Abstract
Sleep disorders are common in patients with neurogenerative diseases and manifest early in the disease process. Among a number of possible mechanisms underlying the sleep disturbances, there is evidence that dysfunction in the circadian system is a contributing factor. Focusing on a mouse model of Huntington’s disease has enabled us to determine that at the onset of symptoms, spontaneous electrical activity of neurons within the central clock is disrupted even though the molecular clockwork is still functional. These findings suggest that the fundamental deficit contributing to disordered sleep is reduced SCN output. The mechanism underlying this deficit is not yet known, but mitochondrial dysfunction and oxidative stress are likely involved. Disruption of circadian output from the SCN would be expected to have wide ranging impact on the body including SCN regulated brain regions and the heart. In fact, there is a great deal of overlap in the non-motor symptoms experienced by HD patients and the consequences of circadian disruption. This raises the possibility that the disordered sleep and circadian function experienced by HD patients may be an integral part of the disease. Furthermore, we speculate that circadian dysfunction may accelerate the pathology underlying HD. If these hypotheses are correct, we should focus on treating circadian misalignment and sleep disruptions early in disease progression.
Keywords: Circadian, Huntington’s disease, Non-motor symptoms of Huntington’s disease, Sleep, Suprachiasmatic nucleus
Introduction
Sleep disorders are common in Europe and the USA with an estimated 30 to 40% of the adult population reporting difficulty falling asleep at night and significant daytime sleepiness as a consequence1,2. The National Health Interview Survey has found that an average night’s sleep is 6.5 hrs with the trend shortening for working adults3. These data suggest that many of us are all too familiar with the symptoms of sleep deprivation, including feelings of fatigue, irritability, and difficulty concentrating4. There is also a growing awareness that sleep deprivation is associated with metabolic imbalances and compromised immune response5-7. Fortunately, these symptoms quickly recover with the restorative powers of a good night’s sleep. Unfortunately, for many patients with chronic diseases of the nervous system, a good night’s sleep is very hard to obtain8. There is increasing evidence that in neurodegenerative disorders such as Alzheimer’s, Huntington’s and Parkinson’s diseases, sleep disruptions are common and occur early in the disease progression9-11. In some cases, patients will experience years of sleep disturbances before the onset of dementia or motor symptoms that characterize the specific disease12,13. These sleep disturbances have significant negative consequences for both the patients and their caregivers. While there are a number of possible mechanisms underlying the sleep disturbances, we propose that dysfunction in the circadian system is a contributing factor.
Molecular clockwork common in cells throughout the body
Output from the master circadian clock regulates physiological rhythms through modifications of the phase and amplitude of “clock gene” rhythms measurable in many tissues. Most cells express robust rhythms in the transcription and translation of key clock genes through an autoregulatory negative feedback loop, which then synchronize with other functionally relevant cells. Over the past two decades, circadian rhythm researchers have identified the basic mechanism underlying the cell-autonomous molecular clockwork that generates and regulates circadian rhythms in gene expression14,15. In short, CLOCK and BMAL1 are basic helix-loop-helix PAS-containing transcription factors that heterodimerize and enhance transcription of Period genes (Per1-3)16-19 and Cryptochrome genes (Cry1-2)20 through E-box elements21-23. Once translated, PER and CRY proteins form complexes that accumulate in the cytoplasm and are later translocated to the nucleus recruiting HDAC complex to repress BMAL/CLOCK-mediated transcription20,24-26. To relieve this repression, CRYs are targeted for proteasomal degradation by an ubiquitin process which allows a new cycle to begin25,27,28. The timing and stability of the core negative feedback loop is regulated by the phosphorylation state of clock proteins directing the protein for degradation or nuclear translocation29-31, and additional stabilizing transcriptional loops regulate Bmal132,33 and other clock genes34,35 through nuclear receptors from the REV-ERB and ROR families. Taken together, the network that drives circadian oscillations in gene expression output is a highly regulated, robust, and resilient timekeeper.
Circadian regulation of gene expression varies with tissue
Each of the major organ systems of the body is made up of a network of circadian oscillators, with each of the major organ systems (ie. brain, heart, liver, pancreas, etc) apparently having its own clockwork to regulate the transcription of genes important to the specific target organ14. Using DNA microarray expression profiling it has been found that approximately 8 to 12% of genes display circadian oscillations36-38. Many of these genes are involved in key rate-limiting steps of biochemical pathways36,39. In the nervous system, a number of genes involved in peptide synthesis, secretion, and oxidative phosphorylation are transcribed with a circadian oscillation likely driven by the molecular clockwork present in each of these organs. For example, the genetic overexpression of REV-ERB alpha specifically in the liver shuts down the molecular clock locally and results in a loss of 90% of the transcriptional rhythms measured in mouse hepatocytes40. These data indicate the predominant driver of gene expression rhythms in the liver is the molecular clockwork within hepatocytes, even though some liver rhythms are imposed by external signals such as cortisol41. In some cases, rhythms in mRNA expression do not lead to rhythms in protein levels42,43 and it is therefore important to confirm gene expression rhythms with protein measurements.
SCN output coordinates an array of rhythmic tissues
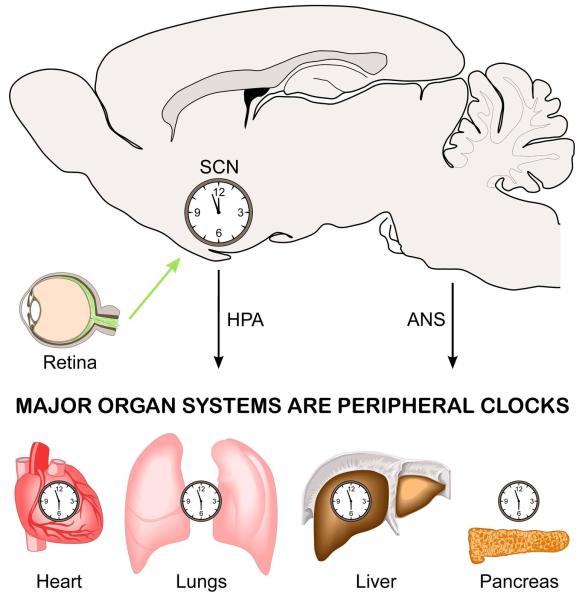
In mammals, the suprachiasmatic nucleus (SCN) of the hypothalamus contains the “master” oscillatory network necessary for coordinating circadian rhythms throughout the body15,44(Fig.1) The SCN is a bilaterally paired nucleus made up of tightly compacted, small-diameter neurons just lateral to the third ventricle atop the optic chiasm45. Anatomical studies generally support the division of the SCN into at least two subdivisions including a ventral (core) and dorsal (shell) region46,47. Ventral neurons are thought integrate environmental irradiance information and from three major input pathways: the retinohypothalamic tract (RHT), the geniculohypothalamic tract from the intergeniculate leaflet of the thalamus, and from the raphe nuclei48. These input processing neurons then communicate environmental information to dorsal SCN neurons. Notably, ventral neurons exhibit relatively low amplitude rhythms in clock gene expression and the neuropeptides vasoactive intestinal peptide (VIP) or gastrin-releasing peptide (GRP) in addition to the neurotransmitter GABA. In contrast, neurons of the dorsal shell appear to generate robust circadian oscillations in gene expression49-51, and express vasopressin (AVP) or prokineticin 2 (PK2) in addition to GABA.
