Pineapples, or self-organized, Taxol-stabilized microtubule assemblies, reveal the richness of self-organizing mechanisms that operate on assembled microtubules during cell division and provide a biochemically tractable system for investigating these mechanisms during meiosis and cytokinesis.
Abstract
Previous study of self-organization of Taxol-stabilized microtubules into asters in Xenopus meiotic extracts revealed motor-dependent organizational mechanisms in the spindle. We revisit this approach using clarified cytosol with glycogen added back to supply energy and reducing equivalents. We added probes for NUMA and Aurora B to reveal microtubule polarity. Taxol and dimethyl sulfoxide promote rapid polymerization of microtubules that slowly self-organize into assemblies with a characteristic morphology consisting of paired lines or open circles of parallel bundles. Minus ends align in NUMA-containing foci on the outside, and plus ends in Aurora B–containing foci on the inside. Assemblies have a well-defined width that depends on initial assembly conditions, but microtubules within them have a broad length distribution. Electron microscopy shows that plus-end foci are coated with electron-dense material and resemble similar foci in monopolar midzones in cells. Functional tests show that two key spindle assembly factors, dynein and kinesin-5, act during assembly as they do in spindles, whereas two key midzone assembly factors, Aurora B and Kif4, act as they do in midzones. These data reveal the richness of self-organizing mechanisms that operate on microtubules after they polymerize in meiotic cytoplasm and provide a biochemically tractable system for investigating plus-end organization in midzones.
INTRODUCTION
Mitosis and cytokinesis in animal cells are orchestrated by two microtubule arrays that assemble sequentially. First, the meiotic spindle congresses and then segregates chromosomes. Next the midzone complex (also called the central spindle) forms between separating chromosomes to position the cleavage furrow. The cleavage plane in many systems is determined by the location of midzone plus ends. Spindles and midzones both self-organize using a combination of spatially regulated polymerization dynamics and sliding forces from motor proteins, but they differ in the molecules used, the geometry of the final assembly, and the rate of microtubule turnover. Spindles and midzones are normally bipolar, but both can also assemble in monopolar form when activity of kinesin-5 (also called Eg5, Kif11, and KSP) is removed (Gaglio et al., 1996; Mayer et al., 1999; Canman et al., 2003; Hu et al., 2008; Shrestha et al., 2012).
Many or all of the conserved proteins required for spindle and midzone assembly have been defined in recent years by a combination of genetics, biochemistry, and imaging (reviewed in Barr and Gruneberg, 2007; Glotzer, 2009; Hutchins et al., 2010; Lee et al., 2012). Here we focus on seven proteins with central roles in spindle or midzone assembly as representative assembly factors: 1) dynein, a minus end–directed motor that aggregates spindle microtubules and clusters minus ends at spindles poles; 2) NUMA, a large, mostly coiled-coil protein, which helps dynein organize spindle poles, which is transported to minus ends by dynein (Merdes et al., 2000) and served as our minus-end polarity marker; 3) kinesin-5 (also known as Eg5, Kif11), a plus end–directed, tetrameric kinesin that promotes antiparallel sliding and pushes spindle poles apart; 4) Aurora B, the kinase subunit of the chromosome passenger complex (CPC; Ruchaud et al., 2007), which localizes near midzone plus ends and plays a central role in midzone assembly; it is transported to plus ends in midzones by Kif20A (Gruneberg et al., 2004) and served as our plus end polarity marker. 5) Kif4 (also known as XKLP1 in Xenopus), a plus end–directed kinesin that inhibits microtubule polymerization and accumulates near midzone plus ends, where it is believed to inhibit their growth (Bieling et al., 2010; Hu et al., 2011); 6) PRC1L, the embryonic isoform in Xenopus (Klein et al., 2002) of the conserved antiparallel cross-linker PRC1 that plays a key role in midzone assembly; and 7) KIF23 (also known as CHO1, MKLP1), part of the centralspindlin complex that signals from midzone plus ends to the cortex to position furrows. How these and other factors work together to promote spindle and midzone assembly and dynamics is a major topic of current research.
One approach to elucidating microtubule-organizing mechanisms is to study them in cell-free extract systems. Compared to living cells, extracts offer a wider range of experimental approaches and easier perturbation of essential processes. Compared to pure protein reactions, they provide access to more complex mixtures and to proteins at physiologically relevant concentrations and states of posttranslational modification. Crude meiotic extracts from Xenopus eggs that contain organelles and ribosomes faithfully recapitulate both polymerization dynamics and spatial organization of microtubules and have been widely used to reconstitute meiosis II spindle assembly (Desai et al., 1999; Maresca and Heald, 2006). However, crude egg extract is almost as complex as a living cell and works best when prepared fresh. Cytosol preparations—extracts cleared of organelles and ribosomes by high-speed centrifugation—are more biochemically tractable, but physiological microtubule polymerization dynamics is not retained in cytosol for unknown reasons.
Spindle and midzone assembly are sufficiently complex that we need to break them into subreactions to understand them. One broad conceptual division is between factors that control where microtubules polymerize or depolymerize and factors that act on assembled microtubules to move or cross-link them. Factors that regulate polymerization dynamics can be bypassed in extract systems by adding Taxol. By artificially forcing nucleation and stabilization, Taxol provides direct access to activities that organize preformed microtubules. The Karsenti group pioneered this approach and used it to discover that minus ends in crude meiotic extracts are clustered by dynein (Verde et al., 1991). The Compton group adapted the Taxol aster approach to meiotic HeLa lysates clarified by centrifugation. They refined our understanding of motor-driven organizational processes in spindle pole assembly (Gaglio et al., 1995, 1996; Chakravarty et al., 2004) and identified new proteins involved in spindle pole assembly simply by cosedimentation with microtubules (Mack and Compton, 2001).
In this study we revisit the Taxol aster approach using undiluted meiotic cytosol from Xenopus eggs. We recently reported this preparation as part of an effort to biochemically reconstitute microtubule nucleation stimulated by Ran.GTP (Groen et al., 2011). A key innovation was to supplement cytosol with glycogen, which is naturally present at ∼30 mg/ml in egg cytoplasm. Glycogen provided energy, reducing equivalents and a mild crowing effect, and significantly improved the physiological state of the cytosol as judged by metabolic measurements and response to Ran.GTP. When we added Taxol and dimethyl sulfoxide (DMSO) to this system we expected to observe Taxol asters as previously reported by the Karsenti and Compton groups and were surprised to see more complex assemblies with minus ends aligned on the outside and plus ends on the inside. We called these “pineapples” because round examples resembled a slice of canned pineapple (Figure 1B). We found that pineapples assemble by a combination of spindle and midzone mechanisms, suggesting that they will provide a useful system for analyzing these mechanisms. Here we describe the system and provide evidence that both spindle and midzone mechanisms are involved.
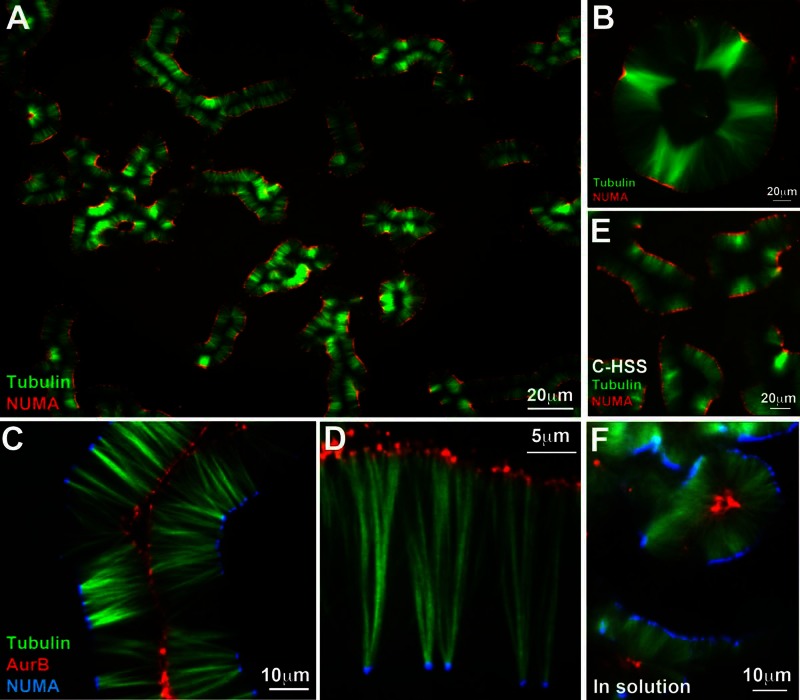
FIGURE 1:
Pineapple assembly. (A) Typical field 1 h after addition of 5 μM Taxol and 5% DMSO to meiotic HSS containing tubulin (green) and NUMA (red) probes; 20× wide-field imaging. Note formation of similar assemblies across the whole field, comprising paired lines of microtubule bundles with NUMA aligned on the outside. (B) Round assemblies with open centers resembled a slice of canned pineapple (∼2 h of assembly, 20× wide field). Tubulin (green), NuMA (red). (C) A 40× confocal image at 90 min showing localization of Aurora B (red) on the inside and NUMA (blue) on the outside. Note global alignment of both types of foci on curved planes, which appear as lines in optical sections. (D) A 100× confocal image taken at 90 min showing one side of an open assembly. NUMA (blue) was on the outside as usual. Both NUMA and Aurora B (red) accumulate at foci. Aurora B foci are smaller and more numerous. (E) Pineapple assembly in C-HSS, which is completely free of organelles; 120 min, 20× wide field. Tubulin (green), NUMA (red). (F) Pineapple assembly in solution. This image was taken by assembly in solution for 1 h, followed by 20× dilution into a Taxol-containing buffer without DMSO, squashing between a slide and coverslip, and imaging within ∼5 min (40× confocal). Other examples are shown in Figure 4. NUMA (blue), Aurora B (red), tubulin (green).
