Abstract
SIMPLE, also known as LITAF, EET1 and PIG7, was originally identified based on its transcriptional upregulation by estrogen, p53, lipopolysaccharide or a microbial cell-wall component. Missense mutations in SIMPLE cause Charcot-Marie-Tooth disease (CMT), and altered SIMPLE expression is associated with cancer, obesity and inflammatory bowel diseases. Despite increasing evidence linking SIMPLE to human diseases, the biological function of SIMPLE is unknown and the pathogenic mechanism of SIMPLE mutations remains elusive. Our recent study reveals that SIMPLE is a functional partner of the endosomal sorting complex required for transport (ESCRT) machinery in the regulation of endosome-to-lysosome trafficking and intracellular signaling. Our results indicate that CMT-linked SIMPLE mutants are loss-of-function mutants which act dominantly to impair endosomal trafficking and signaling attenuation. We propose that endosomal trafficking and signaling dysregulation is a key pathogenic mechanism in CMT and other diseases that involve SIMPLE dysfunction.
Keywords: peripheral neuropathy, Charcot-Marie-Tooth disease, SIMPLE, LITAF, ESCRT, endosome, lysosome, endosomal trafficking, signaling attenuation
Charcot-Marie-Tooth disease (CMT) is the most common hereditary peripheral neuropathy with no effective treatment.1 Human genetic studies reveal that missense mutations in SIMPLE, a protein of unknown function, cause autosomal dominant CMT type 1C (CMT1C).2-6 SIMPLE, also known as LITAF, EET1 and PIG7, was originally identified as a gene product whose transcription is upregulated by estrogen,7 p53 protein,8 lipopolysaccharide (LPS)9 and a microbial cell-wall component.10 Reduced SIMPLE expression is associated with several types of cancer, including breast cancer,11 lymphoma,12 leukemia13 and thyroid carcinoma,14 whereas increased SIMPLE expression is linked to obesity15 and inflammatory bowel diseases, such as Crohn’s disease and ulcerative colitis.16 The connection of SIMPLE to multiple human diseases underscores the importance of understanding the biochemical function and cellular role of this enigmatic protein.
SIMPLE Functions in the Regulation of Endosome-to-Lysosome Trafficking and Cell Signaling
Endosome-to-lysosome trafficking is a crucial cellular process that not only controls protein degradation but also regulates intracellular signaling.17,18 Receptors at cell surface are internalized in response to ligand binding and delivered to the early endosome, where they are either recycled to the cell surface or sorted to intralumenal vesicles of multivesicular bodies for transport to the lysosome for degradation. The core machinery for mediating endosomal cargo sorting to the lysosomal pathway comprises ESCRT-0, -I, -II and -III complexes.17 The ESCRT accessory factors and mechanisms that confer temporal and spatial control to the endosomal trafficking process remain largely unknown.
SIMPLE is a 161-amino-acid protein with widespread expression in a variety of tissues and cells.2,10,19,20 Although a distinct transcript coding for a 228-amino-acid protein was reported to be encoded by the SIMPLE gene,9 it is now clear that this larger transcript is the result of a DNA sequencing error.4,10,21 Accumulating evidence4,10,21 indicates that SIMPLE is unlikely to be a transcription factor as initially proposed.9 The function of SIMPLE is unknown, although it contains binding sites for TSG101 and NEDD4 (Fig. 1A).22 SIMPLE also contains a cysteine-rich (C-rich) domain (Fig. 1A) that was hypothesized to be a putative RING finger with E3 ubiquitin-protein ligase activity.10,23 However, our analysis shows that the SIMPLE C-rich domain is not a RING finger because it lacks a key His residue and is interrupted by an embedded transmembrane domain (Fig. 1A).19 Furthermore, the results of our biochemical experiments reveal that SIMPLE protein has no E3 ligase activity either in vitro or in vivo.24 By using highly specific anti-SIMPLE antibodies, we found that endogenous SIMPLE is an early endosomal membrane protein,19 rather than a nuclear protein9,25 or a lysosomal/late endosomal membrane protein,10 as previously suggested.