Fig. 1.
Schematic illustrating key components of the circadian system. The circadian system consists of a network of circadian oscillators. The central clock is located in the suprachiasmatic nucleus (SCN). Neurons in this cell population receive light information from melanopsin-expressing retinal ganglion cells found in the retina. The axons of these ganglion cells make a direct synaptic connection onto cells in the SCN. These SCN neurons integrate this photic information with other timing cues to generate robust circadian oscillations that are synchronized to the environment. Signals from the SCN travel out via the hypothalamic-pituitary-adrenal (HPA) axis, as well as through the autonomic nervous system (ANS), to coordinate and regulate the independent circadian oscillations found throughout the body.
The fact that many core neuronal projections terminate on shell neurons supports the idea that the interplay between these two regions is responsible for coordinating output of circadian information from the SCN52. SCN outputs from both core and shell subpopulations largely project to other hypothalamic regions, including the subparaventricular zone and other medial hypothalamic structures surrounding the SCN53-55. These hypothalamic relay nuclei send projections throughout the nervous and endocrine systems, providing multiple pathways by which the SCN output conveys temporal information about the environment to the rest of the brain and body14,55. Additionally, output from physiological systems receiving circadian regulation can “feeds-back” to alter the master clock. For example, both sleep states56 and locomotor activity57-59 “feedback” to regulate SCN neural activity recorded in vivo. These physiological studies demonstrate the difficulty in attempting to disentangle the role of the circadian system and sleep on behavior and physiology, because it may not be possible to alter sleep without impacting the circadian system.
Focus on Huntington’s disease
HD is caused by an expanded CAG repeat coding for polyglutamine in the huntingtin (HTT) protein60. Onset of HD is typically in middle age but symptoms can start quite young. Generally, the larger the CAG repeat, the earlier the age of onset and the severity of the symptoms. The classical clinical features of HD are abnormal movements including both involuntary movements (chorea) and difficulty with motor coordination61. Pathologically, HD motor symptoms appear to be due to progressive neuronal cell loss in basal ganglia and cortex62,63. HD patients also exhibit early-onset non-motor symptoms such as sleep disturbances, depressed mood, metabolic disorder, and cognitive dysfunction64-72. In humans, many of these non-motor symptoms begin before the onset of motor symptoms, and are likely due to alterations in hypothalamic and endocrine system function73,74, resulting in a cluster of neurophychiatric symptoms. Common neuropsychiatric aspects of HD include cognitive impairment that progresses to dementia, anxiety, apathy, personality changes, and sleep disorders73,75,76. Sleep disorders are extremely common in HD and have major detrimental effects on daily functioning and the quality of life of patients and their caregivers77-80. Since disruptions in sleep are common and often become apparent years before the onset of motor symptoms, an improved understanding of the mechanism underlying these disruptions has important implications for early diagnosis and treatment of HD.
Temporal components of several prominent non-motor symptoms of HD raise the possibility that SCN coordination of physiological and behavioral rhythms is dysfunctional. Most strikingly, sleep disruptions reported by HD patients include increased sleep latency, decreased sleep maintenance, fragmented sleep, and excessive daytime sleepiness. These symptoms may reflect alterations in the temporal patterning of sleep which can be the result of circadian dysfunction. Similarly, the autonomic dysfunction related to cardiovascular function reported in HD is also consistent with possible circadian dysregulation. In order to explore the possible contribution of circadian dysfunction to HD disease progression, we examined mouse models of HD.
BACHD mouse model of HD
Previous work has shown that the R6/2 CAG150+ mouse model of HD exhibits a progressive and rapid breakdown of the circadian rest/activity cycle that mimics the condition observed in human patients typified by loss of consolidated sleep, increased wakeful activity during the sleep phase, and more sleep during the active/waking phase10,81. We sought to extend this work by examination of bacterial artificial chromosome (BAC) HD transgenic mice that express the entire human HTT gene with stable expression of 97 mixed CAA-CAG repeats in somatic and germline tissues under the control of human HD promoter82. No single mouse model can be expected to recapitulate all aspects of the human disease, and we hence felt that it was important to explore possible circadian dysfunction in a sophisticated transgenic HD mouse model with stable CAA-CAG repeat expansion and the full-length human HTT.
Deficits in BACHD mouse circadian behavior and temporal distribution of sleep
The BACHD line was screened by monitoring daily rhythms of wheel-running activity, the method of choice for screening mutations that influence the circadian system of mammals83. BACHD mice exhibit low amplitude, fragmented rhythms in wheel running behavior under both a light/dark photic environment and constant darkness (Fig. 2). Amplitude and coherence of behavioral rhythms progressively decline over the life of WT and BACHD mice; however, this decline occurs earlier in BACHD mice with young BACHD mice essentially exhibiting the circadian phenotype of an older WT mouse. By young adulthood (3-6 mo), BACHD mice exhibit deficits in circadian rhythm fragmentation, rhythm power, and activity amount as measured by wheel-running activity. With increasing age they show further deterioration in activity amount. Additionally, video recordings were used to measure sleep distribution in BACHD mice as measured by video recording. The effects of the mutation on sleep were subtle, with BACHD mice exhibiting significantly reduced sleep during the beginning of their rest phase. In other words, BACHD mice show deficits in the onset of sleep, a similar sleep phenotype to human HD patients.
Fig. 2.
Circadian dysfunction is a common feature of mouse models of HD. Mice were placed individually in cages with running wheels, and locomotor activity was recorded under different lighting conditions. Each horizontal row represents an activity record for a 24-hr day. Successive days are plotted from top to bottom. The grey shading represents darkness. Mice were initially held in LD (12:12) and then released into DD. Panels show examples of the wheel-running activity recorded from WT (left), BACHD (middle). The average waveform of activity for each genotype (black line =WT; grey line = BACHD) as measured over 10-days in LD is also shown (right panel).The mice were 2-3 mo of age. Data from Kudo and colleagues83.