RESULTS
Pineapple assembly
Addition of Taxol dissolved in DMSO to final concentrations of 5 μM and 5%, respectively, induced the formation of microtubule structures in meiotic high-speed supernatant (HSS) that we termed pineapples. Figure 1, A–D, shows typical fields that illustrate the final morphology at different magnifications. In addition to labeled tubulin, these reactions contained nonperturbing probes to visualize microtubule polarity. Minus ends were localized with an antibody probe for NUMA and plus ends with an antibody probe for Aurora B (both nonperturbing). NUMA is transported to minus ends in spindles by dynein and is a standard reporter for minus ends in Taxol aster reactions (Gaglio et al., 1995). The CPC complex, of which Aurora B is part, localizes to midzone plus ends in both bipolar (Ruchaud et al., 2007) and monopolar (Canman et al., 2003; Hu et al., 2008) midzones. Its accumulation at midzone plus ends depends on the plus end–directed kinesin Kif20A (Gruneberg et al., 2004). Our Aurora B antibody probe presumably reports on whole CPC localization, since the subunits form a tight, stoichiometric complex in Xenopus egg extracts (Sampath et al., 2004). Consistent with this, a different CPC subunit, borealin, also localized to plus ends on the inside, using a labeled antibody probe (data not shown). Because NUMA and Aurora B localize by motor-dependent transport, they accumulate on the most-outward ends in an assembly and do not report on internal ends. In addition, they presumably report on average polarity rather than polarity of all microtubules, so they would miss a small fraction of microtubules oriented opposite to the majority.
NUMA localized to foci on the outside of pineapples and Aurora B to foci on the inside (Figure 1). This was true in 100% of the examples we imaged (n >1000 assemblies from >50 experiments with >10 batches of HSS and clarified HSS [C-HSS]). With the caveats mentioned earlier, we interpret these images as indicating that microtubules are oriented in pineapples with minus ends toward the outside, as in bipolar spindles and midzones. Both NUMA and Aurora B foci had a strong tendency to align globally, which we visualized as lines of dots in optical projections and confocal slices (Figure 1), although focusing showed that these lines were actually planes, or cylindrical envelopes, projected in Z or optically cross sectioned. The extent to which pineapples were planar versus cylindrical depended on their width relative to the separation between coverslips (∼15 μm in most experiments). Pineapple morphology at 60–90 min was highly reproducible both across one coverslip in individual reactions and between reactions. Individual batches of HSS showed some variation in how long the reaction took to achieve this morphology (∼20–40 min) but less in final morphology. Pineapple width at a given time point was sensitive to the precise Taxol concentration (see later discussion).
C-HSS, which was completely free of organelles and particulates (Groen et al., 2011), gave rise to pineapples with similar morphology (Figure 1E), so organelles were not required. Most of our work was done with HSS, which is easier to prepare, and key experiments were repeated with C-HSS.
Pineapples self-organize
Assembly between two glass surfaces generated flattened pineapples that were easy to visualize, but pineapples also assembled without surface constraints. Figure 1F shows a reaction that was assembled in a tube, diluted into Taxol-containing buffer at 60 min, squashed between coverslips, and immediately imaged. Pineapple width, after squashing, was comparable to the same reaction assembled between coverslips. Assemblies formed in solution were presumably cylindrical or spherical and were unpredictably distorted by being flattened for imaging. We also explored different coverslip treatments for assembly between glass surfaces. Assembly was qualitatively similar when performed between coverslips to which casein or bovine serum albumin had been adsorbed, between coverslips covalently modified with poly(ethylene glycol), and in the center between plain glass coverslips (data not shown). These observations showed that pineapples self-organized, and their assembly did not depend on surface-bound factors.
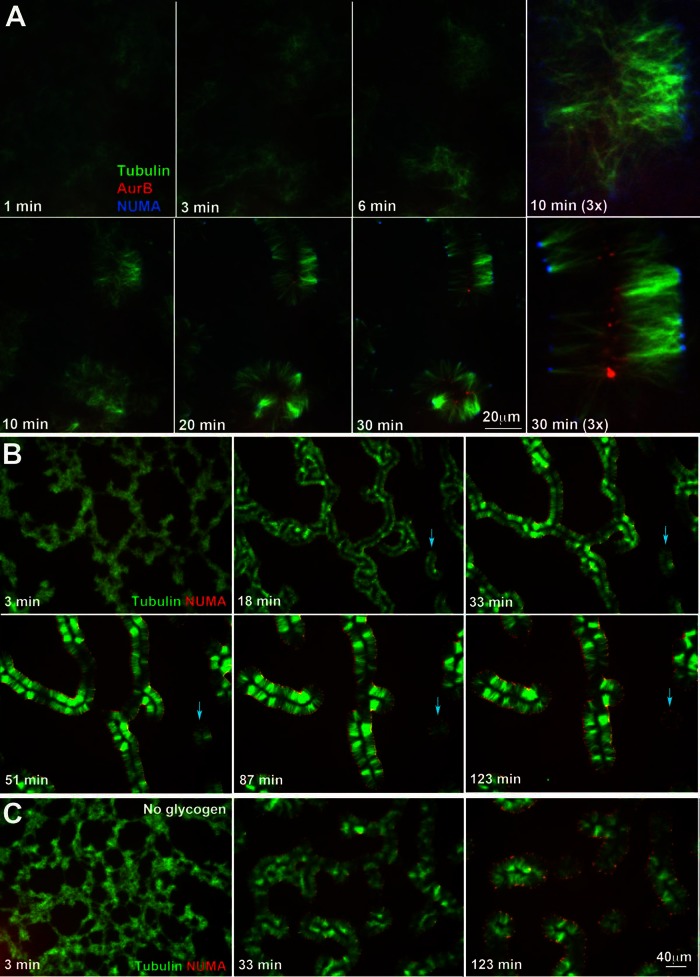
Assembly time course
Figure 2 illustrates typical time courses of assembly, which could be informally broken down into a series of events. At the earliest stages (0–2 min) microtubules polymerized in response to Taxol and DMSO and initially appeared to be distributed approximately uniformly. Between 2 and 10 min they condensed into local clusters. Between 10 and 20 min local clusters seemed to expand a little and to reorganize. Microtubule bundling became pronounced starting at ∼20 min and appeared to play a major role in final morphology. NUMA started to localize toward the outside of clusters around 10 min and Aurora B toward the inside around 20 min. The final morphology, with pronounced parallel bundling and globally aligned foci of microtubule ends that accumulated NUMA on the outside and Aurora B on the inside, was achieved at ∼30 min and was stable for ∼2–3 h.
FIGURE 2:
Time-course of pineapple assembly. (A) Early stages of assembly, followed by 40× confocal imaging. NUMA (blue) started to concentrate on the outside around 10 min (see 3× field). Aurora B (red) started to concentrate on the inside around 20 min. Tubulin (green). (B) Later stages of assembly followed by 20× wide-field imaging. Once formed, pineapple width increased over tens of minutes and individual assemblies contracted lengthwise. Smaller assemblies tended to become round and then gradually disappear (blue arrows). (C) Assembly time course in the absence of glycogen. This reaction was run in parallel to the plus-glycogen reaction shown in B. Pineapple-like structures still formed, but assembly was slower, and final structures were less organized. NUMA (red), tubulin (green).
Over tens of minutes mature pineapple width (distance between lines of NUMA and Aurora B parallel to microtubules) increased, whereas the length of individual assemblies decreased (Figure 2B). In this experiment, width increased at ∼0.15 μm/min or ∼4 tubulin dimers/s, but this rate varied between experiments, depending on the DMSO and Taxol concentrations. Smaller pineapples sometimes became round and sparse and eventually disappeared completely (blue arrow in Figure 2B). In some time courses there was a tendency for the two sides of an assembly to move apart to generate more open, circular assemblies (examples shown later). Whether pineapples formed as long, paired lines of bundles or round, open assemblies depended on the size and shape of the aggregates formed early in assembly; linear aggregates tended to evolve into pairs of lines, whereas round aggregates tended to evolve into open circles.
