Specific basic and aromatic amino acids in the C-terminal tail of Mgm101 play key roles in mediating ssDNA binding, stabilizing the oligomeric rings, and maintaining protein stability in vivo. The C-tail may couple ssDNA binding with ring disassembly, which facilitates the formation of nucleoprotein complexes competent for the repair of mtDNA.
Abstract
Mgm101 is a Rad52-type single-stranded annealing protein (SSAP) required for mitochondrial DNA (mtDNA) repair and maintenance. Structurally, Mgm101 forms large oligomeric rings. Here we determine the function(s) of a 32–amino acid carboxyl-terminal tail (Mgm101238–269) conserved in the Mgm101 family of proteins. Mutagenic analysis shows that Lys-253, Trp-257, Arg-259, and Tyr-268 are essential for mtDNA maintenance. Mutations in Lys-251, Arg-252, Lys-260, and Tyr-266 affect mtDNA stability at 37°C and under oxidative stress. The Y268A mutation severely affects single-stranded DNA (ssDNA) binding without altering the ring structure. Mutations in the Lys-251–Arg-252–Lys-253 positive triad also affect ssDNA binding. Moreover, the C-tail alone is sufficient to mediate ssDNA binding. Finally, we find that the W257A and R259A mutations dramatically affect the conformation and oligomeric state of Mgm101. These structural alterations correlate with protein degradation in vivo. The data thus indicate that the C-tail of Mgm101, likely displayed on the ring surface, is required for ssDNA binding, higher-order structural organization, and protein stability. We speculate that an initial electrostatic and base-stacking interaction with ssDNA could remodel ring organization. This may facilitate the formation of nucleoprotein filaments competent for mtDNA repair. These findings could have broad implications for understanding how SSAPs promote DNA repair and genome maintenance.
INTRODUCTION
The DNA single-stranded annealing proteins (SSAPs) are widespread in organisms from bacteriophages to humans. The best studied is probably the Rad52 protein, which is involved in DNA recombination and repair in the eukaryotic nucleus. Rad52 is a tripartite protein (Figure 1A) with two distinct roles in the recombinational repair of double-stranded breaks (Kowalczykowski et al., 1994; Symington, 2002; San Filippo et al., 2008). In a canonical recombination pathway, the 5′ end at a double-stranded DNA break is first resected by an exonuclease. The exposed single-stranded DNA (ssDNA) is stabilized by the single-stranded binding protein RPA. Rad52 then loads the Rad51 recombinase onto ssDNA by displacing RPA. The resulting Rad51/ssDNA presynaptic complex finally promotes homology search, strand invasion, and homologous pairing within duplex DNA templates. In this regard, Rad52 acts as a recombination mediator through its capabilities to interact with ssDNA, RPA, and Rad51. The N-terminal one-third of Rad52 promotes ssDNA binding (Mortensen et al., 1996; Kagawa et al., 2002; Singleton et al., 2002), and the C-terminal portion is required for the interactions with both Rad51 (Shinohara et al., 1992; Milne and Weaver, 1993) and RPA (Hays et al., 1998; Shinohara and Ogawa, 1998; Sugiyama et al., 1998). A more recent study designated the middle portion between the N- and C-termini as the RPA-interacting domain (Seong et al., 2008). In addition to its recombination mediator function, the N-terminus of Rad52 can independently catalyze single-stranded annealing (SSA), an activity critical for the capture of the second DNA end at the recombination site to generate Holliday junctions (Sugiyama et al., 2006; Shi et al., 2009). The SSA activity also allows Rad52 to catalyze the annealing of RPA-coated complementary single DNA strands, a property that enables recombination between direct DNA repeats independent of Rad51 (Mortensen et al., 1996; Shinohara et al., 1998; Sugiyama et al., 2006). The SSA event is accompanied by the deletion of intervening sequences between the tandem repeats.
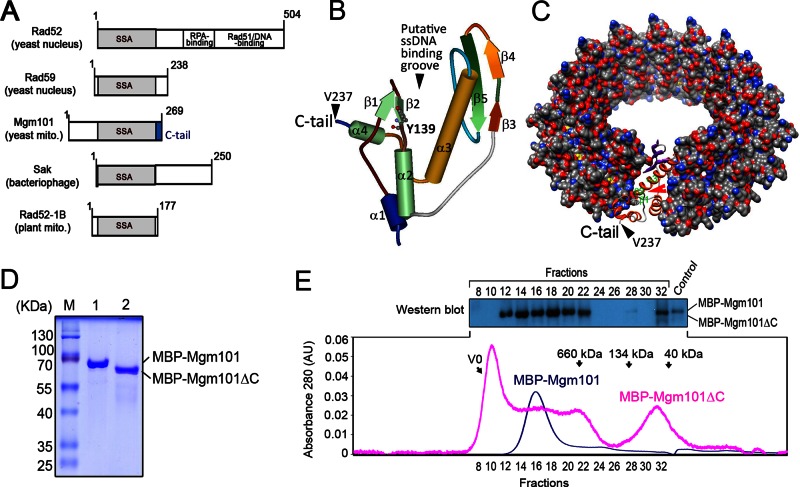
FIGURE 1:
The C-tail deletion destabilizes the oligomeric state of Mgm101. (A) Domain organization of SSAPs from various species. All SSAPs contain the conserved SSA domain (shown in gray). The Mgm101-specific C-tail is shown in blue. (B) Solid ribbon structure of Mgm101115-237 modeled on the N-terminal ssDNA-binding domain of human Rad52 (1H2I). The projected position of Tyr-139 is represented by scaled balls and sticks. (C) Surface representation model of an Mgm101 14-mer ring modeled on the crystal structure of the human Rad52 (1H2I). The red arrow indicates the putative ssDNA-binding groove predicted for Rad52. The black arrow indicates that Val237 is displayed on the surface of the ring, which is followed by the C-tail. (D) SDS–PAGE showing purified Mbp-Mgm101ΔC after amylose resin affinity chromatography (lane 2) compared with a wild-type fusion control (lane 1). (E) Size exclusion chromatography of MBP-Mgm101ΔC (pink) with corresponding fractions analyzed by Western blot as depicted. Wild-type fusion control (blue) is a monodisperse peak of ∼940 kDa, in contrast to the polydisperse MBP-Mgm101ΔC. Note that no signal is detected by Western blot for MBP-Mgm101ΔC present in the void volume, likely due to precipitation of the protein aggregates in the solution.
Although the SSAPs are routinely called Rad52-type recombination proteins, many of them have a molecular architecture that is highly diverged from that of Rad52 (Figure 1A). For instance, the yeast Rad59 protein and the SSAPs from bacteriophages, including Redβ and Erf from the bacteriophages λ and P22, RecT from the prophage rac, and Sak from the lactococcal phage ul36, lack an apparent large C-terminal domain for interactions with Rad51 or RecA (Bai and Symington, 1996; Iyer et al., 2002; Ploquin et al., 2008; Lopes et al., 2010). Like Rad52, all of the proteins catalyze SSA. Rad59 partially overlaps with Rad52 in double-stranded break repair by the classic strand invasion mode, but it also has unique functions, such as the stimulation of Rad52-mediated SSA (Davis and Symington, 2001; Wu et al., 2006; Feng et al., 2007). Similarly, the phage SSAPs are generally believed to promote recombination via the strand-annealing mode independent of RecA. This recombination activity is important for DNA repair (Stahl et al., 1997), genome recircularization between terminally repeated sequences during the infection cycle (Botstein and Matz, 1970; Fenton and Poteete, 1984), and genome replication (Mosig et al., 2001). With regard to DNA repair, an interesting functional feature of the phage SSAPs is that they can promote homologous recombination through the SSA mode without causing deletions in the genome. The Redβ protein has been shown to initiate strand annealing by preferentially targeting to the homologous sequences on the lagging strand of a replication fork to achieve the error-free recombinational repair of phage DNA (Lim et al., 2008; Poteete, 2008).