Disrupted circadian regulation of the cardiovascular system
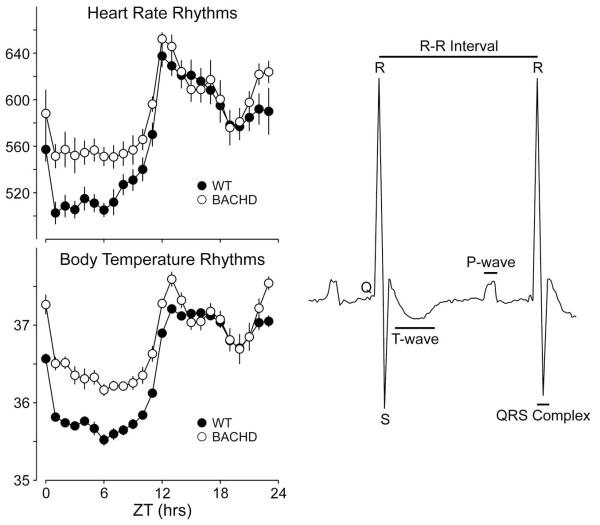
In additional to sleep related non-motor symptom in HD patients, poor cardiovascular health is also observed. Patient cardiac dysfunction is best evidenced by alterations in autonomic tone84-86. Heart rate variability (HRV) is a measure of variation in the beat-to-beat (R-R) interval and reflects the dynamic balance of sympathetic and parasympathetic control of heart function and displays a robust diurnal and circadian rhythm. BACHD mice fail to show day/night differences in HRV suggesting a loss of circadian control, as well as an overall decrease in HRV over a 24hr period when compared to WT controls83. The decrease in HRV and autonomic dysfunction may be responsible for the elevated HR and body temperature measured during the rest phase (daytime) in BACHD mice. Notably, a similar decrease in HRV has also been reported in HD patients beginning during the presymptomatic and early stages of HD progression84,87. Reduced HRV is generally considered an indication of poor cardiovascular health and a predictor for cardiovascular disease and mortality88-90. In this regard, it is worth considering that loss of temporal control and autonomic imbalance may be promoting cardiac failure -- a leading cause of death among HD patients91-92. Electrocardiograph (ECG) parameters were also examined in WT and BACHD mice (Fig. 3). While most parameters were not different between the two genotypes, we detected a loss of day/night differences in the PR interval of BACHD mice, with a significant elongation during the active period when compared to WT mice. The PR interval reflects the time it takes for the cardiac electrical impulse to travel from the sinus node of the atria to the atrioventricular node, which is regulated by a number of factors, one being input from the autonomic system93,94. Duration of the PR interval is also sensitive to K+ handling in cardiomyocytes, which could present as a potential mechanism for the loss of circadian regulation of heart function95. Finally, we measured the baroreceptor reflex, a process dependent on proper autonomic function, in the BACHD mice96. BACHD mouse showed a blunted response of the baroreceptor reflex that suggested dysfunction of both branches of the autonomic nervous system, as well as significantly higher daytime blood pressure in BACHD mice compared to WT controls, consistent with the observed increase in HR during the rest phase. Overall, BACHD mice showed a dramatic decrease in HRV, alterations in the PR interval, and an inability to appropriately decrease heart rate and blood pressure in BACHD mice during sleep, all of which are indications of compromised autonomic function.
Fig. 3.
BACHD mice exhibit a decrease in the amplitude in circadian rhythms as measured by telemetry. Examples of heart rate and body temperature measured from WT (left panels) and littermate BACHD (right panels) mice at ~6 mo of age are shown. Each horizontal row represents an activity record for a 24-hr day. Activity double plotted to aid detection of activity patterns. Successive days are plotted from top to bottom. The average waveform of activity for each genotype as measured over 10-days in LD is also shown (right panels). Representative example of an ECG tracing from a WT mouse labeled with features and intervals of the waveform. The PR interval was significantly increased in the BACHD mice. Data from Kudo and colleagues83.
The molecular clockwork is not obviously altered in the BACHD line
As a first screen for possible deficits in the molecular clockwork responsible for generating circadian oscillation (see above), we examined at PER2 expression in the SCN at peak and trough time-points (Fig. 4), but found no differences in BACHD mice83. In contrast, prior work with the R6/2 line suggests that behavioral impairment is accompanied by disordered in vivo expression of circadian clock genes in the SCN and striatum81,97. Additionally, livers from the R6/2 line lost rhythms in the core clock gene Cry1 and the output gene Dbp, but not of the core clock genes Bmal1 and Per297. Therefore, further work is required to definitively determine whether rhythms in clock gene expression are altered in the BACHD mice.
Fig. 4.
PER2 rhythm within the SCN did not appear to be disrupted in BACHD mice. Mice were held in DD and wheel running activity measured to determine circadian phase. IHC was used to measure PER2 immunoreactivity in the SCN of BACHD and WT controls. Tissue was collected in subjective day (CT 2) or subjective night (CT 14). (top panels) Photomicrographs of SCN tissue of each genotype in low (10×) and higher (40×) magnification. (bottom panel). Data from Kudo and colleagues83.
Reduced daytime firing rate of SCN neurons in BACHD mice during the early stages of HD progression
SCN neurons are spontaneously active and generate action potentials with peak firing rates during the day98. Neuronal output from the SCN regulates physiological rhythms via the regulation of the autonomic nervous system55, as well as other neural and hormonal pathways14. In the daytime (Fig. 5), BACHD mouse SCN neurons showed significantly reduced spontaneous firing rates83. Decreased daytime electrical activity in the SCN would therefore weaken both neural and hormonal outputs. This observation is consistent with the hypothesis that the weakening of electrical output from the SCN is part of the pathology responsible for the circadian behavioral phenotypes observed in the BACHD mice.
Fig. 5.
Daytime spontaneous neural activity is reduced in the SCN of BACHD mice. Using the current-clamp recording technique in the cell-attached configuration, we measured the spontaneous firing rate (SFR) in dorsal SCN neurons during the day and night. The panels show representative examples of firing rate recorded from the WT and BACHD mice at each time point. Data from Kudo and colleagues83.
Reduced daytime firing rates in the SCN of the BACHD line were measured from neurons embedded within a circuit. One mechanism by which HD may alter the function of the circadian system is by reducing the strength of inter-cellular coupling within the SCN circuit. Recent work in a variety of mouse models of neurodevelopmental and psychiatric disorders suggests that alterations in the balance between synaptic excitation and inhibition are at the heart of the pathophysiology99-103. Synaptic alteration in R6/2, and YAC mouse models of HD have been observed in cortex and striatum. During early stages of HD progression increased spontaneous inhibitory post-synaptic currents (IPSC) frequency are followed by late-stage decreases in spontaneous IPSC frequency in cortical pyramidal neurons104. In striatal medium spiny neurons the amplitude of spontaneous excitatory post-synaptic current (EPSC) increases in early stages, then in late stages there are decreased spontaneous EPSC frequencies105. These changes may underlie observed increases in firing rates in both brain regions and decreased synchrony/coupling or correlated firing106-108. While we have no specific evidence that HD alters synaptic transmission or coupling within the SCN circuit, alterations in synaptic transmission remains a very plausible mechanism to explain the disruption in circadian function.
