 ppm mass accuracy, and putative mass matches are filtered based on isotope distribution. Next, the tandem mass spectrum recorded for the unknown signal is compared against that of the putative metabolite in the Metlin database or alternative resources (e.g., online databases and publications). The confidence of assignment is further enhanced by measuring the migration time and tandem mass spectrum of the authentic chemical standard that corresponds to the putative candidate (e.g., see Figure 2b). (Optional) When performing a targeted analysis, this step is skipped.
35| Quantify the compounds of interest in follow-up measurements by establishing a correlation between the CE-ESI-MS signal intensity and analyte concentration using related chemical standards (external calibration) (e.g., see Figure 3c). ▲ CRITICAL STEP Depending on inter-day variances in system performance, it is advisable to perform external calibration on the same day as analysis of the neuron extract(s). (Optional) The standard addition method can be alternatively used to deduce compound concentrations.
● TIMING
Steps 1–3, preparation for neuron isolation: ~2–28 h
Steps 4A i–ii, A. californica CNS ganglion isolation: ~20 min
Steps 4A iii–iv, A. californica ganglion incubation and rinsing: 50 min–3 h
Step 4A v, A. californica neuron isolation: 2–5 min/neuron
Step 4A vi, A. californica cell culturing: variable; 8–18 h
Step 4B, Rat DRG neuron isolation: ~30 min DRG isolation and treatment; 3–5 min/neuron isolation
Steps 5–11, neuron extract preparation: 30–120 s
Steps 12–16, system assembly and cleaning: variable; 2–4 h
Steps 17–27, neuron extract measurement: variable; typically 1 h/measurement
Steps 28–29, system performance test, calibration: variable, depending on tests
Steps 30–33, data processing: variable; 5 min–1 d
Step 34, metabolite identification: variable; 2 h–1 wk
Step 35, metabolite quantitation: variable; 3 h–1 d
Typically for an experiment conducted with multiple single cells, sample stabilization, single-cell isolation, and preparation of the analyte extracts, take a total of approximately 1–4 h. Cell culture time depends on the neuron type and experimental conditions, including the composition and temperature of the culturing medium, as well as the purpose of the study. In our experiments, B1 and B2 neurons of A. californica are successfully cultured over 12–16 h in ASW-antibiotic solution containing HEPES34. Initial setup of the CE-ESI platform requires 2–4 h. CE-ESI-MS measurements take 1 h per sample including cleaning system components. Depending on the quality of the acquired mass spectra, semi-manual data processing entailing mass calibration, finding of molecular features, and integration of ion signal abundances consume about 25 min for ~100 analytes, in part by taking advantage of the automated script shown in Figure S3. In contrast, targeted analysis reduces time demands to ~5 min for each file. Data consolidation and multivariate and statistical data interpretation over multiple cells and cell types usually call for several hours to a day. The most involved portion of the data analysis is analyte identification and quantitation, with an estimated time commitment ranging from several hours to days. Additional CE-ESI-tandem mass spectrometric measurements that facilitate metabolite identification can expand the time frame by days to weeks. This is especially true for compounds having putative identities with no corresponding standards available.
TROUBLESHOOTING
Troubleshooting considerations are provided in Table 1.
Table 1.