The mechanism by which the SSAPs catalyze SSA is not well understood. All of the SSAPs so far studied form large homo-oligomeric rings of 10- to 14-fold symmetry in vitro (Poteete et al., 1983; Passy et al., 1999; Ploquin et al., 2008). Although the native Rad52 is mainly heptameric (Stasiak et al., 2000), the N-terminal SSA domain of Rad52 forms undecameric rings (Kagawa et al., 2002; Singleton et al., 2002). One model posits that ssDNA binds to a large and positively charged groove along the surface of the ring. This allows its presentation to a complementary ssDNA during the ssDNA annealing reaction (Singleton et al., 2002). Data from fluorescence resonance energy transfer analysis support the idea that successive interactions between two separate ssDNA-wrapped rings occur until the formation of a homologous and stable duplex (Rothenberg et al., 2008; Grimme et al., 2010).
We recently showed that the yeast mitochondrial protein Mgm101 is a SSAP of bacteriophage origin. MGM101 was first identified as a nuclear gene essential for the maintenance of the mitochondrial genome (Chen et al., 1993; Clark-Walker and Chen, 1996; Zuo et al., 2002). It is specifically associated with the mitochondrial nucleoids and is required for the tolerance of mitochondrial DNA (mtDNA) to DNA-damaging agents (Meeusen et al., 1999; Kaufman et al., 2000). Mgm101 shares biochemical, structural, and functional similarities with Rad52 and the bacteriophage SSAPs despite very limited similarity in their primary sequences (Mbantenkhu et al., 2011). It forms oligomeric rings with a diameter of ∼200 Å, which preferentially bind to ssDNA versus double-stranded DNA (dsDNA). The ring organization was recently confirmed by a solution structure of Mgm101 resolved by small-angle x-ray scattering analysis (Nardozzi et al., 2012). Furthermore, Mgm101 forms characteristic helical filaments, as seen with the bacteriophage Redβ protein (Passy et al., 1999). These developments strongly suggest the existence of a SSA-based recombination pathway for DNA repair in mitochondria. The presence of a mitochondrial Mgm101-type SSAP has recently been demonstrated in Physarum polycephalum, lower animals, and plants (Itoh et al., 2011; Janicka et al., 2012; Samach et al., 2012).
The essential role of Mgm101 for mtDNA maintenance provides an amenable model system for functional dissection of SSAPs. Previous studies identified a functional core of Mgm101 spanning residues 98–241 (Zuo et al., 2007; Figure 1A). It is this core that is homologous to the Rad52 SSA domain (Zuo et al., 2007). The N-terminal domain of Mgm101 is predicted to be intrinsically disordered and poorly conserved, but this domain is essential for in vivo function (Hayward et al., 2013). It was also shown that Mgm101 lacking the last nine amino acids on the C-terminus (C-tail) loses its ability to complement an mgm101 mutant in vivo. The Mgm101 C-terminus is a relatively small domain of 32 amino acid residues and is rich in basic and aromatic residues. The C-tail is highly conserved among Mgm101 homologues that operate in mitochondria but not in the eukaryotic Rad52-type proteins, suggesting that it must have evolved in Mgm101 to impart unique function(s) in mitochondrial nucleoids. In this article, we find that the short C-tail is critical for ssDNA binding, as well as for stabilization of the ring structure. The data suggest that the C-tail may have arisen from convergent evolution for a successful adaptation of the protein to promote recombination in the specific genetic setting of the mitochondria.
RESULTS
Mgm101 lacking the C-tail destabilizes the protein in vitro
SSAPs are recognized for their remarkable lack of sequence conservation, but nonetheless they adopt similar global structural folds (Iyer et al., 2002; Ploquin et al., 2008; Erler et al., 2009; Lopes et al., 2010). In vitro studies of these proteins revealed similar quaternary structures characterized by the formation of large oligomeric rings. We previously showed that freshly prepared Mgm101 forms rings of ∼14-fold symmetry (Mbantenkhu et al., 2011). The tertiary structure of the Mgm101 core can be readily modeled by the Phyre2 (Protein Homology/Analogy Recognition Engine) program (Kelley and Sternberg, 2009), based on the known crystal structure of the SSA domain of human Rad52 (Kagawa et al., 2002; Singleton et al., 2002). This generates the characteristic β3-β4-β5-α3-α4 fold shared by all the SSAPs (Figure 1B). In the undecameric rings formed by the truncated Rad52, the helix α4 is followed by a flexible linker region that ends with a short α-helix (α5), which extends from the surface to interact with an adjacent subunit in the ring. This region is therefore predicted to play a role in the stabilization of the ring structure. The Mgm101 C-tail, predicted to consist of two β-strands followed by an unstructured stretch of nine amino acids (Figure 2A), is projected on the surface of the 14-fold rings (Figure 1C).
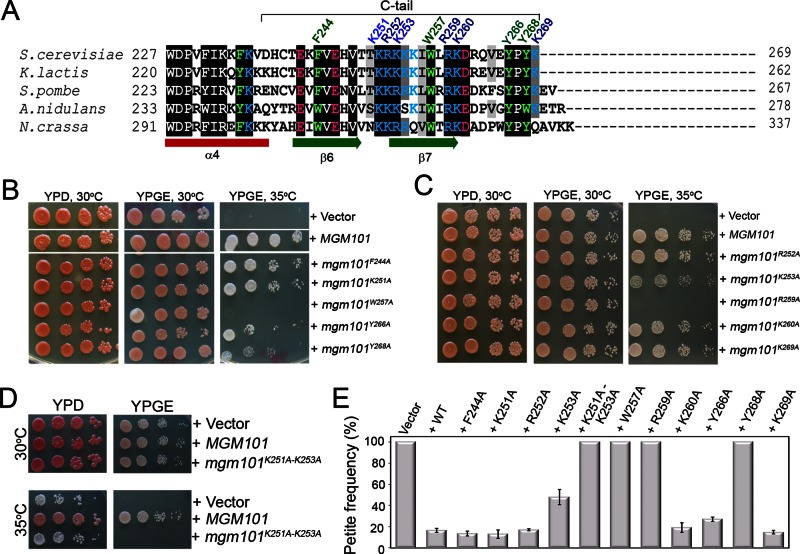
FIGURE 2:
The effect of positively charged and aromatic amino acid C-tail mutants on mtDNA maintenance. (A) Sequence alignment of Mgm101 homologues from fungal species reveals highly conserved basic and aromatic residues. The 10 residues indicated at the top of the alignment represent the ones that are studied by alanine substitution. (B–D) Complementation of the mgm101-1ts allele by mutant C-tail alleles. The mgm101-1ts mutant was transformed with a centromeric vector expressing the C-tail alanine mutants. Ura+ transformants were serially diluted in water and tested for growth on YPGE at 30 and 35°C. (E) Frequency of petite colonies in the mgm101-1ts mutant after transformation with mutant C-tail alleles. The mgm101–1ts mutant was transformed with a centromeric vector expressing the C-tail alanine mutants. Ura+ transformants were plated on YPD medium and incubated at 35°C. Petite frequencies were scored as the ratio of white colonies to the total number of cells growing on that plate. The data are the averaged petite frequency of at least three independent transformants. Error bars indicate average deviations.
To understand the function of the Mgm101 C-tail, we first prepared the Mgm101 variant lacking the 32–amino acid (aa) C-tail (Mgm101ΔC) by using the maltose-binding protein (MBP) tagging strategy. The MBP-Mgm101ΔC fusion can be expressed in bacterial cells (Figure 1D), although the yield is much lower than that of MPB-Mgm101. Once released from MBP by proteolytic cleavage, Mgm101ΔC rapidly precipitated out of solution. Direct analysis of the MBP-Mgm101ΔC fusion by size exclusion column chromatography resulted in poor recovery of the fusion protein. In contrast to the MBP-Mgm101 control, which forms a monodisperse peak of ∼1 MDa, MBP-Mgm101ΔC eluted in polydisperse peaks (Figure 1E). The monomeric form of the protein was detectable. A broad peak that overlaps in the region where the MBP-Mgm101 wild-type fusion would elute was detected. In addition, the highest peak fractions appeared in the void volume (V0). Western blot analysis of these fractions run against a full-length control confirmed the presence of MBP-Mgm101ΔC. No signal was detected in the V0 (fractions 8–10), suggesting that the fusion protein may have precipitated out soon after the elution. Taken together with the difficulty of recovering untagged Mgm101ΔC, the data indicate that the C-tail plays a critical role in maintaining the normal oligomeric structure and stability of Mgm101.