SCN spontaneous electrical activity is controlled by a set of currents that drives daily rhythms in action potentials. During the day, SCN neurons are relatively depolarized with a resting membrane potential (−50 to −55 mV) close to the threshold for generating an action potential (−45 mV). This relatively depolarized resting potential is the result of excitatory drive provided by multiple cation currents109-111. In response, SCN neurons exhibit sustained discharge for 4-6 hours in the subjective day without spike adaptation. Prior work suggests that 3 potassium (K+) currents including fast delayed rectifier (FDR), A-type-K+ current (IA), and large-conductance Ca2+ activated K+ (BK) currents are critical in the regulation of spontaneous action potential firing in SCN neurons during the day98. Reduction in magnitude of the FDR and perhaps the BK currents would consequentially decrease SCN neuronal firing rates. Changes in these currents may therefore underlie the decreased daytime firing we observed in the BACHD mouse SCN.
The molecular basis for decreased firing rate of SCN neurons in HD is unclear
It is unclear if decreased SCN electrical activity is due to changes in synaptic signaling and/or alterations of intrinsic membrane currents; however, there is a third very appealing hypothesis – disrupted mitochondrial function may also reduce neuronal electrical activity112,113. Membrane repolarization following action potential generation is extremely energetically demanding. The ATP-dependent sodium-potassium pump (Na+/K+-ATPase) is critical for maintaining neuronal resting membrane potential. The pump is more active in the day than in the night114, and therefore its activity may be critical to the daily rhythm in membrane potential of SCN neurons. The use of this pump is very energy-expensive and insufficient ATP availability would depolarize membrane potential leading to an inability of neurons to generate action potentials.
Another possible mechanistic candidate causing decreased electrical activity is mutant huntingtin-driven increases in oxidative damage in SCN neurons. Mutant huntingtin aggregation has been found to inhibit mitochondrial trafficking115, and increase reactive oxygen species (ROS) production by decreasing complex I-dependent mitochondrial respiration116, 117, both of which would decrease mitochondrial efficiency and thereby increase ROS byproducts. Notably, circadian transcriptional machinery coordinates anti-oxidant stress by controlling the transcription of anti-oxidant enzymes in many cells118. This suggests that disruption of the intracellular circadian clock may increase susceptibility to oxidative stress and subsequently compromise cellular function and survival119. This is supported by observations of disrupted ROS homeostasis in Clock mutant mice120. Based on observations of disrupted clock gene expression in the R6/2 mouse model, a similar mechanism may cause neuronal dysfunction via reduced ROS homeostasis10. We did not observe deficits in circadian gene expression in BACHD mice, but we still expect to see an increase in ROS levels due to mitochondrial inefficiency. While oxidative stress damages a range of cellular processes, it is particularly relevant to neuronal currents because of K+ channel sensitivity to oxidative damage121,122. Increased daytime firing rates are dependent on K+ channel currents, and therefore oxidative damage to the channels mediating these currents in the SCN may underlie the reduced daytime firing rates observed in the BACHD model.
Based on the literature, the most promising explanation for decreased SCN firing frequency in the BACHD model is decreased mitochondrial function and increased oxidative stress. Future examination of these targets provides new hypotheses to explore the mechanism underlying reduced SCN firing frequency and in turn may provide new insights into disease progression and disease prevention.
Future directions: does decreased SCN output contribute to the symptoms associated with HD?
Altered coupling within the SCN circuit would likely have profound consequences on patient health123-126. Many rhythms researchers believe that robust circadian rhythms depending on a coupled SCN circuit are essential to good health. In recent years, a wide range of studies have demonstrated that disruption of the circadian system leads to a cluster of symptoms including metabolic and cardiovascular disease, as well as cognitive deficits (Table 1). Many of these same symptoms are seen in HD patients. We therefore should consider circadian dysfunction is not just a symptom of HD, but also as a key part of the disease mechanism. We have good reason to think so, because sleep/circadian disruptions occur early in the HD patients and HD mouse models. If this hypothesis is correct, circadian disruption may make the symptoms of HD worse, while stabilizing the rhythm could delay the symptoms (Table 2). Recent work suggests that interventions that stabilize deteriorating daily rhythms can delay cognitive and motor symptom progression in the R6/2 line81. Whether this intervention improves rhythms in SCN output or is effective in other mouse models has yet to be determined.
Table I.
Symptoms due to weakening of circadian output
| - Cognitive dysfunction including memory problems. |
| - Triggering affective disorders. |
| - Metabolic dysfunction including increased risk of Type 2 diabetes. |
| - Cardiovascular disease. |
| - Gastrointestinal disturbances. |
Table II.
How to strengthen daily rhythms in HD.
| - Maintain light/dark cycle with bright sunlight in daytime and darkness during nighttime |
| - Scheduled timing of meals. |
| - Scheduled timing of exercise. |
| - Limit intake of psychostimulants, including caffeine. |
| - Consider short-term use of sedative hypnotics. |
| - Melatonin supplementation. |
| - Cognitive behavioral therapy for insomnia and HD-related mood and/or anxiety disorders. |
Based on present understanding of the pathophysiology of the master oscillator in the BACHD mouse (Fig. 6), future work needs to focus on understanding how mutant huntingtin alters the electrical activity within the SCN and development of interventions that can treat these disruptions. Identifying new therapeutic targets to treat sleep and circadian deficits in HD patients may improve quality of life and potentially slow disease progression for this currently incurable disease.
Fig. 6.
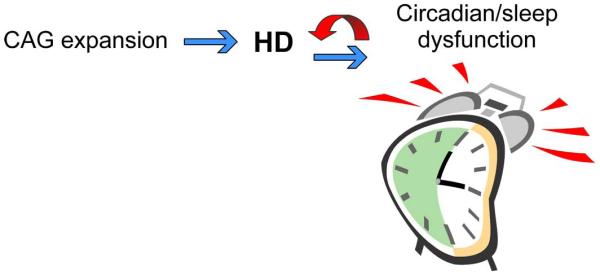
Speculation that circadian dysfunction could accelerate the pathology of HD. Work from several groups indicates that the mouse models of HD show a circadian phenotype. The circadian clockwork regulates mitochondrial function, reactive oxygen species homeostasis, DNA repair and immune response. Dysfunction of this timing system is likely to contribute to chronic inflammation, mitochondrial dysfunction, and DNA damage. These processes are all thought to contribute to the pathology of HD and contribute to age-related changes in the brain. Therefore, we raise the possibility that circadian dysfunction due to genetic or environmental perturbations can accelerate the pathology of HD.