Glycogen was routinely added to assembly reactions at 20–30 mg/ml to approximately reconstitute the physiological concentration in eggs (Groen et al., 2011). Assembly still occurred without glycogen, but it appeared less well organized, with microtubules less bundled and more curved and NUMA foci more widely spaced (Figure 2C). Aurora B was still recruited to microtubules and became enriched away from NUMA foci, but it did not focus sharply (data not shown). Glycogen supplies ATP and reducing equivalents by a physiologically relevant mechanism that helps preserves tubulin behavior in other contexts (Groen et al., 2011), so we did not further explore glycogen-free reactions.
Pineapple length scale depends on initial polymerization conditions
Pineapple width was strikingly constant within each assembly and across a coverslip, although it increased slowly with time (Figure 2B). The source of the initial length scale appears to be the balance between microtubule nucleation and elongation early in the assembly reaction, which depended on the concentrations of Taxol and DMSO. Both Taxol and DMSO promote polymerization and stabilization of microtubules. Taxol works by a well-characterized mechanism that strongly promotes nucleation and also stabilizes assembled microtubules (Schiff et al., 1979; Hamel et al., 1981; Snyder et al., 2001). Actions of DMSO in our system are less clear. DMSO strongly promotes tubulin polymerization (Himes et al., 1976), but it probably also promoted macromolecule aggregation more generally in our system. Theoretically, DMSO could also promote methionine oxidation (Shechter, 1986; Kwak et al., 2009), a possibility we plan to explore in the future using mass spectrometry.
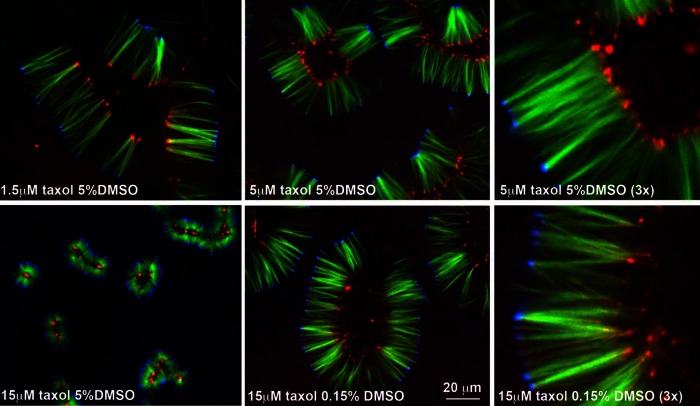
We explored a range of Taxol and DMSO concentrations and show data for a subset in Figure 3. Increasing Taxol at constant DMSO decreased pineapple width (compare 1.5–15 μM Taxol at 5% DMSO in Figure 3). We believe that higher Taxol concentrations promoted faster nucleation and thus shorter microtubules once the soluble tubulin polymerized to a steady state. At 5% DMSO and Taxol concentrations >20 μM microtubules were so short that they were difficult to visualize. At <1 μM they were so long that reorganization by motors appeared to become difficult and locally variable. Increasing DMSO at constant Taxol concentration also shortened microtubules (compare 0.15–5% DMSO at 15 μM Taxol in Figure 2), presumably also by promoting nucleation. Comparing 5 μM Taxol plus 5% DMSO to 15 μM Taxol plus 0.15% DMSO (3× views in Figure 3), we see that the widths of the assemblies are similar, but Aurora B foci are more variable and spread out in the latter condition. Because we were interested in formation and alignment of Aurora B foci, we adopted 5 μM Taxol plus 5% DMSO as our standard condition in all subsequent experiments unless otherwise stated.
FIGURE 3:
Pineapple width depends on Taxol and DMSO concentrations. Concentrations as noted. Images taken from a set of reactions run in parallel and imaged at 90 min (40× confocal). Our standard condition was 5 μM Taxol, 5% DMSO (second and third panels). Tubulin (green), NUMA (blue), Aurora B (red).
Most individual microtubules are shorter than pineapple width
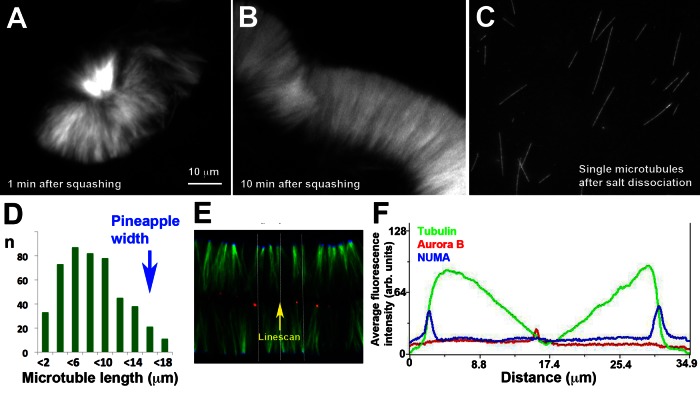
To investigate the length of individual microtubules within pineapples, we took a salt dissociation approach. We assembled pineapples in solution and then treated them with increasing salt concentrations in the presence of Taxol. They dissociated into single microtubules at ∼0.2 M NaCl and higher. Sedimentation and SDS-PAGE showed that most microtubule-binding proteins dissociated at this salt concentration (data not shown). For length measurements we dissociated pineapples by 10× dilution into a buffer containing 0.25 M NaCl and 5 μM Taxol, fixed them with a low concentration of glutaraldehyde, adsorbed single, fixed microtubules to coverslips, and imaged them. We were initially uncertain whether microtubule length would be preserved by dilution into high salt plus Taxol. To test this, we varied the interval between dissociation and fixation from 0.5 to 30 min and found no systematic length change (see Materials and Methods). Thus microtubules were stable in the dissociation buffer, and the length distribution after dissociated is likely to reflect lengths inside pineapples reasonably accurately. The main source of unreliability was measuring the width of whole pineapples assembled in solution, since they were deformed unpredictably by squashing for imaging (e.g., Figures 1E and 4, A and B).
FIGURE 4:
Microtubule length within pineapples. (A) Assembly reaction run in solution for 1 h, squashed, and imaged immediately for width measurement (40× confocal, tubulin probe only). (B) The same reaction as in A imaged 10 min after squashing, which allows time for assemblies to reorganize. (C) Single microtubules from the same reaction as in A and B after salt dissociation and fixation (60× wide-field imaging). For length comparison, A–C are presented at the same magnification. Note that the longest microtubule in C is similar in length to the widest width in B, but most microtubules in C are shorter. (D) Length histogram for dissociated microtubules from the reaction pictured in A–C. Mean pineapple width, estimated from images like those in A and B, is indicated by the blue arrow. (E) Typical pineapple image at 90 min with NUMA (blue) and Aurora B (red) probes selected for intensity profiling (40× confocal). Each color channel was linearly normalized between 0- and 256-Gy levels over the whole image. The white lines indicate a region of 100 pixels wide selected for an intensity profile parallel to the microtubule axis. (F) Fluorescence intensity profile from the line shown in E. Intensity was averaged over 100 pixels normal to the direction of the linescan to smooth out local variation. Note two NUMA peaks on the outside (blue) and an Aurora B peak on the inside (red). Average tubulin (green) intensity rose sharply just inside the two NUMA peaks and then decreased more gradually toward the center.
Figure 4 shows a comparison of an assembly reaction in solution that was split in two for imaging of whole pineapples after squashing (Figure 4, A and B) and for measurement of single microtubules after dissociation (Figure 4C). It was immediately evident that pineapples assembled in solution are built from microtubules with a broad distribution of lengths. Figure 4D shows a length histogram of the dissociated microtubules in this experiment with the estimate of pineapple width for the same reaction superimposed (blue arrow). The average width of whole assemblies was estimated as ∼15 μm (n = 70 measurements from ∼16 pineapples and fragments). The variability in this estimate was at least ∼20% on comparing between individual assemblies and fragments after squashing. The mean length of single microtubules was ∼7 μm, and the top 10% of single microtubules ranged from 13 to 17 μm (Figure 4D). Thus the average width of whole pineapples (measured after squashing) was similar to the length of the longest 10% of microtubules within them (measured after dissociation). Ninety percent of microtubules were shorter than the pineapple width. Evidently pineapples, like meiosis II spindles and cytokinesis midzones, are built from microtubules of varying lengths, most of which are smaller than the assembly as a whole. The defined width of pineapples comes from alignment of microtubule ends, not equalization of their lengths.
To help understand how mostly short microtubules are distributed within pineapples, we measured fluorescence intensity profiles of tubulin along lines parallel to the microtubule axis for pineapples assembled between coverslips. Figure 4, E and F, shows a representative example. Intensity tended to rise sharply moving inward from NUMA foci and then decrease gradually toward a minimum at Aurora B foci. Similar intensity profiles were measured from 20× wide-field images (data not shown). The nearly linear decrease in intensity with distance moving inward seen in Figure 4F was not typical; usually there was some curvature, variability, and lack of symmetry. However, a steep rise near NUMA and gradual fall toward Aurora B were characteristic. This distribution of tubulin fluorescence intensity suggests that minus ends align on the outside more than plus ends align on the inside. Thus there are certainly internal plus ends within pineapples. Whether there are also internal minus ends, or all minus ends align on the outside, is unclear.