Conserved aromatic and basic amino acid residues in the Mgm101 C-tail are required for in vivo function
The destabilizing effect of C-tail deletion suggested an important role of this domain in structural maintenance of Mgm101. To functionally dissect this domain, we replaced the conserved aromatic and basic residues with alanine by site-directed in vitro mutagenesis (Figure 2A). These mutants were F244A, K251A, R252A, K253A, W257A, R259A, K260A, Y266A, Y268A, and K269A. A triple mutant replacing the tribasic KRK251-253 stretch with alanine was also constructed. In the first series of experiments, the mgm101 alleles carried on the centromeric pCXJ22 vector were introduced into an mgm101-1ts conditional mutant. Ura+ transformants were selected and tested for growth on YPD or YPGE (see Materials and Methods for media) at 30 and 35°C. The results showed that mgm101W257A and mgm101R259A were unable to complement the respiratory deficiency at 35°C on YPGE, suggesting that Trp257 and Arg259 are essential for mtDNA maintenance (Figure 2, B and C). Cells expressing the K253A and Y268A alleles exhibited significantly reduced growth under similar conditions. Although cells expressing the K251A and R252A mutations did not significantly compromise cell growth on YPGE at 30 and 35°C, the mgm101[K251-K253]A triple mutant became respiratory deficient, as demonstrated by its inability to complement cell growth on YPGE at 35°C and the formation of white petite colonies on YPD at 35°C (Figure 2D). This indicated an additive or a synergistic effect among the three mutations in the 251–KRK–253 triad. In the second series of experiments, we plated the transformants on YPD and incubated them at 35°C, so that the complementation by the mutant alleles can be quantitatively estimated based on the frequency of the petite colonies in the absence of a counterselection against mtDNA loss (Figure 2E). Consistent with the direct test on YPGE, transformants expressing the W257A and R259A alleles are completely converted into petite colonies. A similar phenotype was found with the Y268A allele, whereas the K253A allele was found to be partially active in mtDNA maintenance in this assay. Incubation at 37 instead of 35°C did not make a significant difference for all the mutants analyzed (data not shown).
Finally, we tested the mutant alleles in a null mgm101 background. Meiotic segregants expressing only the mutant alleles were generated by tetrad dissections. The 11 mutant alleles were marked by URA3 and integrated into the ura3 locus on the chromosome in a diploid strain with one copy of MGM101 disrupted (mgm101Δ::kan). Meiotic spores having the combination of the Ura+ and G418R phenotypes were identified and directly tested for respiratory growth on YPGE medium after replica plating (Figure 3A). We found that the mgm101[K251-K253]A allele is dominant negative under the sporulation conditions, as all the four spores in each tetrad germinated on YPD are petites (data not shown). As expected, the mgm101W257A and mgm101R259A mutants failed to grow on YPGE. Of interest, we also found that the mgm101K253A and mgm101Y268A mutants are respiratory deficient and are therefore unable to maintain mtDNA. The severe defect in mtDNA maintenance is in sharp contrast with the relatively mild phenotype of these two alleles when tested in the mgm101-1ts host cells (Figure 2, B, C, and E). This observation suggested an interallelic complementation between the plasmid-born C-tail mutations and the endogenous mgm101-1ts allele in the host cells.
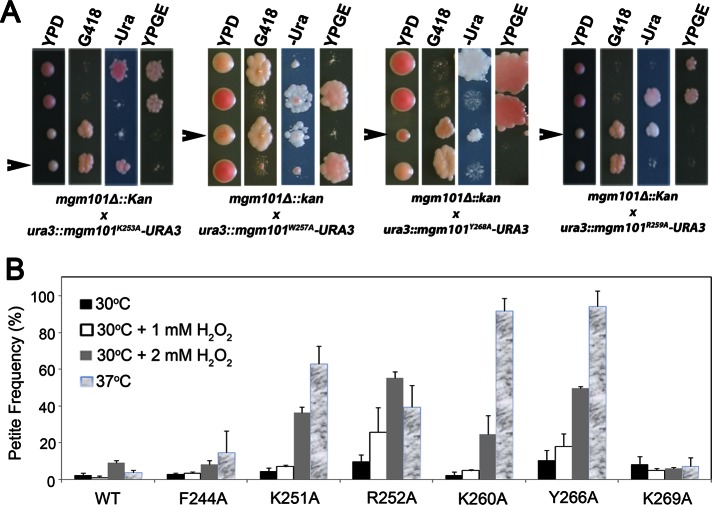
FIGURE 3:
Complementation of an mgm101-null mutant by C-tail mutant alleles. (A) Meiotic analysis showing that mgm101K253A, mgm101W257A, mgm101Y268A, and mgm101R259A failed to complement the null mgm101Δ::kan allele for respiratory growth on ethanol plus glycerol medium at 30°C. A tetrad independently segregating the null mgm101Δ::kan and the mutant alanine alleles was dissected on complete glucose medium. The meiotic segregants were then replica plated onto G418, –Ura, and ethanol plus glycerol medium. G418R marks the presence of the null mgm101Δ::kan allele, and Ura+ marks the mutant alanine alleles. Failure to grow on YPGE indicates the lack of complementation by mutant alanine alleles and the loss of mtDNA. (B) Temperature- and H2O2-induced petite production in the C-tail mutants. Meiotic segregants expressing only the mgm101K251A, mgm101R252A, mgm101K260A, and mgm101Y266A showed significantly increased production of petite colonies when challenged with high temperature or increasing concentrations of H2O2. The haploid cells were first grown in ethanol plus glycerol medium at 30°C. Cells were then inoculated in liquid YPD medium for 24 h at 37 or at 30°C in the presence of 1 or 2 mM H2O2. After dilution in water, cells were plated on YPD and incubated at 30°C for 5 d. Petites were identified as white colonies, and the petite frequency was scored as the percentage of white vs. total number of colonies. The data presented are the averaged petite frequency of at least three independent experiments. Error bars indicate average deviations.
The meiotic segregants expressing only mgm101F244A, mgm101K251A, mgm101R252A, mgm101K260A, mgm101Y266A, and mgm101K269A were respiratory competent on YPGE at 30 and 37°C (data not shown). However, when these cells were subjected to increasing concentrations of H2O2 at 30°C or were challenged by growing at 37°C on YPD medium, the K251A, R252A, K260A, and Y266A mutants showed significant mtDNA instability (Figure 3B).
In summary, the functionality of the 10 amino acids studied can be grouped into three classes: 1) Lys-253, Trp-257, Tyr-268, and Arg-259 are essential for mtDNA maintenance in vivo under physiological conditions; 2) Lys-251, Arg-252, Lys-260, and Tyr-266 are required for mtDNA maintenance under stress conditions, and 3) Phe-244 and Lys-269 are not required for mtDNA maintenance under the conditions tested so far.
Higher-order structural organization of the C-tail mutant proteins
To understand the functional roles of Lys-253, Trp-257, Arg-259, and Tyr-268, we prepared the mutant proteins by the MBP-tagging strategy. The triple (K251–K253)A mutant was also included in the analysis. The MBP-fusion proteins were first analyzed by size exclusion chromatography on a Superose 6 column before proteolytic cleavage between the two moieties. As shown in Figure 4A, MBP-Mgm101K253A, MBP-Mgm101W257A, and MBP-Mgm101Y268A eluted almost like MBP-Mgm101 as a peak of ∼940 kDa. A similar profile was observed with MBP-Mgm101(K251-K253)A, but a shoulder shifting to larger–molecular weight species was noticeable. However, MBP-Mgm101R259A appeared to have an altered oligomeric state, resulting in a sizable fraction in the void volume, which is indicative of protein aggregation (Nardozzi et al., 2012).