Troubleshooting table
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| 4 | Low metabolite signal | Analyte loss due to neuron damage during isolation or culturing because of a) excessive enzymatic digestion; b) cell membrane integrity impacted by mechanical or chemical treatment |
Optimize experimental conditions of cell isolation and culturing for the sample under investigation; use stabilization solutions |
| 18 | Unstable ES in ESI- MS mode. MS signal or spray current exhibits regions of
|
The ES Taylor-cone is destabilized by
|
|
| 19 | Unstable ES in CE- ESI-MS mode. MS signal or spray current has regions of
|
The ES Taylor-cone is destabilized by
|
|
| 23 | Separation capillary inlet end is damaged |
Capillary inlet end has been forced against the sample-loading vial and chips off or breaks (confirm under microscope) |
Avoid bending the capillary during sample loading and separation. Cleave, burn, and clean inlet end of the capillary (Step 15). Alternatively, install a new separation capillary into the platform. Measure capillary length. Note that shortening the capillary will inherently shorten the migration time of analytes, alter separation efficiency, and increase CE current. |
| 25 | Electric sparks audible from the
|
Electrical circuit shorting due to
|
Lower ambient humidity, or
|
| 28 | Poor analytical performance observed for
|
Improper performance in
|
|
ANTICIPATED RESULTS
Electrophoretic separation with ESI-MS detection has been successfully used to measure numerous compounds in single cells. We and others have developed protocols in sample preparation and CE-ESI-MS instrumentation to probe the inherently small sample volumes afforded by individual invertebrate and mammalian neurons31,33,34,40. From a measurement perspective, our single-cell experiments illustrate that the metabolic composition of individual cells can be analyzed, resulting in high quality data. For example, more than 300 compounds the molecular mass range of metabolites were detected in a single A. californica neuron33. Underlying this outcome are system characteristics that allow the user to directly measure endogenous metabolite concentrations; the linear dynamic range of concentration for quantitation spans across 3–4 orders of magnitude, and the limit of detection is ~100 amol for acetylcholine and histamine, among others31. Interestingly, compared with the investigation of multicellular samples, interpretation of SCA results becomes more straightforward as the chemical and structural complexity of the sample is reduced. Single-cell CE-MS, conjoined with metabolic profiling with relatively high analyte coverage, can uncover the biochemical, and in some cases, functional heterogeneity of cellular populations.
Here, we showcase the potential of single-cell CE-ESI-MS for identifying, profiling, and quantifying metabolites in individual molluscan and mammalian neurons, with a cell body of 25–500 µm in diameter (Figure 2a). For example, as shown in Figure 2b, analysis of an MCC neuron extract revealed a molecular feature having a nominal mass of m/z 182 at a 29.63 min migration time. Subsequent calibration of the MS spectrum provided an accurate mass of m/z 182.0810 for the ion (Figure 2b, right insets). A search in the Metlin database with 5 ppm mass accuracy suggests protonated tyrosine as one of the likely candidates. CE-ESI-MS and MS/MS analysis of the tyrosine chemical standard provided corroborative pieces of evidence — the migration times and tandem mass spectra of the endogenous compound and the tyrosine standard are in perfect agreement (Figure 2b, left insets). Measured migration times are transformed using a quadratic equation based on internal CE migration time markers, as described previously33. Targeted metabolic comparisons among single LPl1, MCC, and R15 neurons from A. californica show similar small-molecular profiles for some amino acids (e.g., leucine, isoleucine, and tyrosine) and osmolytes (e.g., glycine betaine), and highlight neuron-characteristic differences based on acetylcholine and serotonin content (Figure 2c). Obtaining such consistent results from identified neurons lends credence to the detection/identification of unknowns from less well-characterized cells as being biologically relevant and not measurement artifacts. Optimized analyte extraction and analysis allowed a 25-µm-diameter rat DRG neuron (Figure 2a) to be successfully measured (Figure 2c), demonstrating the adaptability of this protocol for cells with progressively decreasing dimensions and of different specimen types.
As a means of nontargeted analysis, more than 50 individual neurons from six different neuron types were compared using unsupervised PCA (Figure 3a). In the score plot, LPl1 and R2 neurons, as well as the B1 and B2 cells, appear chemically similar compared with the MCC and R15 neurons. Heterogeneity within each neuron type is also apparent in the same plot. It is worth noting that PCA has been used to document minor differences among the B1 and B2 cells in a targeted study34. Close inspection of the loading plot demonstrates that serotonin and histamine, proline betaine and glycine betaine, and acetylcholine fall into different data clusters, suggesting that these compounds exhibit dissimilar patterns of variation in signal abundance among the samples. Indeed, Student’s t-tests on the corresponding ion abundances confirmed higher amounts of acetylcholine in the LPl1 and an increased level of serotonin in the MCC neurons (Figure 3b). Likewise, the LPl1 and R15 neurons contained significantly more tyrosine. Notably, a similar data analysis scheme with improved sample preparation was recently used to detect metabolic changes in cultured B1 and B2 buccal neurons of the Aplysia californica CNS34, providing an opportunity to follow the modulation of cellular metabolomic and functional states in single-cell experiments using the protocol described here.