Acknowledgments
Our work is supported by the CHDI Foundation, the Oppenheimer Foundation and the American Heart Association. We would also like to thank Ms. Donna Crandall for assistance with the graphics.
References
- 1.Leger D, Guilleminault C, Dreyfus JP, Delahaye C, Paillard M. Prevalence of insomnia in a survey of 12778 adults in France. Journal of Sleep Research. 2000;9:32–42. doi: 10.1046/j.1365-2869.2000.00178.x. [DOI] [PubMed] [Google Scholar]
- 2.Hossain JL, Shapiro CM. The prevalence, cost implications, and management of sleep disorders: an overview. Sleep and Breathing. 2002;6(2):85–102. doi: 10.1007/s11325-002-0085-1. [DOI] [PubMed] [Google Scholar]
- 3.Luckhaupt SE, Tak SW, Calvert GM. The prevalence of short sleep duration by industry and occupation in the National Health Interview Survey. Sleep. 2010;33:149–59. doi: 10.1093/sleep/33.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durmer JS, Dinges DF. Neurocognitive consequences of sleep deprivation. Semin Neurol. 2005;25:117–129. doi: 10.1055/s-2005-867080. [DOI] [PubMed] [Google Scholar]
- 5.Mullington JM, Haack M, Toth M, Serrador JM, Meier Ewert HK. Cardiovascular, inflammatory, and metabolic consequences of sleep deprivation. Progess in Cardiovascular Diseases. 2009;51(4):294–302. doi: 10.1016/j.pcad.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen S, Doyle WJ, Alper CM, Janicki Deverts D, Turner RB. Sleep habits and susceptibility to the common cold. Arch Intern Med. 2009;169(1):62–67. doi: 10.1001/archinternmed.2008.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moldofsky H, Luk WP, Dickstein J. Sleep, health and immunoscompetence. Neuroimmune Biology. 2001;1:255–268. [Google Scholar]
- 8.Foley D, Ancoli Israel S, Britz P, Walsh J. Sleep disturbances and chronic disease in older adults: results of the 2003 National Sleep Foundation Sleep in America survey. Journal of Psychosomatic Research. 2004;56(5):497–502. doi: 10.1016/j.jpsychores.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Prinz PN, Vitaliano PP, Vitiello MV, Bokan J, Raskind M, Peskind E, Gerber C. Sleep, EEG and mental functioni changes in senile dementia of the Alzheimer’s type. Neurobiol Aging. 1982;3:299–309. doi: 10.1016/0197-4580(82)90024-0. [DOI] [PubMed] [Google Scholar]
- 10.Morton A, Wood N, Hastings M, Hurelbrink C, Barker R, Maywood E. Disintegration of the sleep-wake cycle and circadian timing in Huntington’s disease. J. Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partinen M. Sleep disorder related to Parkinson’s disease. J Neurol. 1997;244(Suppl 1):S3–6. doi: 10.1007/BF03160564. [DOI] [PubMed] [Google Scholar]
- 12.Iranzo A, Santamaria J, Serradell M, Marti MJ, Valldeoriola F, Tolosa E. Rapid-eye-movement sleep behaviour disorder as an early marker for a neurodegenerative disorder: a descriptive study. Lancet Neurol. 2006;5(7):572–577. doi: 10.1016/S1474-4422(06)70476-8. [DOI] [PubMed] [Google Scholar]
- 13.Julein C, Thompson J, Wild S, Yardumian P, Snowden J, Turner G, Craufurd D. Psychatric diorders in preclinical Huntington’s disease. J Neurol Neurosurg Psychiatry. 2007;78:939–943. doi: 10.1136/jnnp.2006.103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 15.Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci. 2011;34(7):349–358. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91(7):1055–64. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 17.Shearman LP, ZYlka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 19(6):1261–9. doi: 10.1016/s0896-6273(00)80417-1. 1007. [DOI] [PubMed] [Google Scholar]
- 18.Sun ZS, Albrecht U, Zuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90(6):1003–11. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 19.Tei H, Okamura H, Shigeyoshi Y, Fukuhura C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389(6650):512–6. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 20.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–30. doi: 10.1038/19323. doi:10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 21.Gekakis N, Staknis D, Nguyen H, Davis FC, Wilsbacher L, King D, Takahashi JS, et al. Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. doi:10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 22.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(10):5474–9. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahata S, Sogawa K, Kobayashi A, Ema M, Mimura J, Ozaki N, Fujii Kuriyama Y. Transcriptionally Active Heterodimer Formation of an Arnt-like PAS Protein, Arnt3, with HIF-1a, HLF, and Clock. Biochemical and Biophysical Research Communications. 1998;794(248):789–794. doi: 10.1006/bbrc.1998.9012. [DOI] [PubMed] [Google Scholar]
- 24.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282(5393):1490–4. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 25.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332(6036):1436–9. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 28.Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The afterhours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 29.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Etchegaray JP, Cagampang FRA, Loudon ASI, Reppert S. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107(7):855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 31.Harms E, Kivimae S, Young MW, Saez L. Posstranscriptional and postrranslational regulation of clock genes. J Biol Rhyths. 2004;19(5):361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- 32.Preitner N, Damiola F, Lopez Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REVERB alpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 33.Sato F, Kawamoto T, Fujimoto K, Noshiro M, Honda KK, Honma S, Honma K, Kato Y. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. Eur J Biochem. 2004;271(22):4409–19. doi: 10.1111/j.1432-1033.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- 34.Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J Biol Chem. 2010;285(46):35386–92. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crumbley C, Burris TP. Direct regulation of CLOCK expression by REV-ERB. PLoS One. 2011;6(3):e17290. doi: 10.1371/journal.pone.0017290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. 2002. [DOI] [PubMed] [Google Scholar]
- 37.Storch KF, Lipna OIL, Viswanathan N, Davis F, Wong W, Weitz C. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 38.Hogenesch JB, Panda S, Kay S, Takahashi JS. Circadian transcriptional output in the SCN and liver of the mouse. Novartis Found Symp. 2003;253:171–80. [PubMed] [Google Scholar]