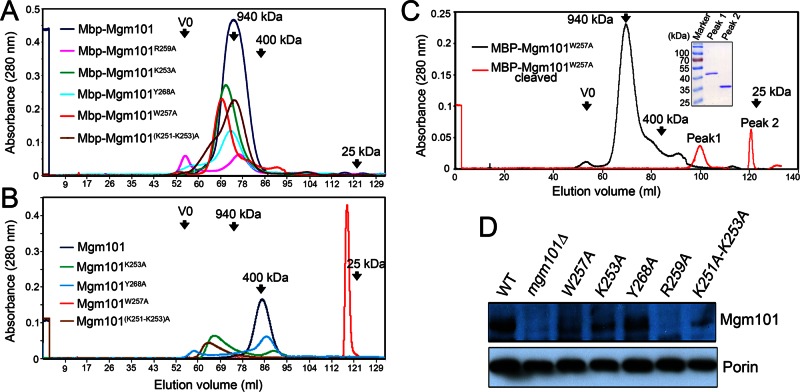
FIGURE 4:
Chromatographic profile, oligomeric state and in vivo stability of mutant Mgm101. (A) Size exclusion chromatography of purified MBP-Mgm101 and its mutant variants on a calibrated Superose 6 column. V0, void volume. (B) Size exclusion chromatography of purified Mgm101 and its C-tail variants. (C) Size exclusion chromatography of Mgm101W257A on a Superose 6 column before and immediately after the release from MBP (red line). Inset shows SDS–PAGE analysis of peaks 1 and 2, which contain MBP and Mgm101, respectively. (D) Western blot analysis showing the levels of the mutant Mgm101 in isolated mitochondria. The mitochondrial outer membrane protein porin is used as a control for calibrating sample loading.
We then cleaved the fusion proteins and removed MBP by cation exchange to generate tag-free mutant Mgm101. Mgm101R259A could not be recovered from solution after the removal of MBP, indicating that Arg-259 is critical for structural organization and solubility of Mgm101. On a size exclusion column, Mgm101Y268A eluted mainly in a peak of ∼400 kDa, like the wild type. In contrast, Mgm101K253A and Mgm101(K251-K253)A are mainly present in fractions larger than the ∼400 kDa expected for the wild-type Mgm101 rings (Figure 4B). Negative-stain transmission electron microscopy revealed that Mgm101K253A, Mgm101(K251-K253)A, and Mgm101Y268A all form oligomeric rings (Figure 5A). Of interest, we observed frequent lateral interactions between the rings, especially in the case of Mgm101(K251-K253)A. The formation of the large ring clusters may explain the early elution of the proteins from the size exclusion column (see Figure 4B).
FIGURE 5:
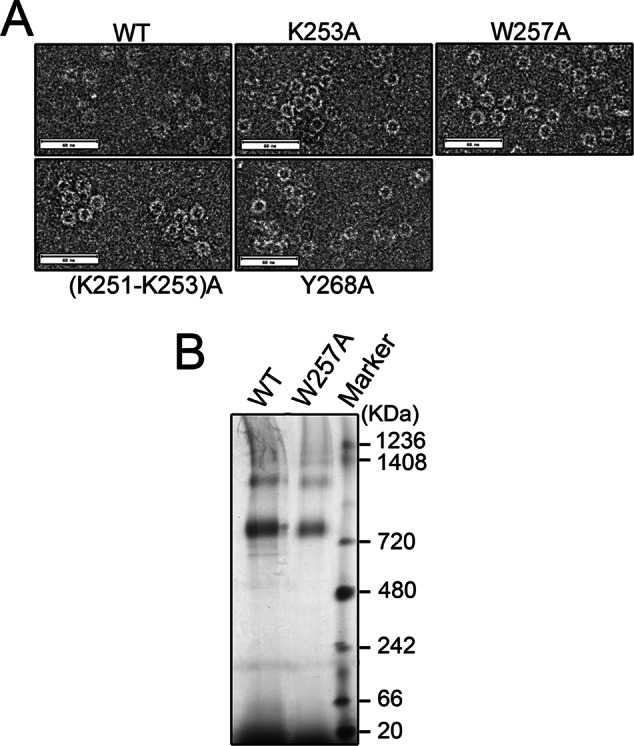
Oligomeric state of mutant Mgm101. (A) Negative-stain transmission electron microscopy showing the ring structure of mutant Mgm101. (B) Blue-native gel electrophoresis of Mgm101 (WT) and Mgm101W257A (W257A).
We observed that the W257A mutation dramatically altered the elution profile in size exclusion chromatography. Mgm101W257A appeared in an unusually sharp peak close to 25 kDa. To confirm this dramatic change, we directly analyzed the cleavage products of MBP-Mgm101W257A by size exclusion chromatography without passing through cation exchange (Figure 4C). Again, the uncleaved MBP-Mgm101W257A eluted as a large peak of ∼940 kDa, with a significant tail of smaller sizes. The cleaved MBP-Mgm101W257A resulted in two distinct peaks. SDS–PAGE confirmed that the large peak corresponds to MBP and the small peak contains Mgm101 (Figure 4C, inset). These data tended to support the idea that Mgm101W257A may exist in a monomeric form in solution. Surprisingly, transmission electron microscopy revealed that Mgm101W257A forms rings with a size similar to that of the wild type (Figure 5A). Using blue-native PAGE, we showed that Mgm101W257A migrates as large oligomers of ∼800 kDa, like the wild type (Figure 5B).
Taking the results together, we found that, except for Mgm101R259A, which drastically destabilizes the protein, the gross ring structure is maintained in Mgm101K253A, Mgm101(K251-K253)A, Mgm101W257A, and Mgm101Y268A. In the case of Mgm101W257A, the alanine substitution may dramatically change the surface property of the protein, which alters its elution profile in size exclusion chromatography (see Discussion). The structural stability appeared to correlate with in vivo stability of the proteins. Western blot analysis showed that Mgm101R259A does not accumulate in mitochondria, and Mgm101W257A is barely detectable (Figure 4D). Mgm101K253A and Mgm101(K251-K253)A have reduced levels compared with the wild type. The level of Mgm101Y268A, which has a size exclusion profile closest to that of the wild type, is little changed.
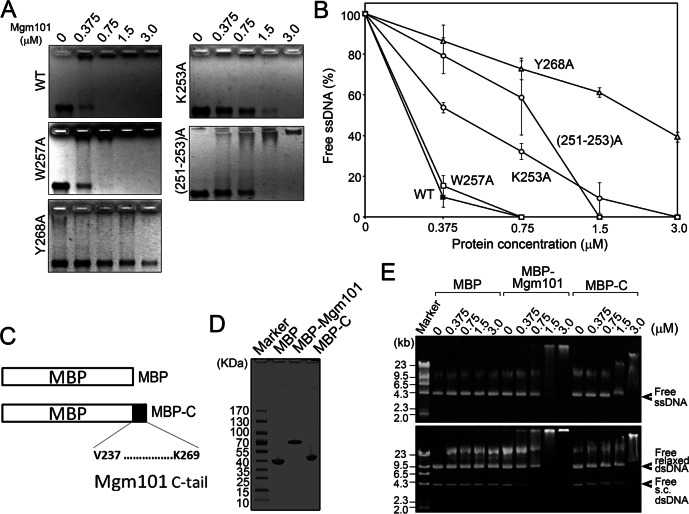
K253A, (K251-K253)A, and Y268A, but not W257A, affect ssDNA binding
We next compared the ssDNA-binding activity of Mgm101K253A, Mgm101(K251-K253)A, Mgm101W257A, and Mgm101Y268A with that of the wild-type Mgm101. Of interest, we found that Mgm101Y268A showed severe defect in binding to M13mp18 ssDNA on a 0.8% native agarose gel (Figure 6A). The ssDNA-binding activity is reduced by approximately ninefold compared with the wild-type control at the protein concentration of 0.375 μM (Figure 6B). This suggests that Tyr268 plays a key role in mediating ssDNA binding. Mgm101K253A and Mgm101(K251-K253)A also exhibited significant reduction in ssDNA-binding activity but to a lesser extent than Mgm101Y268A. In contrast, the Mgm101W257A maintained ssDNA-binding activity comparable to that of the wild-type control.