This protocol can also be used to quantify metabolites in individual neurons. External calibration, here using serine as a standard, helped to determine the solution-phase concentration of this amino acid in extracts of selected R15 and LPl1 neurons. Representative examples are shown in Figure 3c, where differences in serine concentration are apparent between the LPl1 and R15 neurons, with higher concentrations found in the former samples. The results also revealed variation in the analyte levels among individual neurons of the same type. By taking into account the actual size of an analyzed neuron, it becomes possible to determine the intracellular concentration of virtually any metabolite that is detected and available as a chemical standard.
Supplementary Material
Acknowledgments
Theodore Lapainis contributed to the development of the CE-ESI-MS platform, Xiying Wang facilitated the single-neuron preparation work, and Ann M. Knolhoff helped with the interpretation of CE-ESI-MS data in the original CE-MS studies. This work was supported by the National Science Foundation under Award Number CHE 11-11705, the National Institute of Neurological Disorders and Stroke under Award No. NS031609, the National Institute of Dental and Craniofacial Research (NIDCR) and the Office of Director (OD), National Institutes of Health (NIH) under Award No. DE018866, and the National Institute on Drug Abuse under Award No. P30 DA018310. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either actual or implied endorsement of such products by the Department of Health and Human Services. The authors thank Stephanie Baker for assistance in the preparation of this publication, and the machine shop of the School of Chemical Sciences, University of Illinois at Urbana-Champaign, for providing various components for the construction of the CE ESI system, and Miri Kim for help in photography.
Footnotes
- Lapainis, T., Rubakhin, S. S. & Sweedler, J. V. Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal. Chem. 81, 5858–5864 (2009), doi 10.1021/ac900936g
- Nemes, P., Knolhoff, A. M., Rubakhin, S. S. & Sweedler, J. V. Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis-electrospray ionizationmass spectrometry. Anal. Chem. 83, 6810–6817 (2011), doi 10.1021/ac2015855
- Nemes, P., Knolhoff, A. M., Rubakhin, S. S. & Sweedler, J. V. Single-cell metabolomics: changes in the metabolome of freshly isolated and cultured neurons. ACS Chem. Neurosci. 3, 782–792 (2012), doi 10.1021/cn300100u
- Knolhoff, A. M., Nemes, P., Rubakhin, S. S. & Sweedler, J. V., MS-based methodologies for single-cell metabolite detection and identification, Methodologies for Metabolomics 2013, pp 119-139, Experimental Strategies and Techniques, Eds. Wevers, R., Lutz, N. & Sweedler, J. V., Cambridge University Press, New York, NY
Supplementary Methods
Troubleshooting examples via selected-ion electropherograms, a vi (Supplementary Sequence Archive.zip) and the corresponding protocol for controlling the CE HVPS by a personal computer, a script for automating data analysis, and representative metabolite identifications are provided.
Author contributions
The contributions of the authors to the original studies that were used to generate the protocol are indicated in the original cited articles. For the protocols described here, PN generated many of the protocol steps related to CE-MS operation and wrote the data analysis script and vi, JTA modified the protocols and validated them using the DRG neurons, SSR performed the cell isolations and optimized the protocols for cell handling, and JVS modified and clarified the protocol. All authors participated in the writing.
Competing financial interests
The authors declare that they have no competing financial interests.