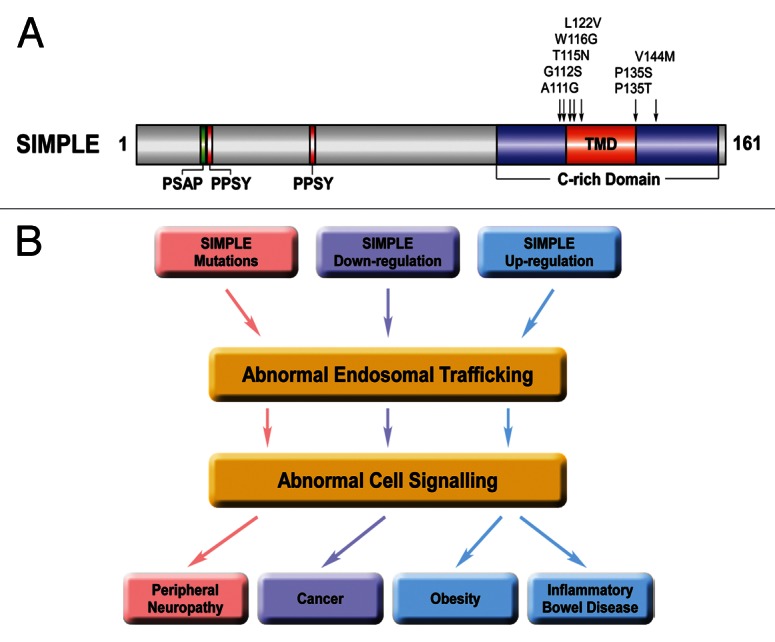
Figure 1. Endosomal trafficking and signaling dysregulation as a potential pathogenic mechanism in CMT and other diseases that involve SIMPLE dysfunction. (A) Domain structure of SIMPLE and mutations found in CMT1C patients. PSAP, predicted TSG101-binding site; PPSY, predicted NEDD4-binding site; C-rich domain, cysteine-rich domain; TMD, predicted transmembrane domain. The locations of CMT1C-linked SIMPLE mutations are indicated on the domain structure. (B) Potential pathogenic roles of SIMPLE dysfunction in CMT, cancer, obesity and inflammatory bowel diseases. Our recent work24 suggests a pathogenic pathway by which CMT1C-linked SIMPLE mutations2-6 cause demyelinating peripheral neuropathy by disrupting endosome-to-lysosome trafficking and signaling attenuation of NRG1-activated ErbB2/ErbB3 receptors and consequently prolonging their signaling to downstream pathways in Schwann cells (colored in pink). Downregulation of SIMPLE expression found in several types of cancer11-14 may contribute to the process of malignant transformation by impairing endosome-to-lysosome trafficking and signaling attenuation of mitogenic signaling receptors (colored in lavender). Upregulation of SIMPLE expression found in obesity15 and inflammatory bowel diseases16 may contribute to the pathogenesis or progression of these diseases by altering endosomal trafficking and intracellular signaling (colored in blue).