FIGURE 6:
ssDNA-binding affinity of Mgm101 C-tail variants. (A) Electrophoretic mobility shift assays showing the ssDNA-binding activity of Mgm101 C-tail variants. (B) Graphic representation of the results from A. Error bars, average deviations of three independent experiments for respective C-tail variants. (C) Schematics showing the fusion of the Mgm101 C-tail with MBP. (D) SDS–PAGE showing the purification of the Mgm101 C-tail fused with MBP (MBP-C). (E) DNA-binding activity of MBP-C in comparison with the fusion of the full-length Mgm101 with MBP (MBP-Mgm101). DNA binding was performed in the absence of Mg2+ and in the presence of 2 mM EDTA.
The C-tail is sufficient to mediate DNA binding
To confirm the ssDNA-interacting ability of the C-tail, we generated a fusion protein in which the last 32 codons of Mgm101 were cloned downstream of MBP (Figure 6C). The fusion protein (MBP-C) was purified to homogeneity by amylose column chromatography (Figure 6D) and tested for DNA-binding activity (Figure 6E). We found that the migration of the ssDNA in an agarose gel is significantly retarded after incubating with MBP-C but not with the MBP control. The results confirm that the C-tail of Mgm101 alone is sufficient to promote ssDNA binding. The titration of free ssDNA by MBP-C is slightly weaker than by MBP-Mgm101 containing the full-length Mgm101. The extent of mobility shift by MBP-C is also much less than with MBP-Mgm101. This may be explained by the monomeric feature of MBP-C in contrast to the oligomeric MBP-Mgm101, which overly reduces the initial affinity to ssDNA in the binding assay. MBP-C also shifted the migration of both supercoiled and relaxed dsDNA. The affinity of the C-tail to these DNA substrates is also lower than for the full length of Mgm101.
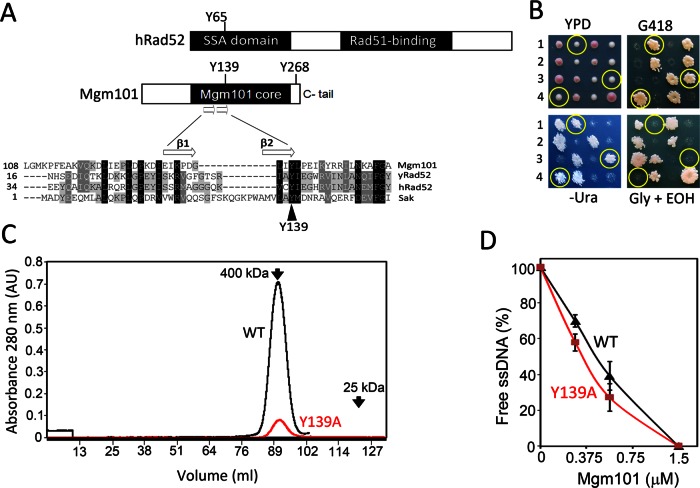
The evolutionarily conserved Tyr-139 in the predicted central groove is essential for mtDNA maintenance but not for ssDNA binding
The foregoing data support a critical role of Tyr-268 in mediating ssDNA binding. This finding also prompted us to examine the role of the Mgm101 core for ssDNA binding. Previous studies showed that mutations in Arg-55, Tyr-65, Lys-152, and Arg-156 in its homologous domain in Rad52, known as the central groove, affect ssDNA binding (see Figure 1B; Kagawa et al., 2002; Singleton et al., 2002). Among these amino acids, only Tyr-65 is conserved in SSAPs of prokaryotic and viral origin, which corresponds to Tyr-42 in Sak of the lactococcal phage ul36 and Tyr-139 in Mgm101 (Figure 7A). In support of a critical function of this amino acid, we found that the Y139A is unable to maintain mtDNA and support respiratory growth among the meiotic segregants (Figure 7B).
FIGURE 7:
Mgm101Y139A is functionally defective in vivo but competent in ssDNA binding in vitro. (A) Sequence alignment of Mgm101 with human Rad52. Shown in the Mgm101 functional core is Y139, which is equivalent to Y47 in yRad52 (yeast), Y65 in hRad52 (humans), and Y42 in Sak (lactococcal ul36). (B) Meiotic analysis showing that the mgm101Y139A is unable to complement the mgm101Δ::kan allele for growth on complete medium containing glycerol and ethanol (Gly+EtOH) as carbon sources. Four complete tetrads are shown, in which the segregants cosegregating mgm101Δ::kan (G418R) and the mgm101Y139A allele (Ura+) are circled. (C) Size exclusion chromatography of purified Mgm101Y139A on a Superose 6 column showing that the mutant variant coelutes with the wild-type protein. (D) ssDNA-binding activity of Mgm101Y139A compared with wild type. Error bars, average deviations of three independent experiments.
To know whether Tyr-139 is required for ssDNA binding, we prepared Mgm101Y139A. Size exclusion chromatography showed an oligomeric peak overlapping with the wild-type Mgm101 (Figure 7C). Thus Y139A has little effect on the oligomeric state of the protein. More importantly, Mgm101Y139A maintained ssDNA-binding activity comparable to that of the wild-type protein (Figure 7D). The data suggest that the evolutionarily conserved Tyr-139 in Mgm101 affects mtDNA maintenance by a mechanism other than ssDNA binding.
DISCUSSION
The mechanism by which SSAPs catalyze SSA is not well understood. Among the pertinent questions are the following: 1) How is ssDNA binding initiated? 2) Is the ring structure required for ssDNA binding? 3) How is the homology search promoted? Studies of the eukaryotic Rad52 protein support the model that ssDNA binds to the central groove along the surface of the ring (Singleton et al., 2002). Homology search is believed to proceed by successive interaction between two separate ssDNA-wrapped rings until the formation of a stable duplex. The force generated by the zippering of annealed DNA duplexes may drive ssDNA release from the overlapping nucleoprotein complexes (Rothenberg et al., 2008; Grimme et al., 2010). However, it remains perplexing that Rad52 often forms nucleoprotein filaments of 10-nm thickness on ssDNA, which suggests that the ring structure is disrupted upon DNA binding (Kagawa et al., 2001). Although several mutations in the presumptive central binding groove affect ssDNA-binding activity (Kagawa et al., 2002; Lloyd et al., 2005), these data need to be validated by solving the structure of Rad52/ssDNA cocrystals. On the other hand, study of Redβ by atomic force microscopy revealed that the ring/helix organization is clearly disrupted when the phage protein interacts with ssDNA (Erler et al., 2009). In this case, monomeric structures are also detected in the Redβ–ssDNA complexes. It was proposed that it is the interaction between two ssDNA-bound Redβ monomers that facilitates the annealing of the ssDNA molecules. The latter observations point to the possibility that the SSAPs from various sources may have different modes of action in binding to ssDNA and in catalyzing strand annealing.