- 39.Baggs JE, Hogenesch JB. Genomics and systems approaches in the mammalian circadian clock. Curr Opin Genet Dev. 2010;20(6):581–7. doi: 10.1016/j.gde.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lejeune Lenain C, Van Cauter E, Désir D, Beyloos M, Frackson JR. Control of circadian and episodic variations of adrenal androgens secretion in man. J Endocrinol Invest. 1987;10(3):267–76. doi: 10.1007/BF03348129. [DOI] [PubMed] [Google Scholar]
- 42.Deery MJ, Maywood ES, Chesham JE, Sladek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS, Hastings MH. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol. 2009;19:2031–6. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 43.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 44.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–77. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van den Pol AN. The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J Comp Neurol. 1980;191(4):661–702. doi: 10.1002/cne.901910410. [DOI] [PubMed] [Google Scholar]
- 46.Antle MC, Smith VM, Sterniczuk R, Yamakawa GR, Rakai BD. Physiological responses of the circadian clock to acute light exposure at night. 2009;10(4):279–91. doi: 10.1007/s11154-009-9116-6. [DOI] [PubMed] [Google Scholar]
- 47.Golombek DA, Rosenstei RE. Physiology of circadian entrainment. Physiol Rev. 2010;90(3):1063–102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 48.Morin LP, Allen CN. The circadian visual system, 2005. Brain Res Rev. 2006;51(1):1–60. doi: 10.1016/j.brainresrev.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. 2004;19(7):1741–8. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25(23):5481–7. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15(7):1153–62. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- 52.Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;18(3):145–51. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervations, intrinsic organization and efferent projections. Brain Res. 2001;6(102):172–91. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 54.Kriegsfeld LJ, Leak RK, Yackulic CB, LeSauter J, Silver R. Organization of suprachiasmatic nucleus projections in Syrian hamsters (Mesocricetus auratus): an anterograde and retrograde analysis. J Comp Neurol. 2004;468(3):361–79. doi: 10.1002/cne.10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalsbeek A, Palm IF, LaFleur SE, Scheer FAJL, Perreau Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J. Biol. Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 56.Deboer T, Vansteensel MJ, Detari L, Meijer JH. Sleep states alter activity of suprachiasmatic nucleus neurons. Nature Neuroscience. 2003;6:1086–1090. doi: 10.1038/nn1122. [DOI] [PubMed] [Google Scholar]
- 57.Meijer JH, Schaap J, Watanabe K, Albus H. Multiunit activity recordings in the suprachiasmatic nuclei: in vivo versus in vitro models. Brain Research. 1997;753:322–327. doi: 10.1016/s0006-8993(97)00150-9. [DOI] [PubMed] [Google Scholar]
- 58.Yamazaki S, Kerbeshian MC, Hocker CG, Block GD, Menaker M. Rhythmic properties of the hamster suprachiasmatic nucleus in vivo. J. Neurosci. 1998;18:10709–10723. doi: 10.1523/JNEUROSCI.18-24-10709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaap J, Meijer JH. Opposing effects of behavioral activity and light on neurons of the suprachiasmatic nucleus. Eur. J. Neurosci. 2001;13:1955–1962. doi: 10.1046/j.0953-816x.2001.01561.x. [DOI] [PubMed] [Google Scholar]
- 60.Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleuotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 61.Bonelli RM, Beal MF. Huntington’s disease. Handb Clin Neurol. 2012;106:507–26. doi: 10.1016/B978-0-444-52002-9.00030-9. [DOI] [PubMed] [Google Scholar]
- 62.Margolis RL, Ross CA. Diagnosis of Huntington disease. Clin Chem. 2003;49(10):1726–32. doi: 10.1373/49.10.1726. [DOI] [PubMed] [Google Scholar]
- 63.Schulte J, Littleton JT. The biological function of the Huntingtin protein and its relevance to Huntington’s Disease pathology. Curr Trends Neurol. 2011;5:65–78. [PMC free article] [PubMed] [Google Scholar]
- 64.Petersén A, Gil J, Maat-Schieman M, Björkqvist M, Tanila H, Araújo I, Smith R, Popovic N, Wierup N, Norlén P, Li J, Roos R, Sundler F, Mulder H, Brundin P. Orexin loss in Huntington’s disease. Hum Mol Genet. 2005;14:39–47. doi: 10.1093/hmg/ddi004. [DOI] [PubMed] [Google Scholar]
- 65.Aziz NA, Pijl H, Frolich M, Schroder-vander Elst JP, van der Bent C, Roelfsema F, Roos RA. Growth hormone and ghrelin secretion are associated with clinical severity in Huntington’s disease. Eur J Neurol. 2010;17(2):280–8. doi: 10.1111/j.1468-1331.2009.02798.x. [DOI] [PubMed] [Google Scholar]
- 66.Petersén A, Hult S, Kirik D. Huntington’s disease – new perspectives based on neuroendocrine changes in rodents models. Neurodegener Dis. 2009;6(4):154–64. doi: 10.1159/000225377. [DOI] [PubMed] [Google Scholar]
- 67.Kremer H, Roos R, Dingjan G, Marani E, Bots G. Atrophy of the hypothalamic lateral tuberal nucleus in Huntington’s disease. J Neuropathol Exp Neurol. 1990;49:371–382. doi: 10.1097/00005072-199007000-00002. [DOI] [PubMed] [Google Scholar]
- 68.Lavin P, Bone I, Sheridan P. Studies of hypothalamic function in Huntington’s chorea. J Neurol Neurosurg Psychiatry. 1981;44:414–418. doi: 10.1136/jnnp.44.5.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood NI, Goodman AO, van der Burg JM, Gazeau V, Brundin P, Bjorkqvist M, Petersen A, Tabrizi SJ, Barker RA, Morton AJ. Increarsed thirst and drinking in Huntington’s disease and the R6/2 mouse. Brain Res Bull. 2008;76(102):70–9. doi: 10.1016/j.brainresbull.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Hinton SC, Paulsen JS, Hoffmann RG, Reynolds NC, Zimbelman JL, Rao SM. Motor timing variability increases in preclinical Huntington’s disease patients as estimated onset of motor symptoms approaches. J Int Neuropsychol Soc. 2007;13:539–543. doi: 10.1017/S1355617707070671. [DOI] [PubMed] [Google Scholar]