Thin-section electron microscopy reveals midzone-like foci at plus ends
For thin-section electron microscopy (EM) analysis we fixed pineapples in perfusion chambers and flat embedded them. Figure 5 shows representative images. Figure 5, A–D, shows control, and Figure 5, E–I, shows kinesin-5–inhibited, reactions (see Figure 6 for light-level analysis of kinesin-5 inhibition). Minus- and plus-end foci were distinguished in EM images by their location in the whole assembly and characteristic morphology differences. The position of the two types of focus relative to the whole assembly was particularly clear in kinesin-5–inhibited reactions (Figure 5, E–I), where plus-end foci, marked at the light level by Aurora B accumulation (Figure 6B), were always on the outside.
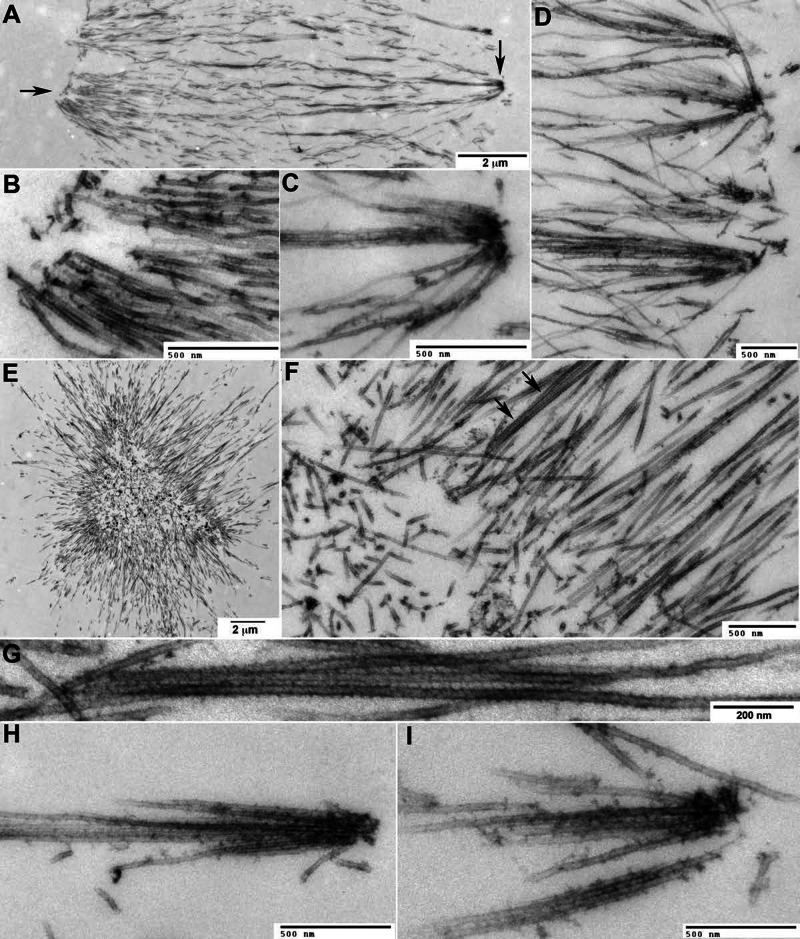
FIGURE 5:
Thin-section electron microscopy. Pineapples were assembled in perfusion chambers, fixed, stained with heavy metals, embedded, and thin sectioned. (A–D) Images from a control reaction. (E–I) Images from a reaction assembled in the presence of STLC to inhibit kinesin-5. (A) One side of a control pineapple with minus-end foci oriented to the left and plus end foci to the right. Arrows indicate regions where higher magnification views are shown in B and C. (B) Typical minus-end focus. Note the bundling of large numbers of approximately parallel microtubules. Minus-end foci lacked the electron-dense coating characteristic of plus-end foci, but we observed accumulation of small, dark puncta and filamentous material in some images. (C) Typical plus-end focus. Note fewer microtubules than in B and electron-dense coating near plus ends. Microtubules join the focus from a range of angles. (D) A line of plus-end foci from a different pineapple. Microtubules join each focus from a range of angles, mostly terminating there. A few appear to make side-on interactions with electron-dense material and bridge between foci. (E) Kinesin-5–inhibited pineapple. Note aster-like shape. From light-level images (Figure 6B) we know that plus-end foci are on the outside. (F) Inside of the pineapple shown in E. Note the dense microtubule bundles. Arrows indicate a bundle shown in higher magnification in G. (G) Bundle from the image in F rotated and enlarged. Note the periodic decoration of microtubules. (H, I) Examples of plus-end foci on the outside of kinesin-5 inhibited pineapples. Morphology and electron-dense coating near plus ends are similar to those for plus-end foci in control pineapples (C, D). One microtubule in I appears to make a side-on interaction.
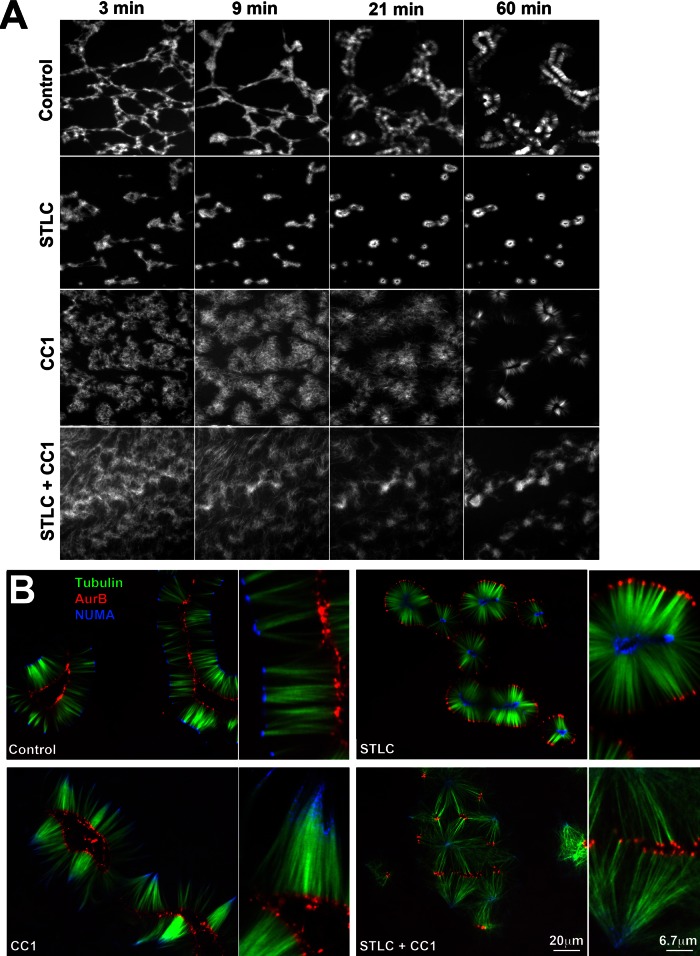
FIGURE 6:
Role of spindle assembly motors. (A) Time-course of assembly when kinesin-5 (Eg5), dynein or both motors were inhibited in parallel reactions (20× wide field; tubulin probe only shown). Kinesin-5 was inhibited with 100 μM STLC. Dynein was inhibited with the CC1 fragment of dynactin (1 mg/ml). Note that STLC alone slightly accelerated initial aggregation, whereas CC1 slowed it considerably. (B) Final assemblies at 120 min from the reactions shown in A (40× confocal). In STLC alone, NUMA (blue) foci were on the inside and Aurora B (red) foci on the inside. In CC1 alone, minus ends and NUMA were less focused. In both inhibitors clusters of Aurora B foci were the dominant organizing feature. These combined with loose NUMA aggregates to organize microtubules into quasi-hexagonal arrays in some parts of the coverslip. The second panel in each pair is a 3× view of a region in the first panel. Each panel was normalized to 8 bits in each color channel to facilitate visualization of structure. This normalization overestimates local NUMA intensity in CC1 and CC1 + STLC, where it was much weaker than control and STLC alone. Aurora B foci were of similar intensity in all conditions.
Minus-end foci formed from large numbers of densely packed, nearly parallel microtubule bundles terminating close together on the outside of conventional pineapples (Figure 5B) or the inside of kinesin-5–inhibited pineapples (Figure 5F). Small puncta of electron-dense material and fibrous material were sometimes seen near presumptive minus ends (Figure 5B). Plus-end foci formed from fewer microtubules, which joined an individual focus from a wide range of angles (Figure 5, C, D, H, and I). Most microtubules terminated in or near a plus-end focus, but some appeared to make lateral rather than end-on interactions (Figure 5, D and I). In all plus-end foci examined by EM (>50) microtubules were coated with characteristic electron-dense material (more correctly, heavy metal stain–accumulating material). This material started to coat microtubules ∼500 nm from a focus and was densest where plus ends terminated. It resembled the characteristic electron-dense material that accumulates near the center of midzones in animal cells (McIntosh and Landis, 1971; Mullins and Biesele, 1977) and at the plus ends of parallel bundles in monopolar midzones in HeLa cells (Hu et al., 2008).