Mgm101 forms condensed nucleoprotein filaments on ssDNA, like the bacteriophage SSAPs, such as the Sak protein from the lactococcal phage ul36 (Ploquin et al., 2008; Mbantenkhu et al., 2011). It also forms highly compressed helical filaments, like Redβ from the bacteriophage λ (Passy et al., 1999). These molecular features are not seen in Rad52. The data from the present study further suggest that Mgm101, and probably also the bacteriophage SSAPs, may have a DNA-binding mode different from that proposed for Rad52. Mutation in Tyr-65 of Rad52 reduces ssDNA binding by fourfold (Kagawa et al., 2002; Lloyd et al., 2005). Tyr-65 is the only conserved residue in several SSAPs among the amino acids predicted to bind to ssDNA in the putative central binding groove. This residue corresponds to Tyr-139 in Mgm101 and Tyr-42 in Sak (Figures 1B and 7A). We provided evidence that Tyr-139 is essential for the mtDNA maintenance function of Mgm101 in vivo, but the Y139A mutation does not affect ssDNA binding in vitro. The similar Y42A mutation in Sak was also been to have little effect on DNA binding under similar conditions (Ploquin et al., 2008). It is noteworthy that Tyr-139 is not conserved in some SSAPs, including Redβ (Lopes et al., 2010). These observations raise the possibility that the central core of Mgm101 may not play a major role, if any, in ssDNA binding. Mgm101 may have evolved a novel mode of interaction with ssDNA substrates.
Mutagenesis analysis combined with in vivo functional assay and in vitro biochemical characterization revealed a robust ssDNA-binding site at the short C-tail. The C-tail is highly conserved among the Mgm101 homologues that function in mitochondria (Figure 2A). This particular domain of 32 amino acids in length is not conserved in Rad52, which operates in the eukaryotic nucleus. The C-tail contains highly conserved aromatic and basic amino acids that are predicted to form two β-sheets followed by an unstructured end. Among the 10 amino acids analyzed, Lys-253, Trp-257, Arg-259, and Tyr-268 were found to be essential for mtDNA maintenance in vivo. Mutations in Lys-251, Lys-252, Lys-260, and Tyr-266 affected mtDNA maintenance only under stress conditions. Petite colony formation was dramatically increased in these mutants when cells were incubated at high temperature (37°C). Increased petite formation was also seen when cells were treated with hydrogen peroxide, which is consistent with the previously reported role of Mgm101 in repair of oxidatively damaged mtDNA (Meeusen et al., 1999). The most intriguing finding is perhaps that mutations in some of these residues affect ssDNA binding. We found that the gross ring structure of Mgm101Y268A, Mgm101K253A, and Mgm101(K251-K253)A is unaltered, as confirmed by single-particle imaging using transmission electron microscopy. There is apparently an increase in lateral interaction between the rings, which changes the elution profile of the proteins in size exclusion chromatography. However, ssDNA binding by these mutant proteins is severely affected. ssDNA-binding activity of Mgm101Y268A, which eluted mainly like the wild type in size exclusion chromatography, is reduced ninefold. ssDNA-binding activity of Mgm101K253A is reduced 5.5-fold. ssDNA-binding activity of the triple substitution mutant Mgm101(K251-K253)A is reduced 8.2-fold, indicating that Lys-251 and/or Arg-252 also contribute to the interaction with ssDNA. A confirmation for the ssDNA-binding activity of the C-tail comes from an experiment showing that the C-tail alone, when fused to MBP, is sufficient to mediate ssDNA binding. The data indicate that the C-tail of Mgm101 is capable of binding to ssDNA, which could be critical for mtDNA repair and maintenance in vivo. Although the in vitro data suggest that the 251–KRK–253 triad may contribute to the interaction with ssDNA, this may not fully account for the defect in mtDNA maintenance in vivo (Figure 3A). The Mgm101K253A and Mgm101(K251-K253)A proteins are partially unstable in vivo. It is possible that ssDNA-binding defect and protein instability both contribute to mtDNA instability. We previously reported the C240A mutation in the C-tail. This mutation does not affect mtDNA maintenance in vivo (Mbantenkhu et al., 2011), and the ssDNA-binding activity of the mutant protein is not affected compared with the wild type (our unpublished data).
The mechanism of ssDNA binding has been extensively studied in oligonucleotide/oligosaccharide-binding fold (OB-fold) proteins. In these cases, ssDNA interacts with a rather limited surface on the proteins primarily via stacking interactions with aromatic amino acid chains (Theobald et al., 2003). The base-stacking interactions allow the protein to distinguish ssDNA from dsDNA, whose base has less accessibility. Although Mgm101 does not belong to the OB-fold protein family, its C-tail may represent a novel module for ssDNA recognition. We speculate that Tyr-268 may play a pivotal role in mediating interaction with ssDNA by stacking interactions. The second aromatic residue, Tyr-266, which also affects mtDNA stability in response to high temperature and oxidative stress, may also promote stacking and assist the stabilization of the interactions with ssDNA. On the other hand, the 251–KRK–253 triad may attract ssDNA to the ring surface through electrostatic interactions, which facilitates its interactions with Tyr-268 and Tyr-266.
Biochemical analysis of Mgm101 with alanine substitution also revealed an important role of the Mgm101 C-tail in structural stabilization. Even in the MBP-fused form, mutation in Arg-259 resulted in an unstable protein that tends to elute in the void volume in size exclusion chromatography, which is suggestive of aggregate formation. The mutant protein after cleavage from MBP was not recoverable. A similar property was observed when Mgm101 lacking the entire C-tail was analyzed. In the latter case, a sizable MBP-fused monomeric peak was noticeable in addition to a major peak in the void volume. These data suggest that the C-tail is important for oligomerization and stabilization of the protein. Arg-259 may play a key role in mediating interactions between the adjacent subunits in the ring by facilitating salt bridge formation. The mutation may disassemble the rings into monomeric Mgm101, which is unstable in solution. A rapid conversion of monomeric Mgm101 to insoluble aggregates has been speculated (Mbantenkhu et al., 2011; Nardozzi et al., 2012). Although it can only be transiently detected, the monomeric peak of MBP-Mgm101ΔC is the only unoligomerized Mgm101 species detected so far. The presence of MBP likely contributes to the partial stabilization of the mutant form of monomeric Mgm101 in solution.
A role of the C-tail in maintaining an intact ring could be analogous to the C-terminal end of the SSA domain of Rad52 (or Rad521-209). The C-terminal end of this truncated form of Rad52 is characterized by a flexible linker, which is followed the α-helix 5, which swaps across the subunit interface and interacts with an adjacent subunit in the undecameric ring (Kagawa et al., 2002; Singleton et al., 2002). This “helix-swapping” strategy promotes subunit interactions, thereby stabilizing the ring structure. Whether a similar strategy is used for oligomerization in the full-length heptameric Rad52 is unknown. In place of α-helix 5 in Rad521-209, the C-tail of Mgm101 is predicted to form two β strands followed by a short, unstructured C-terminal end (β6 and β7; see Figure 2A). These β strands are likely involved in interactions with adjacent subunits in the ring. Arg-259 may be required for stabilizing the β6–β7 interaction or for direct interaction with adjacent subunits. As a consequence, mutation in Arg-259 prevents ring formation.
Trp-257 is another aromatic amino acid on the C-tail of Mgm101 essential for its function. However, mutation in Trp-257 does not have a detectable defect in ssDNA binding. Mgm101W257A has a peculiar elution profile on the Superose 6 column. The protein was eluted as a sharp peak corresponding to the molecular weight of monomeric Mgm101. However, blue-native gel showed that the mutant protein forms oligomeric structures indistinguishable from the wild type. Inspection by transmission electron microscopy did not show an obvious difference in the shape and size of the rings formed by Mgm101W257A and the wild-type protein. Furthermore, Mgm101W257A also remained stable in solution like the wild type. It is likely that Mgm101W257A exists as oligomeric rings rather than in a monomeric form. It is possible that the W257A mutation may change the surface property of the rings, which increases interactions with the column matrix and causes the dramatic retardation of the protein on the column. This novel property is probably corrected or masked by the tagging of the 42-kDa MBP on the N-terminus, which permitted an elution profile close to that of the wild-type protein (see Figure 4, A and C).