In a recent study,24 we examined the cellular function of SIMPLE and found that SIMPLE is a novel regulator of endosome-to-lysosome trafficking. Our results indicate that SIMPLE participates in the recruitment of ESCRT components STAM1, Hrs and TSG101 to the early endosomal membrane and functions with the ESCRT machinery in controlling endosomal sorting and lysosomal degradation of cargo proteins, such as ErbB receptors. In addition, we found that SIMPLE is required for efficient attenuation of signaling events downstream of ligand-activated ErbB receptors. Our data support that SIMPLE regulates cell signaling by promoting endosome-to-lysosome trafficking and degradation of signaling receptors.24 Given its widespread expression pattern,2,10,19,20 our findings suggest that SIMPLE may regulate endosomal trafficking and signaling processes in many different cells, including LPS-induced inflammatory signaling in macrophages.26
Endosomal Trafficking and Signaling Dysregulation: A Key Mechanism in CMT Pathogenesis
Despite the identification of eight distinct point mutations in SIMPLE (Fig. 1A) as the genetic defects for causing CMT1C,2-6 the pathogenic mechanisms of these mutations are unknown. We have shown that SIMPLE is a post-translationally inserted, C-tail-anchored membrane protein that uses its TMD for anchoring to the early endosomal membrane.19,24 Interestingly, all of the identified disease-causing SIMPLE mutations map in and around the TMD (Fig. 1A). We found that CMT1C-linked SIMPLE W116G and P135T mutations promote SIMPLE misfolding and impair its membrane insertion, causing SIMPLE to mislocalize from the endosomal membrane to the cytosol.19,27 Our recent results indicate that SIMPLE W116G and P135T are loss-of-function mutants that exert dominant pathogenic effects to impair endosome-to-lysosome trafficking and signaling attenuation in cells.24 The dominant pathogenic role of SIMPLE mutation is further supported by our finding of a CMT1C-like peripheral neuropathy phenotype in transgenic mice expressing SIMPLE W116G mutant28 and the lack of a neuropathy phenotype in SIMPLE knockout mice.20
Our work reveals a critical role of dysregulated endosome-to-lysosome trafficking in the pathogenesis of demyelinating CMT1C.24 Previous studies have shown that mutations in MTMR2 and MTMR13, which are also involved in regulation of endosomal trafficking, cause demyelinating CMT4B1 and CMT4B2.1,29 Thus, endosomal trafficking dysregulation appears to be a common pathogenic mechanism in several demyelinating CMT diseases. The fact that mutations in these ubiquitously expressed proteins cause demyelinating peripheral neuropathy suggests that, compared with other cell types, Schwann cells are particularly susceptible to defects in endosomal trafficking. We found that SIMPLE is highly abundant in Schwann cells19,28 and that SIMPLE W116G and P135T mutations cause dysregulation of neuregulin-1 (NRG1)-ErbB2/ErbB3 signaling,24 a key pathway for controlling peripheral nerve myelination by Schwann cells.30 In our transgenic CMT1C mouse model, SIMPLE W116G mutation-induced peripheral neuropathy is accompanied by myelin infolding—the focally infolded myelin loops that protrude into the axons.28 Together, our findings suggest a pathogenic pathway (Fig. 1B) by which SIMPLE mutation disrupts endosome-to-lysosome trafficking and signaling attenuation of NRG1-activated ErbB2/ErbB3 receptors in Schwann cells, causing prolonged activation of downstream signaling pathways, thereby leading to myelin infolding and demyelinating peripheral neuropathy.
Conclusions
Our recent study revealing the function of SIMPLE as a regulator of endosome-to-lysosome trafficking and intracellular signaling has provided new insights into the mechanisms of SIMPLE action in health and disease. The evidence obtained from our work indicates that SIMPLE mutation-induced endosomal trafficking and signaling dysregulation in Schwann cells play a key role in CMT1C pathogenesis. Our findings also raised the possibility that altered SIMPLE expression found in cancer,11-14 obesity15 and inflammatory bowel diseases16 may contribute to the pathogenesis or progression of these diseases by altering SIMPLE-dependent endosomal trafficking and signaling (Fig. 1B). Future studies to examine this possibility and identify trafficking and signaling defects caused by SIMPLE dysfunction should enhance our understanding of the pathogenic mechanisms involved in CMT and other diseases and may provide new strategies for therapeutic intervention.
Acknowledgments
This work was supported in part by the National Institutes of Health (AG034126 to L.S.C. and ES015813 and GM103613 to L.L.).