EM views reinforced our impression from light-level images that microtubules are densely bundled throughout pineapples, especially near minus-end foci. Given NUMA and Aurora B localizations, these are presumably parallel bundles. Periodic striations were often visible on the walls of bundled pineapple microtubules (Figure 5G), presumably reflecting a regular periodic arrangement of microtubule-binding or -bundling factors.
Dynein and kinesin-5 play spindle-like roles in pineapple assembly
Previous work implicated these meiotic spindle motors as the main engines of microtubule organization when Taxol was added to meiotic crude extract or cytosol (Verde et al., 1991; Gaglio et al., 1996). We inhibited kinesin-5 (also known as Kif11, Eg5) using S-trityl-L-cysteine (STLC; DeBonis et al., 2004). This caused initial aggregation of microtubules into local aggregates to occur somewhat faster and more strongly than controls. This tended to generate smaller, completely disconnected initial aggregates that matured into aster-shaped or circular assemblies (Figure 6A, compare first and second rows). Remarkably, the inside–outside polarity was completely reversed in STLC-containing reactions (Figure 6B). NUMA and Aurora B still condensed into foci, but NUMA was on the inside and Aurora B on the outside in 100% of individual assemblies (n > 200 assemblies in >10 experiments). The distance between NUMA and Aurora B foci was similar between control and STLC reactions. Thus kinesin-5 activity determined inside–outside polarity, presumably by sliding antiparallel microtubules apart so as to push minus ends to the outside as it does in spindles. It did not seem to participate in aligning minus or plus ends into foci or determining pineapple width between NUMA and Aurora B foci.
Dynein was inhibited using the CC1 fragment of dynactin (King et al., 2003) added as a bacterially expressed protein or orthovanadate. Both inhibitors had similar effects; only CC1 data are shown. Inhibition of dynein strongly inhibited initial aggregation of microtubules (Figure 6A, compare first and third rows). Eventually, discrete structures formed with plus ends focused on the inside, and Aurora B accumulated there (Figure 6B). Minus ends never focused well, and microtubules were less bundled throughout the reaction. NUMA accumulation was strongly inhibited. Eventually NUMA did accumulate toward the outside (Figure 6B), presumably due to residual dynein activity, but it never focused well. Note that intensity levels are normalized to maximum intensity in each panel in Figure 6B, so the local NUMA density is strongly overestimated in CC1 compared with control.
Simultaneous inhibition of Eg5 and dynein also caused strong inhibition of initial aggregation, with microtubules appearing disorganized for 30 min or more (Figure 6A, fourth row). After 30–60 min, Aurora B foci emerged that resembled those in the control reaction, and these tended to locally cluster and align (Figure 6B). NUMA also partially focused at late time points, similar to the CC1-only reaction. The combination of Aurora B clustering and residual dynein activity sometimes organized microtubule bundles into an approximately hexagonal network of bundles (Figure 6B).
We conclude that kinesin-5 and dynein function in pineapple assembly as they do in spindles. Our data largely confirm conclusions of previous studies (Verde et al., 1991; Gaglio et al., 1996), although the definitely inside-out nature of kinesin-5–inhibited pineapples is novel and remarkable. A second novel finding is that plus ends clustered and aligned in Aurora B–containing foci even when both spindle motors were inhibited (Figure 6B). These data suggest that plus-end alignment does not depend on kinesin-5 or dynein, although it normally occurs in the context of an assembly that has been shaped by these two central spindle assembly motors.
Midzone-like properties
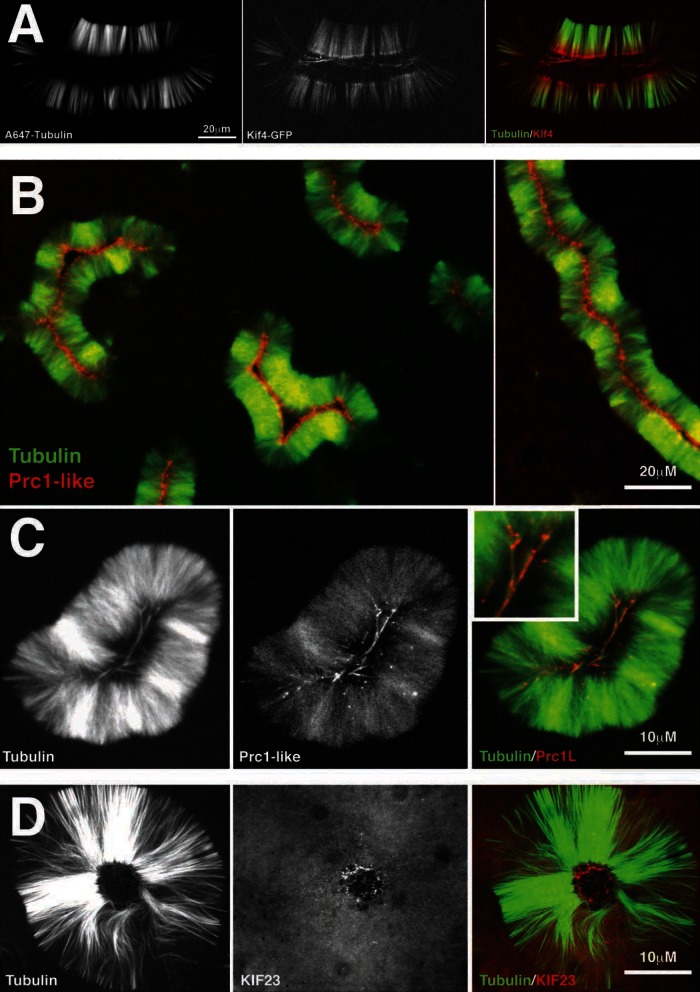
The localization of Aurora B and the EM morphology of pineapples suggested a resemblance to cytokinesis midzones. To explore this, we localized additional midzone proteins (Figure 7). We found that the conserved midzone proteins Kif4, PRC1L (the embryo isoform of PRC1 in our system), and Kif23 all localized at or near plus ends. Additional midzone proteins, including Racgap1 (the partner of Kif23 in centralspindlin), Kif20A, and RCC2, were present in pineapples by proteomics (data not shown), but we do not yet have imaging reagents for them. Kif4 and PRC1L were both localized by adding low, nonperturbing concentration of purified, green fluorescent protein (GFP)–tagged proteins. PRC1L localization was similar when imaged using directly labeled antibody raised to the C-terminus (data not shown). Both localized faintly throughout the pineapple structure and were concentrated in the center, near plus ends (Figure 7, A and B). At higher magnification (Figure 7C), PRC1L strongly localized to a few microtubule bundles in the center. We suspect that these may be antiparallel bundles that selectively recruit the antiparallel cross-linker. Kif23, part of centralspindlin, was localized using a directly labeled peptide antibody only, so the data are less reliable. It was concentrated in distinct foci near the centers of the pineapple and not precisely on microtubule plus ends (Figure 7D). These may be equivalent to the electron-dense aggregates observed by EM (Figure 5).
FIGURE 7:
Cytokinesis midzone markers localize to pineapples. (A) Kif4 localization. A low, nonperturbing concentration of a functional Xenopus Kif4-GFP fusion (Bieling et al. 2010) was added to a pineapple assembly reaction. Shown is typical localization at 60 min (40× confocal). Kif4-GFP (red) was recruited to microtubules and accumulated at plus ends. (B) PRC1-like localization using a functional GFP fusion (see Materials and Methods). Shown is typical localization at 60 min (20× wide field). Note that PRC1L (red) localizes on the inside of the microtubule (green) structure. (C) Higher-magnification image of PRC1L localization. Shown is typical localization at 60 min (40× confocal). Note the microtubules (green) in the center of the pineapple staining for PRC1L (red). Inset, ∼2× of the colored image. (C) KIF23 (red) localization using labeled antibody raised to a C terminal peptide. Shown is typical localization at 60 min (40× confocal).
Aurora B and Kif4 are required for plus-end alignment
The localizations of Kif4, PRC1L, Aurora B, and KIF23 suggested that midzone assembly pathways might play a role in pineapple assembly. To test this, we depleted or inhibited three midzone assembly factors, Aurora B, Kif4, and PRC1L. Immunodepletion of PRC1L (>90%) had no obvious effect (data not shown). Immunodepletion of >90% of Aurora B and Kif4 was readily achieved with antibody-coated magnetic beads (Figure 8A), and functional Kif4-GFP was used for addbacks. An advantage of working with HSS compared with crude extract is that immunodepleted HSS could be refrozen without loss of activity (data not shown).