We observed that Mgm101R259A and Mgm101W257A not only are structurally unstable in vitro, but are also highly unstable in vivo. This provides an explanation for the rapid loss of mtDNA in the mutants. It seems that the maintenance of an appropriately configured ring structure is important for protein stability in vivo. Monomerization or changes to the ring surface properties may trigger protein degradation. For future studies, it would be interesting to determine which protease is involved in degrading disassembled and improperly assembled Mgm101 structures in the mtDNA nucleoids.
In summary, we found that the 32-aa C-tail of Mgm101 is required for ssDNA binding, structural organization, and protein stability in vivo. It can be speculated that the positively charged 251–KRK-253 triad may initiate the contact with the phosphate backbone of ssDNA by electrostatic interactions. The highly conserved 266–YPY–268 motif at the extreme end of the protein may stabilize the interactions by base stacking. These interactions could disrupt the ring structure maintained by salt bridge interactions involving Arg-259. This may facilitate the deployment of additional subunits in the ring to spread the interactions with ssDNA. The model is consistent with the idea that the ring structure serves as a store in the mitochondrial nucleoids so that the protein can be rapidly mobilized after mtDNA damage and the initial interaction with DNA. This ultimately generates recombination-competent nucleoprotein filaments for the repair of double-stranded breaks in mtDNA, in concert with other repair proteins, including those involved in homologous pairing and nonhomologous end joining (Ling et al., 1995; Ling and Shibata, 2002; Kalifa et al., 2012). Future studies are necessary to test this model.
MATERIALS AND METHODS
Growth media and strains
Complete medium for growth of yeast (YPD) contains 1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose. YPGE medium is prepared by adding 2% glycerol plus 2% ethanol instead of glucose. Minimal medium (YNBD) contains 0.17% Difco yeast nitrogen base without amino acids, 0.5% ammonium sulfate, and 2% glucose. Nutrients essential for auxotrophic strains were added to YNBD at 25 μg/ml for bases and 50 μg/ml for amino acids. The yeast strains used in this study are listed in Table 1.
TABLE 1:
Genotypes and sources of yeast strains used in this study.
| Strain name | Genotype | Source |
|---|---|---|
| Haploid | ||
| M2915-6A | MATa, ade2, leu2, ura3 | This laboratory |
| M2915-7C | MATa, ade2, leu2, his4, ura3, mgm101-1ts | Chen et al. (1993) |
| W303-1B | MATα, ade2, trp1, his3, leu2, ura3 | R. Rothstein (Columbia University) |
| W303-1B/A | MATa, ade2, trp1, his3, leu2, ura3 | R. Rothstein (Columbia University) |
| MM3/2-2B | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101F244A | This study | |
| MM4/1-2D | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101K251A | This study | |
| MM5/2-1C | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101W257A | This study | |
| MM6/1-2B | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101Y266A | This study | |
| MM7/2-4C | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101Y268A | This study | |
| MM8/4-4B | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101R252A | This study | |
| MM9/1-6D | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101K253A | This study | |
| MM10/1-4C | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101R259A | This study | |
| MM11/2-2D | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101K260A | This study | |
| MM12/2-6D | W303 background, mgm101Δ::kan, | |
| ura3::pUC-URA3/4-mgm101K269A | This study | |
| MM13/1-1D | W303 background, mgm101Δ::kan, | |
| ura3:: pUC-URA3/4-mgm101K252A, R252A, K253A | This study | |
| Diploid | ||
| W303/a/α | MATa/MATα, ade2/ade2, his3/his3, trp1/trp1, leu2/leu2, | |
| ura3/ura3, | R. Rothstein (Columbia University) | |
| CS1638/1 | As W303/a/α, but +/mgm101Δ::kan | This study |
| MM3/2 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101F244A | This study |
| MM4/1 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101K251A | This study |
| MM5/2 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101W257A | This study |
| MM6/1 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101Y266A | This study |
| MM7 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101Y268A | This study |
| MM8/4 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101R252A | This study |
| MM9/1 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101K253A | This study |
| MM10/1 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101R259A | This study |
| MM11/2 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101K260A | This study |
| MM12/2 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101K269A | This study |
| MM13/1 | As CS1638/1, | |
| but ura3/ura3::pUC-URA3/4-mgm101K252A, R252A, K253A | This study | |
| MM14/1 | As CS1638/1, but ura3/ura3::pUC-URA3/4-mgm101Y139A | This study |
Construction and expression of mutant mgm101 alleles
The QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) was used to generate mutant mgm101 alleles. The oligonucleotides used for in vitro mutagenesis are listed in Supplemental Table S1. The plasmid pCXJ22-MGM101 was used as a template for generating constructs for complementation in the temperature-sensitive mgm101 mutant M2915-7C. Correct mutagenesis was confirmed by sequencing the entire open reading frame of MGM101 using MGM101P2 (5′-AATCTAGCAGCAAACGC-3′) and MGM101P5 (5′-CGCTAACCCTGAAATAG-3′) as primers. For chromosomal integration, mutations were introduced in the plasmid pUC-URA3/4-MGM101. The resulting plasmids were linearized by digesting with EcoRV within the URA3 gene and integrated into the ura3 locus of the diploid strain CS1638/1 by selection for Ura+ transformants. This allowed the determination of the functionality of the mutant alleles in meiotic segregants with the endogenous wild-type MGM101 gene disrupted. All the strains used carry the ade2 mutation, which turn red under respiring conditions in the presence of a functional mitochondrial genome. The petite frequency was measured by determining the percentage of white colonies as a function of the total number of colonies.
Expression and purification of mutant Mgm101
All the Mgm101 variants were expressed and purified from Escherichia coli as previously described (Mbantenkhu et al., 2011). The plasmid pMalC2e-MGM101, expressing the MBP-MGM101 fusion, was used as a template to introduce the mgm101 mutations by in vitro site-directed mutagenesis. The oligonucleotides used for introducing the mutant alleles are listed in Supplemental Table 1. The MBP-Mgm101 fusions lack the 22–amino acid mitochondrial targeting signal of Mgm101 and contain a cleavage site for Prescission protease (GE Healthcare, Piscataway, NJ) for releasing MBP. The plasmids were then introduced into the strain BL21-Codon+(DE3)-RIL (E. coli B F− ompT hsdS(rB− mB−) dcm+ Tetr gal λ(DE3) endA Hte (argU ileY leuW Camr); Stratagene). Expression of the fusion proteins was induced with isopropyl β-d-1-thiogalactopyranoside at 30°C for 5 h. After sonication of the cells, DNase I (50 μg/ml) was used to digest DNA. The fusion proteins were purified on an amylose column and digested with the Prescission protease to release Mgm101. MBP and Mgm101 were separated by cation exchange chromatography using a Bio-Scale Mini Prep UNOsphere cartridge (Bio-Rad, Hercules, CA). Mgm101 was further purified by size exclusion chromatography on a calibrated Superose 6 column equilibrated with 20 mM 3-(N-morpholino)propanesulfonic acid, pH 7.0, 150 mM NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA, and 0.2 mM phenylmethanesulfonyl fluoride.
The mgm101 allele lacking the C-tail (MGM101ΔC) was amplified by PCR using the primers MGM101P1 (5′-CGGGGATCCGTGGTGAGTACCGGCACTAG-3′) and MGM101ΔCP2 (5′-AGGGCTGCAGCTACACTTTGAATTTCTTTATGAAAACAG-3′). After digestion with BamHI and PstI, the PCR DNA was placed downstream of the malE sequence of pMal-c2e. This created the fusion MBP-MGM01ΔC, which, after cleavage by Prescission Protease, releases Mgm101 lacking the last 32 amino acids.
To purify the MBP-C-tail fusion, we used the oligonucleotide MGM101PCs (5′-CGGGGATCCGTGGATCATTGTACTGAGAAG-3′) and MGM101MP2 (5′-AAAACTGCAGCTATTTATAAGGATATTCAACTTG-3′) to amplify the last 32 codons of MGM101. The DNA segment was then digested with BamHI and PstI and placed downstream of the malE sequence in the plasmid pMal-c2e. The fusion protein was then purified by amylose column affinity chromatography.