Glossary
Abbreviations:
- CMT
Charcot-Marie-Tooth disease
- CMT1C
CMT type 1C
- C-rich
cysteine-rich
- ESCRT
endosomal sorting complex required for transport
- NRG1
neuregulin-1
- TMD
transmembrane domain
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/24214
References
- 1.Patzkó A, Shy ME. Update on Charcot-Marie-Tooth disease. Curr Neurol Neurosci Rep. 2011;11:78–88. doi: 10.1007/s11910-010-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Street VA, Bennett CL, Goldy JD, Shirk AJ, Kleopa KA, Tempel BL, et al. Mutation of a putative protein degradation gene LITAF/SIMPLE in Charcot-Marie-Tooth disease 1C. Neurology. 2003;60:22–6. doi: 10.1212/WNL.60.1.22. [DOI] [PubMed] [Google Scholar]
- 3.Bennett CL, Shirk AJ, Huynh HM, Street VA, Nelis E, Van Maldergem L, et al. SIMPLE mutation in demyelinating neuropathy and distribution in sciatic nerve. Ann Neurol. 2004;55:713–20. doi: 10.1002/ana.20094. [DOI] [PubMed] [Google Scholar]
- 4.Saifi GM, Szigeti K, Wiszniewski W, Shy ME, Krajewski K, Hausmanowa-Petrusewicz I, et al. SIMPLE mutations in Charcot-Marie-Tooth disease and the potential role of its protein product in protein degradation. Hum Mutat. 2005;25:372–83. doi: 10.1002/humu.20153. [DOI] [PubMed] [Google Scholar]
- 5.Latour P, Gonnaud PM, Ollagnon E, Chan V, Perelman S, Stojkovic T, et al. SIMPLE mutation analysis in dominant demyelinating Charcot-Marie-Tooth disease: three novel mutations. J Peripher Nerv Syst. 2006;11:148–55. doi: 10.1111/j.1085-9489.2006.00080.x. [DOI] [PubMed] [Google Scholar]
- 6.Gerding WM, Koetting J, Epplen JT, Neusch C. Hereditary motor and sensory neuropathy caused by a novel mutation in LITAF. Neuromuscul Disord. 2009;19:701–3. doi: 10.1016/j.nmd.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Everett LM, Li A, Devaraju G, Caperell-Grant A, Bigsby RM. A novel estrogen-enhanced transcript identified in the rat uterus by differential display. Endocrinology. 1997;138:3836–41. doi: 10.1210/en.138.9.3836. [DOI] [PubMed] [Google Scholar]
- 8.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–5. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 9.Myokai F, Takashiba S, Lebo R, Amar S. A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor alpha gene expression: molecular cloning, sequencing, characterization, and chromosomal assignment. Proc Natl Acad Sci USA. 1999;96:4518–23. doi: 10.1073/pnas.96.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriwaki Y, Begum NA, Kobayashi M, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis Bacillus Calmette-Guerin and its cell wall complex induce a novel lysosomal membrane protein, SIMPLE, that bridges the missing link between lipopolysaccharide and p53-inducible gene, LITAF(PIG7), and estrogen-inducible gene, EET-1. J Biol Chem. 2001;276:23065–76. doi: 10.1074/jbc.M011660200. [DOI] [PubMed] [Google Scholar]
- 11.Abba MC, Drake JA, Hawkins KA, Hu Y, Sun H, Notcovich C, et al. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6:R499–513. doi: 10.1186/bcr899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–80. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Liu J, Tang K, Xu Z, Xiong X, Rao Q, et al. Expression of pig7 gene in acute leukemia and its potential to modulate the chemosensitivity of leukemic cells. Leuk Res. 2009;33:28–38. doi: 10.1016/j.leukres.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Lui WO, Foukakis T, Lidén J, Thoppe SR, Dwight T, Höög A, et al. Expression profiling reveals a distinct transcription signature in follicular thyroid carcinomas with a PAX8-PPAR(gamma) fusion oncogene. Oncogene. 2005;24:1467–76. doi: 10.1038/sj.onc.1208135. [DOI] [PubMed] [Google Scholar]