References
- 1.Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens DJ, Allan VJ. Light microscopy techniques for live cell Imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 3.Fischer RS, Wu YC, Kanchanawong P, Shroff H, Waterman CM. Microscopy in 3D: a biologist's toolbox. Trends Cell Biol. 2011;21:682–691. doi: 10.1016/j.tcb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 5.Planchon TA, et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods. 2011;8:U417–U468. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan RX, et al. Nanowire-based single-cell endoscopy. Nat. Nanotechnol. 2012;7:191–196. doi: 10.1038/nnano.2011.226. [DOI] [PubMed] [Google Scholar]
- 7.Wang DJ, Bodovitz S. Single cell analysis: the new frontier in 'omics'. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amantonico A, Urban PL, Zenobi R. Analytical techniques for single-cell metabolomics: state of the art and trends. Anal. Bioanal. Chem. 2010;398:2493–2504. doi: 10.1007/s00216-010-3850-1. [DOI] [PubMed] [Google Scholar]
- 9.Rubakhin SS, Romanova E, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells Nat . Methods. 2011;8:S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YQ, Trouillon R, Safina G, Ewing AG. Chemical analysis of single cells. Anal. Chem. 2011;83:4369–4392. doi: 10.1021/ac2009838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemes P, Vertes A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. Trac-Trends Anal. Chem. 2012;34:22–34. [Google Scholar]
- 12.Lee YJ, Perdian DC, Song ZH, Yeung ES, Nikolau BJ. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012;70:81–95. doi: 10.1111/j.1365-313X.2012.04899.x. [DOI] [PubMed] [Google Scholar]
- 13.Rubakhin SS, Lanni EJ, Sweedler JV. . Curr. Opin. Biotechnol. 2012;24:1–10. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal. Chem. 2006;78:7267–7272. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 16.Neupert S, Rubakhin SS, Sweedler JV. Targeted single cell microchemical analysis: MS-based peptidomics of individual paraformaldehyde-fixed immunolabeled neurons. Chem. Biol. 2012;19:1010–1019. doi: 10.1016/j.chembiol.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat. Protoc. 2007;2:1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 18.Northen TR, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–U1033. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 19.Greving MP, Patti GJ, Siuzdak G. Nanostructure-initiator mass spectrometry metabolite analysis and imaging. Anal. Chem. 2011;83:2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban PL, Amantonico A, Fagerer SR, Gehrig P, Zenobi R. Mass spectrometric method incorporating enzymatic amplification for attomole-level analysis of target metabolites in biological samples. Chem. Commun. 2010;46:2212–2214. doi: 10.1039/b925433a. [DOI] [PubMed] [Google Scholar]
- 21.Stolee JA, Walker BN, Zorba V, Russo RE, Vertes A. Laser-nanostructure interactions for ion production. PCCP. 2012;14:8453–8471. doi: 10.1039/c2cp00038e. [DOI] [PubMed] [Google Scholar]
- 22.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 2005;127:12152–12153. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 23.Yang HJ, et al. Detection of characteristic distributions of phospholipid head groups and fatty acids on neurite surface by time-of-flight secondary ion mass spectrometry. Med. Mol. Morphol. 2010;43:158–164. doi: 10.1007/s00795-009-0487-2. [DOI] [PubMed] [Google Scholar]
- 24.Kurczy ME, et al. Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2751–2756. doi: 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuyama N, Mizuno H, Masujima T. Mass spectrometry for cellular and tissue analyses in a very small region. Anal. Sci. 2011;27:163–170. doi: 10.2116/analsci.27.163. [DOI] [PubMed] [Google Scholar]
- 26.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 27.Shrestha B, Patt JM, Vertes A. In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal. Chem. 2011;83:2947–2955. doi: 10.1021/ac102958x. [DOI] [PubMed] [Google Scholar]
- 28.Coello Y, Jones AD, Gunaratne TC, Dantus M. Atmospheric-pressure femtosecond laser imaging mass spectrometry. Anal. Chem. 2010;82:2753–2758. doi: 10.1021/ac9026466. [DOI] [PubMed] [Google Scholar]