Other procedures
For DNA-binding assays, M13mp18 ssDNA or dsDNA (250 ng) was incubated for 40 min at 37°C with increasing concentrations of Mgm101 or its mutant variants (0–3 μM) in the DNA-binding buffer (150 mM Tris-HCl, pH 7, 4% glycerol, 10 mM MgAc). The reaction mix was then loaded onto a 0.8% agarose gel and run in 1× Tris-acetate-EDTA buffer at 80 V on ice. Blue-native PAGE was performed using the Bio-Rad Mini Protean III cell system as recommended by the manufacturer. Transmission electron microscopy was performed as previously described (Mbantenkhu et al., 2011). Mgm101 (with or without DNA) was diluted to between 10 and 50 μg/ml and spread on carbon-coated copper grids. After negative staining with 1% uranyl acetate, Mgm101 oligomers were visualized in a JEM-2100 transmission electron microscope (JEOL, Peabody, MA) operating at 200 kV. Electron micrographs were recorded on a 4096 × 4096–pixel charge-coupled device camera (TVIPS F415-MP) in minimum-dose mode at an electron optical magnification of 40,000× and a defocus of −1.5 μm.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01AG023731.
Abbreviations used:
- dsDNA
double-stranded DNA
- MBP
maltose-binding protein
- mtDNA
mitochondrial DNA
- SSA
single-stranded annealing
- SSAP
single-stranded annealing protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0006) on March 27, 2013.
*Present address: Genetics and Biochemistry Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892.
REFERENCES
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Botstein D, Matz MJ. A recombination function essential to the growth of bacteriophage P22. J Mol Biol. 1970;54:417–440. doi: 10.1016/0022-2836(70)90119-1. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Guan MX, Clark-Walker GD. MGM101, a nuclear gene involved in maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3473–3477. doi: 10.1093/nar/21.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker GD, Chen XJ. A vital function for mitochondrial DNA in the petite-negative yeast Kluyveromyces lactis. Mol Gen Genet. 1996;252:746–750. doi: 10.1007/BF02173982. [DOI] [PubMed] [Google Scholar]
- Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159:515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler A, Wegmann S, Elie-Caille C, Bradshaw CR, Maresca M, Seidel R, Habermann B, Muller DJ, Stewart AF. Conformational adaptability of Redbeta during DNA annealing and implications for its structural relationship with Rad52. J Mol Biol. 2009;391:586–598. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Feng Q, During L, de Mayolo AA, Lettier G, Lisby M, Erdeniz N, Mortensen UH, Rothstein R. Rad52 and Rad59 exhibit both overlapping and distinct functions. DNA Repair (Amst) 2007;6:27–37. doi: 10.1016/j.dnarep.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Fenton AC, Poteete AR. Genetic analysis of the erf region of the bacteriophage P22 chromosome. Virology. 1984;134:148–160. doi: 10.1016/0042-6822(84)90280-0. [DOI] [PubMed] [Google Scholar]
- Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010;38:2917–2930. doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Massey P, Banerjee R, Berg P. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol Cell Biol. 1998;18:4400–4406. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward DC, Dosztanyi Z, Clark-Walker GD. The N-terminal intrinsically disordered domain of Mgm101p is localized to the mitochondrial nucleoid. PLoS One. 2013;8:e56465. doi: 10.1371/journal.pone.0056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, et al. DNA packaging proteins Glom and Glom2 coordinately organize the mitochondrial nucleoid of Physarum polycephalum. Mitochondrion. 2011;11:575–586. doi: 10.1016/j.mito.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicka S, Kuhn K, Le Ret M, Bonnard G, Imbault P, Augustyniak H, Gualberto JM. A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J. 2012;72:423–435. doi: 10.1111/j.1365-313X.2012.05097.x. [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Homologous pairing promoted by the human Rad52 protein. J Biol Chem. 2001;276:35201–35208. doi: 10.1074/jbc.M104938200. [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- Kalifa L, Quintana DF, Schiraldi LK, Phadnis N, Coles GL, Sia RA, Sia EA. Mitochondrial genome maintenance: roles for nuclear nonhomologous end joining proteins in Saccharomyces cerevisiae. Genetics. 2012;190:951–964. doi: 10.1534/genetics.111.138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc Natl Acad Sci USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SI, Min BE, Jung GY. Lagging strand-biased initiation of red recombination by linear double-stranded DNAs. J Mol Biol. 2008;384:1098–1105. doi: 10.1016/j.jmb.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Ling F, Makishima F, Morishima N, Shibata T. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 1995;14:4090–4101. doi: 10.1002/j.1460-2075.1995.tb00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Shibata T. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 2002;21:4730–4740. doi: 10.1093/emboj/cdf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JA, McGrew DA, Knight KL. Identification of residues important for DNA binding in the full-length human Rad52 protein. J Mol Biol. 2005;345:239–249. doi: 10.1016/j.jmb.2004.10.065. [DOI] [PubMed] [Google Scholar]
- Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38:3952–3962. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbantenkhu M, Wang X, Nardozzi JD, Wilkens S, Hoffman E, Patel A, Cosgrove MS, Chen XJ. Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J Biol Chem. 2011;286:42360–42370. doi: 10.1074/jbc.M111.307512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, Tieu Q, Wong E, Weiss E, Schieltz D, Yates JR, Nunnari J. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne GT, Weaver DT. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G, Gewin J, Luder A, Colowick N, Vo D. Two recombination-dependent DNA replication pathways of bacteriophage T4, and their roles in mutagenesis and horizontal gene transfer. Proc Natl Acad Sci USA. 2001;98:8306–8311. doi: 10.1073/pnas.131007398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozzi JD, Wang X, Mbantenkhu M, Wilkens S, Chen XJ. A properly configured ring structure is critical for the function of the mitochondrial DNA recombination protein, Mgm101. J Biol Chem. 2012;287:37259–37268. doi: 10.1074/jbc.M112.389965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson JY. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr Biol. 2008;18:1142–1146. doi: 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR. Involvement of DNA replication in phage lambda Red-mediated homologous recombination. Mol Microbiol. 2008;68:66–74. doi: 10.1111/j.1365-2958.2008.06133.x. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Sauer RT, Hendrix RW. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J Mol Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- Rothenberg E, Grimme JM, Spies M, Ha T. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc Natl Acad Sci USA. 2008;105:20274–20279. doi: 10.1073/pnas.0810317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Melamed-Bessudo C, Avivi-Ragolski N, Pietrokovski S, Levy AA. Identification of Plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell. 2012;23:4266–4279. doi: 10.1105/tpc.111.091744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P, Krejci L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J Biol Chem. 2008;283:12166–12174. doi: 10.1074/jbc.M800763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi I, Hallwyl SC, Seong C, Mortensen U, Rothstein R, Sung P. Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J Biol Chem. 2009;284:33275–33284. doi: 10.1074/jbc.M109.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing vs. invasion in phage lambda recombination. Genetics. 1997;147:961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH. The human Rad52 protein exists as a heptameric ring. Curr Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sugiyama T, Kowalczykowski SC. DNA annealing mediated by Rad52 and Rad59 proteins. J Biol Chem. 2006;281:15441–15449. doi: 10.1074/jbc.M601827200. [DOI] [PubMed] [Google Scholar]
- Zuo X, Xue D, Li N, Clark-Walker GD. A functional core of the mitochondrial genome maintenance protein Mgm101p in Saccharomyces cerevisiae determined with a temperature-conditional allele. FEMS Yeast Res. 2007;7:131–140. doi: 10.1111/j.1567-1364.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- Zuo XM, Clark-Walker GD, Chen XJ. The mitochondrial nucleoid protein, Mgm101p, of Saccharomyces cerevisiae is involved in the maintenance of rho+ and ori/rep-devoid petite genomes but is not required for hypersuppressive rho− mtDNA. Genetics. 2002;160:1389–1400. doi: 10.1093/genetics/160.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.