- 15.Ji ZZ, Dai Z, Xu YC. A new tumor necrosis factor (TNF)-α regulator, lipopolysaccharides-induced TNF-α factor, is associated with obesity and insulin resistance. Chin Med J (Engl) 2011;124:177–82. [PubMed] [Google Scholar]
- 16.Stucchi A, Reed K, O’Brien M, Cerda S, Andrews C, Gower A, et al. A new transcription factor that regulates TNF-alpha gene expression, LITAF, is increased in intestinal tissues from patients with CD and UC. Inflamm Bowel Dis. 2006;12:581–7. doi: 10.1097/01.MIB.0000225338.14356.d5. [DOI] [PubMed] [Google Scholar]
- 17.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Wegner CS, Rodahl LM, Stenmark H. ESCRT proteins and cell signalling. Traffic. 2011;12:1291–7. doi: 10.1111/j.1600-0854.2011.01210.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee SM, Olzmann JA, Chin LS, Li L. Mutations associated with Charcot-Marie-Tooth disease cause SIMPLE protein mislocalization and degradation by the proteasome and aggresome-autophagy pathways. J Cell Sci. 2011;124:3319–31. doi: 10.1242/jcs.087114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somandin C, Gerber D, Pereira JA, Horn M, Suter U. LITAF (SIMPLE) regulates Wallerian degeneration after injury but is not essential for peripheral nerve development and maintenance: implications for Charcot-Marie-Tooth disease. Glia. 2012;60:1518–28. doi: 10.1002/glia.22371. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Bennett CL. Litaf/Simple protein is increased in intestinal tissues from patients with CD and UC, but is unlikely to function as a transcription factor. Inflamm Bowel Dis. 2007;13:120–1. doi: 10.1002/ibd.20010. [DOI] [PubMed] [Google Scholar]
- 22.Shirk AJ, Anderson SK, Hashemi SH, Chance PF, Bennett CL. SIMPLE interacts with NEDD4 and TSG101: evidence for a role in lysosomal sorting and implications for Charcot-Marie-Tooth disease. J Neurosci Res. 2005;82:43–50. doi: 10.1002/jnr.20628. [DOI] [PubMed] [Google Scholar]
- 23.Berger P, Niemann A, Suter U. Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease) Glia. 2006;54:243–57. doi: 10.1002/glia.20386. [DOI] [PubMed] [Google Scholar]
- 24.Lee SM, Chin LS, Li L. Charcot-Marie-Tooth disease-linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. J Cell Biol. 2012;199:799–816. doi: 10.1083/jcb.201204137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X, Fenton MJ, Amar S. Identification and functional characterization of a novel binding site on TNF-alpha promoter. Proc Natl Acad Sci USA. 2003;100:4096–101. doi: 10.1073/pnas.0630562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X, Metzger D, Leeman S, Amar S. LPS-induced TNF-alpha factor (LITAF)-deficient mice express reduced LPS-induced cytokine: Evidence for LITAF-dependent LPS signaling pathways. Proc Natl Acad Sci USA. 2006;103:13777–82. doi: 10.1073/pnas.0605988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SM, Chin LS, Li L. Protein misfolding and clearance in demyelinating peripheral neuropathies: Therapeutic implications. Commun Integr Biol. 2012;5:107–10. doi: 10.4161/cib.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SM, Sha D, Mohammed AA, Asress S, Glass JD, Chin LS, et al. Motor and sensory neuropathy due to myelin infolding and paranodal damage in a transgenic mouse model of Charcot-Marie-Tooth disease type 1C. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt022. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SM, Chin LS, Li L. Therapeutic implications of protein homeostasis in demyelinating peripheral neuropathies. Expert Rev Neurother. 2012;12:1041–3. doi: 10.1586/ern.12.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintes S, Goebbels S, Saher G, Schwab MH, Nave KA. Neuron-glia signaling and the protection of axon function by Schwann cells. J Peripher Nerv Syst. 2010;15:10–6. doi: 10.1111/j.1529-8027.2010.00247.x. [DOI] [PubMed] [Google Scholar]