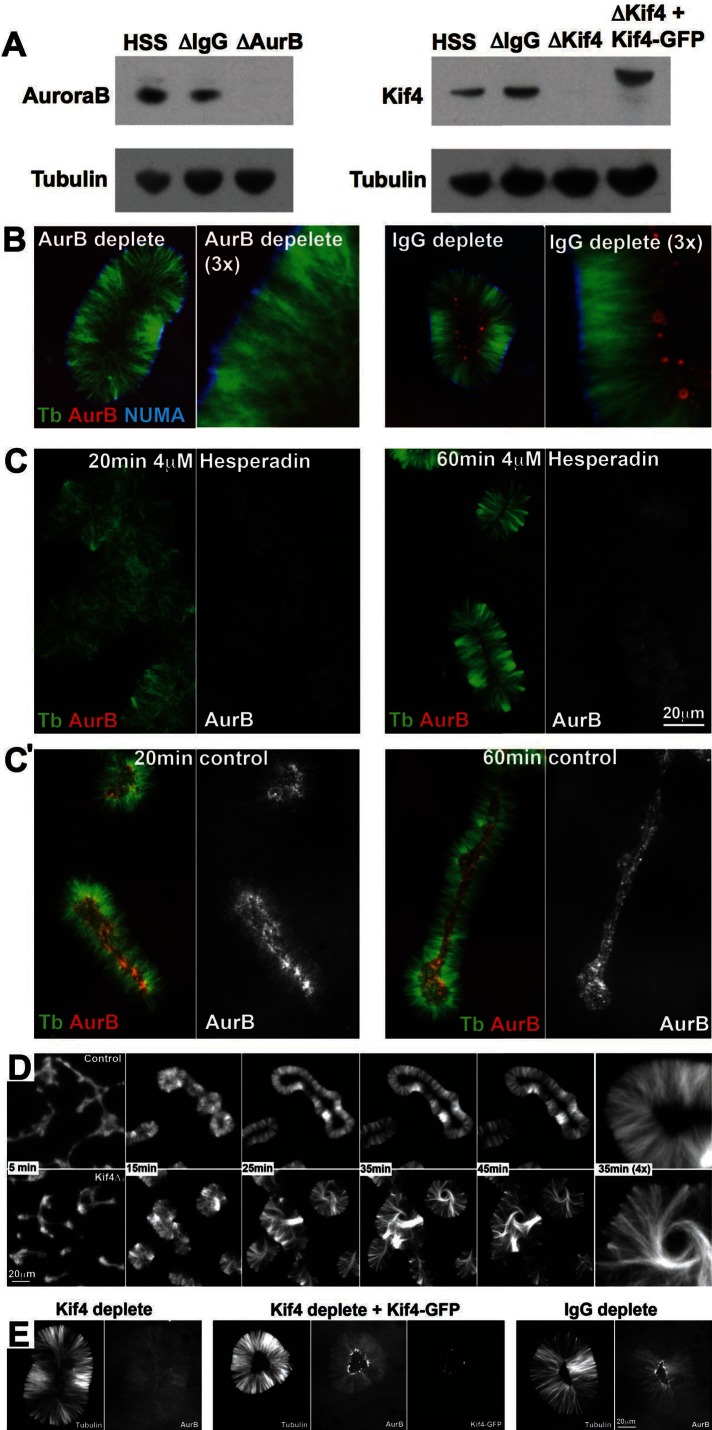
FIGURE 8:
Aurora B and Kif4 are required for plus-end alignment. (A) Aurora B and Kif4 depletion. HSS was depleted with anti-Aurora B, anti-Kif4, or control IgG and analyzed by immunoblotting. Depletion was >90% in both cases. In some experiments Kif4-GFP was added back to approximately endogenous levels. (B) Aurora B (red)–depleted compared with control IgG-depleted pineapples at 60 min (40× confocal). Note that plus-end alignment is lost after Aurora B depletion, and the antibody probe to Aurora B (red) did not localize. NUMA (blue) localization was not affected by Aurora B depletion. (C) Aurora B inhibition. Aurora B activity was inhibited with 4 μM hesperadin in this experiment. At 20 min hesperadin-treated reactions were poorly organized compared with control. At 60 min minus ends always aligned and recruited NUMA. Plus ends aligned in some assemblies as shown, although others resembled Aurora B (red)–depleted assemblies. Hesperadin inhibited, but did not completely block, recruitment of the Aurora B probe to microtubules (compare black and white panels in B and C, which were scaled the same). Plus-end accumulation of the probe was strongly inhibited. Similar results were obtained at 1.4 and 16 μM Hesperadin and with other small-molecule inhibitors (see text). (C′) Control reaction for C run in parallel with no drug. (D) Kif4 depletion. HSS was depleted with anti-Kif4 or control IgG, and pineapple assembly was run in parallel (20×, wide field, tubulin probe only). Depletion of Kif4 (bottom) had no effect on initial aggregation or minus-end alignment. Plus ends in the center appeared to grow longer than control and failed to align, resulting in assemblies that did not hollow out. Microtubules accumulated in whorls in the center in Kif4-depleted pineapples (35 min, 4× panel), suggesting disregulated plus-end growth. (E) Kif4-GFP addback (40× confocal, 100-min time point). Kif4 depletion (left) blocked plus-end focusing and alignment as in B. Aurora B was still recruited to microtubules but did not accumulate in obvious foci, probably because plus ends were distributed. Addback of Kif4-GFP (middle) rescued plus-end alignment and Aurora B accumulation and accumulated at plus ends as in A. Depletion with random IgG (right) had no effect.
In Aurora B–depleted HSS initial formation and aggregation of microtubules was similar to that in immunoglobulin (IgG)-depleted control. However, plus-end clustering and alignment were disrupted, and pineapples did not hollow out in the center as in control (Figure 8B). This was true for 100% of assemblies (>100 assemblies in two experiments). There was no localization of the Aurora B antibody probe, as expected. Thus Aurora B is required for plus-end clustering and alignment in pineapples as in bipolar and monopolar midzones in cells (Ruchaud et al., 2007; Hu et al., 2008).
Aurora B kinase activity was inhibited using three small molecules, all of which compete at the ATP-binding active site: hesperadin (Hauf et al., 2003), ZM447439 (Ditchfield et al., 2003), and VX680 (Harrington et al., 2004). We obtained similar effects with each of these compounds, albeit with varying EC50 values, and show data only for hesperadin. Hesperadin was the most potent, with an EC50 in our system of ∼0.5 μM, compared with ∼10 μM for ZM447439 and ∼50 μM for VX680 (rough estimates based on inhibition of Aurora B localization). These are much higher than reported EC50 values in tissue culture cells, which is often the case for drugs in egg extract experiments, presumably because much of the added drug binds nonspecifically to protein or lipid. Hesperadin blocked Aurora B accumulation at foci on the inside, although we still observed a low level of diffuse recruitment (Figure 7C). Plus-end alignment on the inside was inhibited but not completely blocked. At times when plus ends were already well aligned in control (20 min in Figure 7B), Aurora B–inhibited reactions were poorly organized, and plus ends were not aligned. Later, in a subset of assemblies, relatively normal pineapples formed with aligned plus ends (60 min; Figure 8C). Alignment was observed even at high drug concentrations (16 μM hesperadin, ∼30 times the EC50), although it was not robust. At all hesperadin concentrations >1 μM we observed a mixture of well-aligned and poorly aligned assemblies, and the fraction varied between runs of the experiment. Thus kinase activity of Aurora B is required for Aurora B accumulation at plus ends, and it plays a role in pineapple assembly and plus-end alignment, although it may not be completely required. It is possible that residual kinase activity even at high drug concentration is sufficient to align ends, but we doubt this, since we saw no dose dependence for inhibition of plus-end alignment at late times. Instead, we believe that physically removing the CPC has a stronger effect on plus-end alignment than inhibiting its kinase activity, suggesting that it plays a structural role in addition to acting as a kinase. Alternatively, Aurora B depletion may have codepleted a different essential factor. Distinguishing these possibilities will require pure, functional CPC for addback experiments.
Kif4 depletion caused an effect on plus-end alignment that was reminiscent of Aurora B depletion but more severe. At the time when control pineapples started to align plus ends and hollow out, Kif4-depleted assemblies started to exhibit increased microtubule density in the center (Figure 8D). The excess microtubules often organized into whorls in the center (Figure 8D, 3× panel). Plus-end alignment was strongly perturbed in 100% of Kif4-depleted assemblies (n = >100 in four experiments with three batches of HSS). Increased pineapple width and presence of central whorls suggested that plus-end growth was disregulated in Kif4-depleted pineapples, as was the case in midzones in Kif4-depleted HeLa cells (Hu et al., 2011). Adding back Kif4-GFP to near endogenous levels (see blot in Figure 8A) restored plus-end alignment (Figure 8E, middle), indicating that no other essential factors had been codepleted.
DISCUSSION
Our observations on self-organization of Taxol-stabilized microtubules in meiotic cytosol confirm, and significantly extend, those made by the Karsenti and Compton groups (Verde et al., 1991; Gaglio et al., 1995, 1996; Chakravarty et al. 2004). We confirm a central role of dynein, and presumably NUMA, in aggregating microtubules and clustering minus ends. Under our conditions microtubules did not organize into simple asters with minus ends at the center, but instead formed paired or rounded assemblies of parallel bundles with minus ends at the outside. Kinesin-5 activity was responsible for pushing minus ends to the outside (Figure 6B), presumably using its antiparallel sliding activity. In this sense pineapples modeled bipolar spindles better than the simple asters seen in previous studies.