- 29.Cecala C, Sweedler JV. Sampling techniques for single-cell electrophoresis. Analyst. 2012;137:2922–2929. doi: 10.1039/c2an16211c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapainis T, Sweedler JV. Contributions of capillary electrophoresis to neuroscience. J. Chromatogr. 2008;1184:144–158. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 31.Lapainis T, Rubakhin SS, Sweedler JV. Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal. Chem. 2009;81:5858–5864. doi: 10.1021/ac900936g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page JS, Rubakhin SS, Sweedler JV. Single-neuron analysis using CE combined with MALDI MS and radionuclide detection. Anal. Chem. 2002;74:497–503. doi: 10.1021/ac0156621. [DOI] [PubMed] [Google Scholar]
- 33.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis-electrospray ionization-mass spectrometry. Anal. Chem. 2011;83:6810–6817. doi: 10.1021/ac2015855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Single-cell metabolomics: Changes in the metabolome of freshly isolated and cultured neurons. ACS Chem. Neurosci. 2012;3:782–792. doi: 10.1021/cn300100u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonner RF, et al. Cell sampling-laser capture microdissection: Molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 36.Espina V, et al. Laser-capture microdissection. Nat. Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 37.Miao J, Cui LW. Rapid isolation of single malaria parasite-infected red blood cells by cell sorting. Nature Protocols. 2011;6:140–146. doi: 10.1038/nprot.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen D, et al. Chemical cytometry: Fluorescence-based single-cell analysis. Annu. Rev. Anal. Chem. 2008;1:165–190. doi: 10.1146/annurev.anchem.1.031207.113104. [DOI] [PubMed] [Google Scholar]
- 39.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:U858–U889. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal. Chem. 2010;82:967–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenherr RM, Ye ML, Vannatta M, Dovichi NJ. CE-microreactor-CE-MS/MS for protein analysis. Anal. Chem. 2007;79:2230–2238. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbula GK, Safi S, Chingin K, Perry RH, Zare RN. Interfacing capillary-based separations to mass spectrometry using desorption electrospray ionization. Anal. Chem. 2011;83:1955–1959. doi: 10.1021/ac102648k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris GA, Nyadong L, Fernandez FM. Recent developments in ambient ionization techniques for analytical mass spectrometry. Analyst. 2008;133:1297–1301. doi: 10.1039/b806810k. [DOI] [PubMed] [Google Scholar]
- 44.Knolhoff AM, Nemes P, Rubakhin SS, Sweedler JV. In: Methodologies for metabolomics: Experimental strategies and techniques. Lutz Norbert, Sweedler Jonathan V, Wevers Ron A., editors. Cambridge University Press; 2013. p. 119.p. 139. [Google Scholar]
- 45.Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 2012;7:508–516. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baran R.et alMathDAMP: a package for differential analysis of metabolite profiles BMC Bioinformatics 20067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith CA, et al. METLIN - A metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 50.Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006;34:W267–W272. doi: 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geigl JB, Speicher MR. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat. Protoc. 2007;2:3173–3184. doi: 10.1038/nprot.2007.476. [DOI] [PubMed] [Google Scholar]
- 52.Neupert S, Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem. Biophys. Res. Commun. 2005;327:640–645. doi: 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- 53.Eyer K, Kuhn P, Hanke C, Dittrich PS. A microchamber array for single cell isolation and analysis of intracellular biomolecules. Lab Chip. 2012;12:765–772. doi: 10.1039/c2lc20876h. [DOI] [PubMed] [Google Scholar]
- 54.De Sousa BN, Horrocks LA. Development of rat spinal cord. I. Weight and length with a method for rapid removal. Dev. Neurosci. 1979;2:115–121. [Google Scholar]
- 55.Nemes P, Marginean I, Vertes A. Spraying mode effect on droplet formation and ion chemistry in electrosprays. Anal. Chem. 2007;79:3105–3116. doi: 10.1021/ac062382i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.