- 71.Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, McDonald M, Beglinger LJ, Duff K, Kayson E, Biglan K, Shoulson I, Oakes D, Hayden M. Predict HD investigators and coordinators of the Huntington Study Group Detection of Huntington’s disease decades before diagnosis: The Predict HD study. J Neurol Neurosurg Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Videnovic A, Leurgans S, Fan W, Jaglin J, Shannon KM. Daytime somnolence and nocturnal sleep disturbances in Huntington disease. Parkinsonism Relat Disord. 2009;15(6):471–4. doi: 10.1016/j.parkreldis.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 73.Duff K, Paulsen JS, Beglinger LJ, Langbehn DR, Stout JC. Predict-HD Investigators of the Huntington Study Group Psychiatric symptoms in Huntington’s disease before diagnosis: the Predict-HD study. Biol Psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 74.Soneson C, Fontes M, Zhou Y, Denisov V, Paulsen JS, Kirik D, Petersén A, Huntington Study Group PREDICT-HD investigators Early changes in the hypothalamic region in prodromal Huntington disease revealed by MRI analysis. Neurobiol Dis. 2010;40(3):531–43. doi: 10.1016/j.nbd.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peavy GM, Jacobson MW, Goldstein JL, Hamilton JM, Kane A, Gamst AC, Lessig SL, Lee JC, Corey-Bloom J. Cognitive and functional decline in Huntington’s disease: dementia criteria revisited. Mov Disord. 2010;25(9):1163–9. doi: 10.1002/mds.22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaccarino AL, Sills T, Anderson KE, Bachoud-Lévi AC, Borowsky B, Craudurd D, Duff K, Giuliano J, Groves M, Guttman M, Kupchak P, Ho AK, Paulsen JS, Pedersen KF, van Duijn E, van Kammen DP, Evans K. Assessment of depression, anxiety and apathy in prodromal and early Huntington disease. PLoS Curr. 2011;3:RRN1242. doi: 10.1371/currents.RRN1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodman AO, Rogers L, Pilsworth S, McAllister CJ, Shneerson JM, Morton AJ, Barker RA. Asymptomatic sleep abnormalities are a common early feature in patients with Huntington’s disease. Curr Neurol Neurosci Rep. 2011;11(2):211–7. doi: 10.1007/s11910-010-0163-x. [DOI] [PubMed] [Google Scholar]
- 78.Goodman AO, Barker RA. How vital is sleep in Huntington’s disease? J Neurol. 2010;257(6):882–97. doi: 10.1007/s00415-010-5517-4. [DOI] [PubMed] [Google Scholar]
- 79.Aziz NA, Anguelova GV, Marinus J, Lammers GJ, Roos RA. Sleep and circadian rhythm alterations correlate with depression and cognitive impairment in Huntington’s disease. Parkinsonism Relat Disord. 2010;16(5):345–50. doi: 10.1016/j.parkreldis.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 80.Cuturic M, Abramson RK, Vallini D, Frank EM, Shamsnia M. Sleep patterns in patients with Huntington’s disease and their unaffected first-degree relatives: a brief report. Behav Sleep Med. 2009;7(4):245–54. doi: 10.1080/15402000903190215. [DOI] [PubMed] [Google Scholar]
- 81.Pallier P, Maywood E, Zheng Z, Chesham J, Inyushkin A, Dyball R, Hastings M, Morton A. Pharmacological imposition of sleep slows cognitive decline and reverses dysregulation of circadian gene expression in a transgenic mouse model of Huntington’s disease. J Neurosci. 2007;27:7869–7878. doi: 10.1523/JNEUROSCI.0649-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, Li XJ, Levine MS, Yang XW. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28(24):6182–95. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kudo T, Schroeder A, Loh DH, Kuljis D, Jordan MC, Roos KP, Colwell CS. Dysfunctions in circadian behavior and physiology in mouse models of Huntington’s disease. Exp. Neurol. 2011;228(1):80–90. doi: 10.1016/j.expneurol.2010.12.011. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andrich J, Schmitz T, Saft C, Pastert T, Kraus P, Epplen JT, Przuntek H, Agelink MW. Autonomic nervous system function in Huntington’s disease. J Neurol Neurosurg Psychiatry. 2002;72(6):726–31. doi: 10.1136/jnnp.72.6.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bär KJ, Boettger MK, andrich J, Epplen JT, Fischer F, Cordes J, Hoschke M, Agelink MW. Cadriovascular modulation upon postural change is altered in Huntington’s disease. Eur J Neurol. 2008;15(8):869–71. doi: 10.1111/j.1468-1331.2008.02173.x. [DOI] [PubMed] [Google Scholar]
- 86.Kobal J, Meglic B, Mesec A, Peterlin B. Early sympathetic hyperactivity in Huntington’s disease. Eur J Neurol. 2004;11(12):842–8. doi: 10.1111/j.1468-1331.2004.00894.x. [DOI] [PubMed] [Google Scholar]
- 87.Kobal J, Melik Z, Cankar K, Bajrovic FF, Meglic B, Peterlin B, Zaletel M. Autonomic dysfunction in presymptomatic and early symptomatic Huntington’s disease. Acta Neurol Scand. 2010;121(6):392–9. doi: 10.1111/j.1600-0404.2009.01251.x. [DOI] [PubMed] [Google Scholar]
- 88.Bigger JT, Jr, Fleiss JL, Steinman RC, Rolnitzky LM, Kleiger RE, Rottman JN. Frequency domain measures of heart period variability and mortality after myocardial infarction. Circulation. 1992;85(1):164–71. doi: 10.1161/01.cir.85.1.164. [DOI] [PubMed] [Google Scholar]
- 89.Buccelletti E, Gilardi E, Scaini E, Galiuto L, Persiani R, Biondi A, Basile D, Silveri NG. Heart rate variability and myocardial infarction: systemic literature review and metanalysis. Eur Rev Med Pharmacol Sci. 2009;13(4):299–307. [PubMed] [Google Scholar]
- 90.Thayer JF, Yamamoto SS, Brosschot JF. The relationship of autonomic imbalance, heart rate variability and cardiovascular disease risk factors. Int J Cardiol. 2010;141(2):122–31. doi: 10.1016/j.ijcard.2009.09.543. [DOI] [PubMed] [Google Scholar]
- 91.Chiu E, Alexander L. Causes of death in Huntington’s disease. Med J Aust. 1982;1(4):153. doi: 10.5694/j.1326-5377.1982.tb132224.x. [DOI] [PubMed] [Google Scholar]
- 92.Lanska DJ, Lavine L, Lanska MJ, Schoenberg BS. Huntington’s disease mortality in the United States. Neurology. 1988;38(5):769–72. doi: 10.1212/wnl.38.5.769. [DOI] [PubMed] [Google Scholar]
- 93.Carruthers SG, McCall B, Cordell BA, Wu R. Relationships between heart rate and PR interval during physiological and pharmacological interventions. Br J Clin Pharmacol. 1987;23(3):259–65. doi: 10.1111/j.1365-2125.1987.tb03043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wallick DW, Martin PJ, Masuda Y, Levy MN. Effects of autonomic activity and changes in heart rate on atrioventricular conduction. Am J Physiol. 1982;243(4):H523–7. doi: 10.1152/ajpheart.1982.243.4.H523. [DOI] [PubMed] [Google Scholar]
- 95.Akita M, Kuwahara M, Tsubone H, Sugano S. ECG changes during furosemide-induced hypokalemia in the rat. J Electrocardiol. 1998;31(1):45–9. doi: 10.1016/s0022-0736(98)90006-1. [DOI] [PubMed] [Google Scholar]