Pineapples assemble by both spindle and midzone mechanisms
Figure 9 illustrates our thinking on how pineapples assemble using both meiotic spindle and cytokinesis midzone pathways. Dynein clusters minus ends, whereas kinesin-5 pushes antiparallel microtubules apart and thus sorts microtubules into bundles of uniform polarity. Both mechanisms are believed to be central to meiosis II spindle assembly (Burbank et al., 2007). We propose that a combination of minus end–directed sliding and lateral bundling causes the dynein–NUMA aggregates to self-organize into a coherent plane on the outside of pineapples. Midzone factors are recruited to pineapple plus ends by kinesin-dependent transport, and Kif4 then acts to restrain plus-end polymerization. Loss of Kif4 caused overgrowth of plus ends in pineapples (Figure 8), as it does in midzones in cells (Hu et al., 2011). We propose that midzone assembly factors aggregate laterally (zigzag line in Figure 9). When combined with plus end–directed transport and plus end–capping activity, this lateral aggregation serves to self-organize midzone factors into a coherent plane. The plane of midzone factors contains only the subset of plus ends that have grow to meet it, whereas the plane of spindle pole factors contains many or all the minus ends in the system, since they are actively concentrated there by dynein's sliding activity. These two planes, and the parallel microtubules that run between them, define the organization of pineapples.
FIGURE 9:
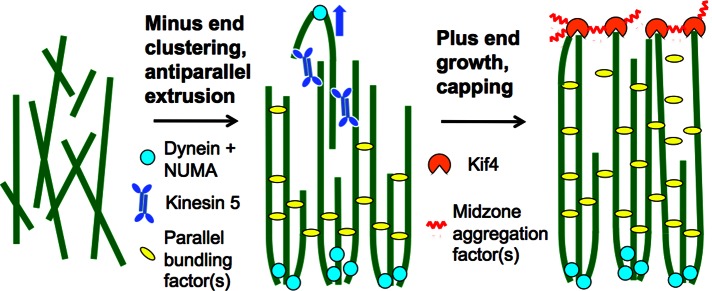
Model for pineapple assembly. Randomly oriented, Taxol-stabilized microtubules (left) recruit the spindle assembly factors dynein, NUMA, and kinesin-5. Dynein and NUMA cluster minus ends, whereas kinesin-5 slides antiparallel microtubules apart (middle, blue arrow). Together with parallel bundling factors, these activities cause the microtubules to sort into two parallel arrays with minus ends clustered on the outside (middle). Only the lower array is shown in the right for clarity. Plus ends recruit midzone assembly factors, including the CPC and Kif4. Plus ends tend to grow, but Kif4 restrains this growth. Lateral aggregation of midzone factors (purple zigzags) causes them to self-organize into a single plane (right). All these processes probably occur simultaneously and are only separated for clarity.
It was puzzling to see midzone assembly processes in cytosol with high Cdk1 activity, since midzones normally assemble in cells only when the Cdk1 level drops after anaphase. However, low Cdk1 activity is not necessary for midzone assembly in cells, since inhibition of Plk1 triggers premature midzone assembly in HeLa cells while Cdk1 activity is high (Hu et al., 2012). Our data suggest that high Cdk1 activity does not prevent the CPC and Kif4 from performing their midzone functions. We suspect that the high concentration of stable microtubules in our system may recruit and activate the CPC, and this triggers midzone assembly processes despite high Cdk1 activity.
Role of parallel bundling
Bundling interactions between presumably parallel microtubules was a dominant feature of pineapple morphology at the light microscopy and EM levels. At the EM level we often observed periodic decoration of bundled microtubules (Figure 5G). This periodicity suggests that a specific bundling factor, or set of interacting factors, might predominate. We suspect that binding energy from interactions between microtubules and bundling factors is an important driver of pineapple morphogenesis. A thermodynamic drive to increase bundling might be responsible for the slow increase of pineapple width, decrease in length, and loss of small pineapples that we observed as ongoing dynamics after assembly was complete (Figure 2B). This slow dynamics may reflect “Ostwald ripening,” a common process in physical chemistry, in which large aggregates grow at the expense of smaller ones due to higher energy of surface relative to internal subunits. Parallel bundling interactions could lower the free energy of internal parts of the microtubule lattice. A drive toward parallel bundling could explain why small, highly curved pineapples were unstable over long times (Figure 1A). The dominant role of parallel bundling in pineapple assembly suggests that it may also play important roles in spindle and midzone assembly in cells. Attention of the midzone field has tended to focus on antiparallel bundling, but tight, parallel bundles are observed in normal midzones and midbodies in regions that flank the central overlap (McIntosh and Landis, 1971; Mullins and Biesele, 1977; Mastronarde et al., 1993), and in monopolar midzones (Canman et al., 2003; Hu et al., 2008). Parallel bundling deserves more attention as a candidate organizing mechanism in spindles and midzones.
Toward biochemistry
A major advantage of HSS and C-HSS is their biochemical tractability compared with crude egg extracts that contain organelles. They can be frozen and thawed without loss of activity and loaded onto columns, and microtubules can easily be sedimented for biochemical analysis of binding proteins (Mack and Compton, 2001). In this initial article we only made use of the convenience of freezing. In preliminary experiments we have begun to characterize the pineapple proteome by mass spectrometry. Simply spinning out pineapples and washing them yielded a fairly clean list of microtubule-binding proteins that were characteristic of both spindles and midzones (Wühr, Gigy, Groen, and Mitchison, unpublished results). We will report the microtubule-bound proteome in pineapples, and how it changes as a function of perturbations that alter morphology, in a future article.
MATERIALS AND METHODS
Materials
Chemicals were from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Kinase inhibitors were from Selleck (Houston, TX). Commercial glycogen from oysters was purified by dissolving in water, precipitating with two volumes of ethanol, washing with ethanol, and drying. It was dissolved in water to a final concentration of 25–30% (wt/vol) and stored frozen.
Extract preparation
Meiotic HSS containing cytochalasin D to prevent actin polymerization and cyclinB-Δ90 to stabilize Cdk1 activity was prepared from unfertilized Xenopus eggs as described (Groen et al., 2011). HSS is free of ribosomes, glycogen, and most organelles. C-HSS was prepared by 10-fold dilution of HSS, recentrifugation, and concentration back to the original volume with a membrane filter and is completely free of organelles. Glycogen was added back to physiological concentrations (20–30 mg/ml) after thawing. This resulted in ATP/ADP ratios and Cdk1 activities that were high and stable for at least 2 h at 20°C (Groen et al., 2011), as well as supply of reducing equivalents that may be important to prevent protein oxidation (Jackson et al., 1983). Meiotic HSS stabilized with cyclin-B Δ90 and C-HSS were prepared from unfertilized Xenopus eggs as described (Groen et al., 2011) and stored as frozen aliquots at −80°C. It was important to use a phosphate-containing buffer for washing the eggs and for diluting HSS to make C-HSS, as previously reported. The meiotic state of the extract was evaluated by immunoblotting with MPM2 (Millipore, Billerica, CA), an antibody that recognizes Cdk1 epitopes, and did not change over 2 h at 20°C.
Probes and inhibitors
Tubulin was labeled with Alexa Fluor NHS esters (Invitrogen, Carlsbad, CA) as described (Hyman et al., 1991). All antibodies were affinity-purified rabbit polyclonals. Only nonperturbing antibodies were used as probes in functional experiments (all except Figure 6), as determined by tubulin imaging with and without probe. Antibodies used for preliminary localization in Figure 6 were not characterized with respect to functional perturbation. Anti-NUMA was raised to a C-terminal peptide (Mitchison et al., 2005). Anti–Aurora B and anti–Kif4 were raised to fusion proteins. Antibody to Xenopus KIF23 and the PRC1L (the embryo isoform of PRC1; Klein et al., 2002) were raised to C-terminal peptides (GenScript, Piscataway, NJ). All antibodies were raised in rabbits and affinity purified. Antibodies were labeled with Alexa Fluor NHS esters as recommended by the manufacturer, both in solution and while attached to protein A beads. The bacterial expression construct for HIS-GFP-PRC1L was subcloned from synthesized codon optimized (for bacterial expression) cDNA for Xenopus PRC1L (GenScript). PRC1L was purified following conventional methods. It was functional, as assayed by depletion-addback experiments in a new midzone assemble system in egg extracts (Mitchison et al., 2012; and data not shown). Taxol was dissolved in DMSO and stored frozen. The CC1 fragment of dynactin was expressed in bacteria and purified as described (King et al., 2003). It was added to a final concentration of 1 mg/ml to inhibit dynein. Sodium orthovanadate was added at 100–200 μM and produced similar results. Xenopus Kif4-GFP (previously called Xklp1-GFP) was prepared and characterized as reported (Bieling et al., 2010).
Coverslips
Casein-coated coverslips we prepared by taking #1.5 coverslips (various vendors) directly from the box and incubating them on one side with 0.4% casein (sodium salt) for at least 10 min at 20°C. They were washed with water, dried by aspiration, and used within a few minutes. A 22-mm-square coverslip was affixed to a metal slide over a 20-mm round hole with melted VALAP (1:1:1 mixture of beeswax, paraffin wax, and lanolin). An 18-mm-square coverslip was used to cover the reaction. The squash was sealed with VALAP for oil immersion imaging; sealing was not necessary for dry imaging.
Typical assembly reaction
We mixed 9.5 μl of HSS supplemented with glycogen and fluorescent probes with 0.5 μl of DMSO containing 100 μM Taxol. Of this mix 5 μl was immediately squashed between casein coated coverslips for a nominal reaction thickness of ∼15 μm. The air temperature was maintained at 20 ± 2°C during assembly and imaging. Reactions were imaged using Nikon microscopes (Nikon, Melville, NY) equipped for wide-field fluorescence with a 20×/0.8 numerical aperture (NA) air objective or confocal imaging with a 40×/1.3 NA or 100×/1.4 NA oil objectives and a spinning disk head (Yokogawa, Tokyo, Japan). Up to four reactions were followed in parallel using a metal slide with four holes and a microscope with an automated xyz stage (dry lens only). Assembly in solution was performed the same way but without spreading the sample. Drugs were added to either the DMSO or the HSS before final mixing, depending on their solubility. Immunodepletions were performed using IgG (20 μg per 50 μl of bead slurry) bound to 2.2-μm magnetic beads coated with protein A (Dynabeads; Invitrogen). These were incubated with HSS for 30 min at 4°C and then removed with a magnet, twice in succession.
Dissociation protocol
Pineapples were assembled in solution for 1 h under standard conditions (5 μM Taxol, 5% DMSO) unless otherwise noted. One aliquot was diluted in 80 mM BRB80 (80 mM K 1,4-piperazinediethanesulfonic acid 6.8, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA]) plus 10 μM Taxol and squashed for whole-assembly imaging; another was removed for dissociation. For dissociation 5 μl of the reaction was added to 50 μl of 250 mM NaCl, 10 mM K 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.7, 1 mM MgCl2, 1 mM EGTA, and 20 μM Taxol and mixed gently. After dissociation for 0.5–30 min (2 min was standard) the resulting single microtubules and a few remaining bundles were fixed by addition of 100 μl of 0.1% glutaraldehyde in BRB80 + 60% (vol/vol) glycerol. Five minutes later the reaction was diluted with 1 ml of BRB80 + 60% (vol/vol) glycerol without glutaraldehyde and mixed, and 2 μl was spread between a slide and a 22-mm2 coverslip (both uncoated). Microtubules were allowed to adhere to the coverslip for at least 30 min before imaging by wide-field fluorescence with a 60×/1.4 NA objective. Microtubule lengths were measured with MetaMorph software (Molecular Devices, Sunnyvale, CA). In one experiment we measured mean microtubule length at different time intervals between dissociation and fixation. Mean lengths at 0.5, 1.5, 4, 12, and 30 min were 7.3, 6.8, 7.0, 6.7, and 7.2 μm, respectively. SD values were 5–6 μm (n > 250 for each time point). We concluded that microtubule length is stable for 30 min in 0.25 M NaCl buffer plus Taxol and used 2 min as our standard dissociation time before fixation.
Electron microscopy
Pineapples were assembled in a flow chamber in which one surface was a square of Acklar that had been glow discharged, soaked in polylysine, rinsed, and dried (Coughlin et al., 2007) and the other surface was a glass coverslip covalently coated with poly(ethylene glycol). Pineapples adhered only to the polylysine-coated surface. After 1–2 h of assembly, the chamber was perfused once with BRB80 + 10 μM Taxol for ∼30 s, then perfused with fixative (1.5% glutaraldehyde + 30 mM lysine in BRB80, mixed immediately before use). Assemblies were fixed for 10–15 min in the glutaraldehyde/lysine mixture and then for 30 min to 1 h in 0.5% glutaraldehyde alone in BRB80. Postfixation with OsO4, block staining with uranyl acetate, and embedding to generate a thin, transparent block were performed as described (Coughlin et al., 2007). Pineapples were identified in the block by phase contrast microscopy, trimmed out, remounted, thin sectioned, and grid stained using standard procedures.
Acknowledgments
This project was initiated as part of the Marine Biological Laboratory physiology course in 2008, and we thank the students who participated. Our work was funded primarily by National Institutes of Health Grant GM23928. We benefited from fellowships from the Marine Biological Laboratory, where several of the authors worked as Whitman investigators in 2009–2012. We thank Nikon, Inc., for outstanding microscopy support at the Marine Biological Laboratory. We thank Ryoma (Puck) Ohi (Vanderbilt University, Nashville, TN) and Todd Stuckenberg (University of Virginia at Charlottesville, Charlottesville, VA) for generously providing antibodies. We thank present and former members of the Mitchison and Salmon groups (University of North Carolina at Chapel Hill, Chapel Hill, NC) for support and lively discussion.
Abbreviations used:
- C-HSS
clarified high-speed supernatant
- HSS
high-speed supernatant
- STLC
S-trityl-l-cysteine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0850) on March 20, 2013.
REFERENCES
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- Chakravarty A, Howard L, Compton DA. A mechanistic model for the organization of microtubule asters by motor and non-motor proteins in a mammalian meiotic extract. Mol Biol Cell. 2004;15:2116–2132. doi: 10.1091/mbc.E03-08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin M, Brieher WM, Ohi R. Cell-free extract systems and the cytoskeleton: preparation of biochemical experiments for transmission electron microscopy. Methods Mol Biol. 2007;369:199–212. doi: 10.1007/978-1-59745-294-6_10. [DOI] [PubMed] [Google Scholar]
- DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. In vitro screening for inhibitors of the human meiotic kinesin Eg5 with antimeiotic and antitumor activities. Mol Cancer Ther. 2004;3:1079–1090. [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study meiotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian meiotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like meiotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Coughlin M, Mitchison TJ. Microtubule assembly in meiotic extract requires glycogen. Mol Biol Cell. 2011;22:3139–3151. doi: 10.1091/mbc.E11-02-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel E, del Campo AA, Lowe MC, Lin CM. Interactions of Taxol, microtubule-associated proteins, and guanine nucleotides in tubulin polymerization. J Biol Chem. 1981;256:11887–11894. [PubMed] [Google Scholar]
- Harrington EA, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes RH, Burton PR, Kersey RN, Pierson GB. Brain tubulin polymerization in the absence of “microtubule-associated proteins.”. Proc Natl Acad Sci USA. 1976;73:4397–4399. doi: 10.1073/pnas.73.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. Kif4 regulates midzone length during cytokinesis. Curr Biol. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CK, Ozlü N, Coughlin M, Steen JJ, Mitchison TJ. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol Biol Cell. 2012;23:2702–2711. doi: 10.1091/mbc.E12-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Campbell EA, Herbert P, Hunt T. The preparation and properties of gel-filtered rabbit-reticulocyte lysate protein-synthesis systems. Eur J Biochem. 1983;131:289–301. doi: 10.1111/j.1432-1033.1983.tb07262.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: the NIH Xenopus Initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Kwak GH, Choi SH, Kim JR, Kim HY. Inhibition of methionine sulfoxide reduction by dimethyl sulfoxide. BMB Rep. 2009;42:580–585. doi: 10.5483/bmbrep.2009.42.9.580. [DOI] [PubMed] [Google Scholar]
- Lee KY, Davies T, Mishima M. Cytokinesis microtubule organisers at a glance. J Cell Sci. 2012;125:3495–3500. doi: 10.1242/jcs.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack GJ, Compton DA. Analysis of meiotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc Natl Acad Sci USA. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of meiotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Landis SC. The distribution of spindle microtubules during mitosis in cultured human cells. J Cell Biol. 1971;49:468–497. doi: 10.1083/jcb.49.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Wuhr M, Nguyen P, Ishihara K, Groen A, Field CM. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken) 2012;69:738–750. doi: 10.1002/cm.21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Shechter Y. Selective oxidation and reduction of methionine residues in peptides and proteins by oxygen exchange between sulfoxide and sulfide. J Biol Chem. 1986;261:66–70. [PubMed] [Google Scholar]
- Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by Taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- Shrestha S, Wilmeth LJ, Eyer J, Shuster CB. PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol Biol Cell. 2012;23:1196–1207. doi: 10.1091/mbc.E11-12-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JP, Nettles JH, Cornett B, Downing KH, Nogales E. The binding conformation of Taxol in beta-tubulin: a model based on electron crystallographic density. Proc Natl Acad Sci USA. 2001;98:5312–5316. doi: 10.1073/pnas.051309398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in meiotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]