- 96.Schroeder AM, Loh DH, Jordan MC, Roos KP, Colwell CS. Baroreceptor reflex dysfunction in the BACHD mouse model of Hungtinton’s disease. PLoS Curr. 2011;3:RRN1266. doi: 10.1371/currents.RRN1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maywood E, Fraenkel E, McAllister C, Wood N, Reddy A, Hastings M, Morton A. Disruption of peripheral circadian timekeeping in a mouse model of Huntington’s disease and its restoration by temporally scheduled feeding. J. Neurosci. 2010;30:10199–10204. doi: 10.1523/JNEUROSCI.1694-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci. 2011;12(10):553–69. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2005;102(35):12560–5. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gogolla N, Leblanc JJ, Quast KB, Sudhof TC, Fagiolini M, Hensch TK. Common circuit defect of excitatory-inhibitory balance in mouse models of autism. J Neurodev Disord. 2009;1(2):172–81. doi: 10.1007/s11689-009-9023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Milnerwood AG, Raymond LA. Early synaptic pathophysiology in neurodegeneration: insights from Huntington’s disease. Trends Neurosci. 2010;33(11):513–23. doi: 10.1016/j.tins.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 102.Nelson SB, Turrigiano GG. Strength through diversity. Neuron. 2008;60(3):477–82. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shepherd GM, Katz DM. Synaptic microcircuit dysfunction in genetic models of neurodevelopmental disorders: focus on Mecp2 and Met. Curr Opin Neurobiol. 2011;21(6):827–33. doi: 10.1016/j.conb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cummings DM, Andre VM, Uzgil BO, Gee SM, Fisher YE, Cepeda C, Levine MS. Alterations in cortical excitation and inhibition in genetic mouse modeles of Huntington’s disease. J Neurosci. 2009;29(33):10371–86. doi: 10.1523/JNEUROSCI.1592-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cepeda C, Hurst RS, Calvert CR, Hernandex-Escheagaray E, Nguyen OK, Jocoy E, Christian LJ, Ariano MA, Levine MS. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J Neurosci. 2003;23(3):961–9. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walker AG, Miller BR, Fritsch JN, Barton SJ, Rebec GV. Altered information processing in the prefrontal cortex of Huntington’s disease mouse models. J Neurosci. 2008;28(36):8973–82. doi: 10.1523/JNEUROSCI.2804-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rebec GV, Conroy SK, Barton SJ. Hyperactive striatal neurons in symptomatic Hungtinton R6/2 mice: variations with behavioral state and repeated ascorbate treatment. Neuroscience. 2006;137(1):327–36. doi: 10.1016/j.neuroscience.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 108.Miller BR, Walker AG, Shah AS, Barton SJ, Rebec GV. Dysregulated information processing by medium spiny neurons in striatum of freely behaving mouse models of Huntington’s disease. J Neurophysiol. 2008;100(4):2205–16. doi: 10.1152/jn.90606.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pennartz CM, Bierlaagh MA, Geurtsen AM. Cellular mechanisms underlying spontaneous firing in rat suprachiasmatic nucleus: involvement of a slowly inactivation component of sodium current. J Neurophysiol. 1997;78(4):1811–25. doi: 10.1152/jn.1997.78.4.1811. [DOI] [PubMed] [Google Scholar]
- 110.Jackson AC, Yao GL, Bean BP. Mechanism of spontaneous firing in dorsomedial suprachiasmatic nucleus neurons. J Neurosci. 2004;24(37):7985–98. doi: 10.1523/JNEUROSCI.2146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kononenko NI, Medina I, Dudek FE. Persistent subthreshold voltage-dependent cation single channels in suprachiasmatic nucleus neurons. Neuroscience. 2004;129(1):85–92. doi: 10.1016/j.neuroscience.2004.06.080. [DOI] [PubMed] [Google Scholar]
- 112.Correia SC, Santos RX, Perry G, Zhu X, Moreira PI, Smith MA. Mitochondrial importance in Alzheimer’s, Huntington’s and Parkinson’s diseases. Adv Exp Med Biol. 2012;724:205–21. doi: 10.1007/978-1-4614-0653-2_16. [DOI] [PubMed] [Google Scholar]
- 113.Schapira AH. Mitochondrial diseases. Lancet. 2012;379(9828):1825–34. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- 114.Wang YC, Huang RC. Effects of sodium pump activity on spontaneous firing in neurons of the rat suprachiasmatic nucleus. J Neurophysiol. 2006;96(1):109–18. doi: 10.1152/jn.01369.2005. [DOI] [PubMed] [Google Scholar]
- 115.Trushina E, Dyer RB, Badger JD, 2nd, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, Doyle K, Bender A, Chacko C, McMurray CT. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24(18):8195–209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, Ishiguro H, Sakoe K, Ooshima T, Sato A, Ikeuchi T, Oyake M, Sato T, Aoyagi Y, Jozumi I, Nagatsu T, Takiyama Y, Nishizawa M, Goto J, Kanazawa I, Davidson I, Tanese N, Takahashi H, Tsuji S. Nat Genet. 2000;26(1):29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 117.Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington’s disease that contributes to polyglutamine pathogenesis. J Biol Chem. 2004;279(6):4988–99. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- 118.Hardeland R, Coto-Montes A, Poeggeler B. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int. 2003;20(6):921–62. doi: 10.1081/cbi-120025245. [DOI] [PubMed] [Google Scholar]
- 119.Antoch MP, Kondratov RV. Circadian proteins and genotoxic stress response. Circ Res. 2010;106(1):68–78. doi: 10.1161/CIRCRESAHA.109.207076. [DOI] [PubMed] [Google Scholar]
- 120.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cotella D, Hernandez-Enriquez B, Wu X, Li R, Pan Z, Leveille J, Link CD, Oddo S, Sesti F. Toxic role of K+ channel oxidation in mammalian brain. J. Neurosci. 2012;32(12):4133–44. doi: 10.1523/JNEUROSCI.6153-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sesti F, Liu S, Cai SQ. Oxidation of potassium channels by ROS: a general mechanism of aging and neurodegeneration. Trends Cell Biol. 2010;20(1):45–51. doi: 10.1016/j.tcb.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 123.Hastings M, Reddy A, Maywood E. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 124.Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20(1):36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takahashi J, Hong H, Ko C, McDearmon E. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bechtold DA, Gibbs JE, Loudon AS. Circadian dysfunction in disease. Trends Pharmacol Sci. 2010;31(5):191–8. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]